Abstract
Background: The incidence of malignant melanoma (MM) continues to increase annually, and tumour invasiveness is a main prognostic factor. Single-nucleotide polymorphisms (SNPs) have become key tools in the study of cancer genetics, influencing susceptibility and prognosis. Methods: In the present study, we analysed the relationship between five SNPs on the PDCDL1 gene (rs822336, rs822337, rs822338, rs229736, rs4143815) with prognosis as well as primary tumour invasiveness characteristics in 377 whole blood samples from MM individuals. Results: Patients who presented the rs822336 CG or GG genotypes (OR = 3.01, 95% CI = 1.53–5.92; p = 0.0017), TA or TT in rs822337 (OR = 2.45, 95% CI = 1.22–4.93; p = 0.0098), and CT or CC of rs822338 (OR = 2.23, 95% CI = 1.05–4.73; p = 0.028) were at an increased risk of developing invasive melanomas. Cases with the AG or GG genotype in rs2297136 presented a lower risk (OR = 0.29, 95% CI = 0.11–0.75; p = 0.0038) of invasive MM. The genetic analysis at the haplotype level resulted in similar findings (OR: 2.95, 95% CI: 1.08–8.10), p = 0.036). Furthermore, patients carrying the homozygous AA genotype in rs2297136 had thicker tumours than those harbouring the AG or GG (1.4 mm vs. 1.0 and 0.8 mm; p = 0.030). No significant association was found between the studied SNPs and melanoma-specific survival (MSS) nor progression-free survival (PFS). Conclusions: Current results suggest that SNPs rs822336, rs822337, rs822338, and rs2297136 genotypes in the PDCDL1 gene are associated with the risk of tumour invasiveness and tumour thickness in MM. Further studies on SNPs considering genetic and epigenetic factors are needed for a better understanding of malignant melanoma susceptibility and its prognosis.
1. Introduction
The incidence of malignant melanoma (MM), both in situ and invasive, has increased significantly over the past 5 decades [1]. In situ MM is potentially curable with early surgery as the malignant cells do not spread beyond the epidermis. Spain accounts for 16% of MM [1,2]. When neoplastic cells invade the dermis or deeper layers, MM is considered invasive. Overall, the prognosis of invasive MM is favourable because more than half of the patients are diagnosed in the early stages of the disease [1]. However, the prognosis deteriorates significantly from stage II onward [3]. Breslow index (tumour thickness) and ulceration in the primary tumour are the prognostic factors for survival and define the T category in the AJCC 8th edition [3,4]. T category, together with nodal status (N) and visceral status (M), enables determining the clinical or pathological stage of the patient, thus establishing an estimation of the patient’s prognosis. As the stage increases, the survival of the patients worsens, shifting from a 5-year survival of 99% for stage IA patients to 32% for stage IIID patients [3].
So far, blood LDH levels are the only included serological prognostic biomarker in the AJCC 8th edition [5]. However, multiple prognostic biomarkers have been suggested [4]. Improved tests will likely be available soon for a more accurate, more individualized, and less aggressive estimation of the prognosis [6].
The presence of certain immune system regulators during the growth of various cancers deserves extensive studies. One of the most important immune system regulators is the axis PD1-PDL1. This interaction exerts an immune homeostasis, vital for developing the immune system and regulating peripheral and central tolerance [7]. This immune homeostasis can be altered in the tumour microenvironment due to mutations or signalling pathway activation (among others) that drive the overexpression of PD-L1 in tumour cells, preventing them from being recognised by the immune system [8]. The release of IFN-γ by T lymphocytes and NK cells, together with the JAK/STAT, MAPK, and NF-KB pathways, is the main precursor of PD-L1 activation [9,10]. PD-L1 is expressed on different tumour cells such as head and neck squamous cell carcinoma, MM, and carcinomas of the oesophagus, lung, breast, etc. [11]. Tumour cells take advantage of the immune system of individuals to express PD-L1 and generate an immunosuppressive tumour microenvironment. PD-L1 overexpression in the primary tumour confers a worse prognosis in multiple neoplasms [12,13].
In MM, there is limited and conflicting information on PD-L1 and its relationship with prognosis. The prevalence of PD-L1 expression in melanoma varies from 24% to 49%, although it has been seen in approximately 60% of tumours of chronically sun-damaged skin [14]. Furthermore, PD-L1 independently predicts a worse prognosis as it is correlated with tumour thickness and lymphatic and visceral dissemination [14]. On the other hand, there is evidence of favourable responses (prior to treatment) in MM without PDL1 expression [15]. This is why there is great controversy about the use and effectiveness of immunotherapeutic treatments based on PD-L1 expression in the tumour.
PD-L1 is encoded by the PDCDL1 gene located on p24.1 of chromosome 9 [16] and consists of seven exons that encode a type 1 transmembrane protein of 40 kDa and 290 amino acids [14]. Various single-nucleotide polymorphisms (SNPs) have been identified in PDCDL1 that increase susceptibility to developing various tumours, such as lung [17], gastric [18], or oesophageal cancer [19]. The development of high-throughput sequencing technologies made it easier to identify various SNPs in PDCDL1, leading to the discovery of new potential biomarkers for cancer. Some SNPs are specifically correlated with various tumour behaviours. It has been suggested that rs822336 and rs822337 are associated with unfavourable prognoses in triple-negative breast cancer patients [20]. A meta-analysis by Zou et al. [18], reported that the variant rs4143815 C > G increased the susceptibility to gastric and bladder cancers [18]. Furthermore, single-nucleotide polymorphisms in PDCDL1 could be important regarding the response to treatment. Nomizo et al. [21] observed that individuals with advanced non-small-cell lung cancer carrying the rs4143815 CC genotype showed an improvement in the response to nivolumab [21]. Thus, in lung cancer, rs822336 [22], rs822337 [22], and rs4143815 [23] act as prognostic markers once the primary tumour has been removed. Another important PDCDL1 SNP is rs2297136; it has been reported that patients carrying the genotype AG vs. GG were at increased risk of lung cancer [17]. In addition, patients with epithelial ovarian cancer carriers of the G allele of rs2297136 presented a shorter progression-free survival (PFS) than those patients harbouring the AA genotypes [24].
There is a lack of knowledge regarding the effect of SNPs present in the PDCDL1 gene in patients with MM, as well as their potential association with the prognosis of the disease. The search for biomarkers has become a highly relevant aspect of personalising the prognosis of each MM patient and is crucial in the early stages of the disease. The present study aimed to define the associations between specific SNPs in the PDCDL1 gene (rs822336, rs822337, rs822338, rs229736, rs4143815) and the main MM features and course. This would substantially improve the management of patients with melanoma.
2. Materials and Methods
2.1. Patients and Study Samples
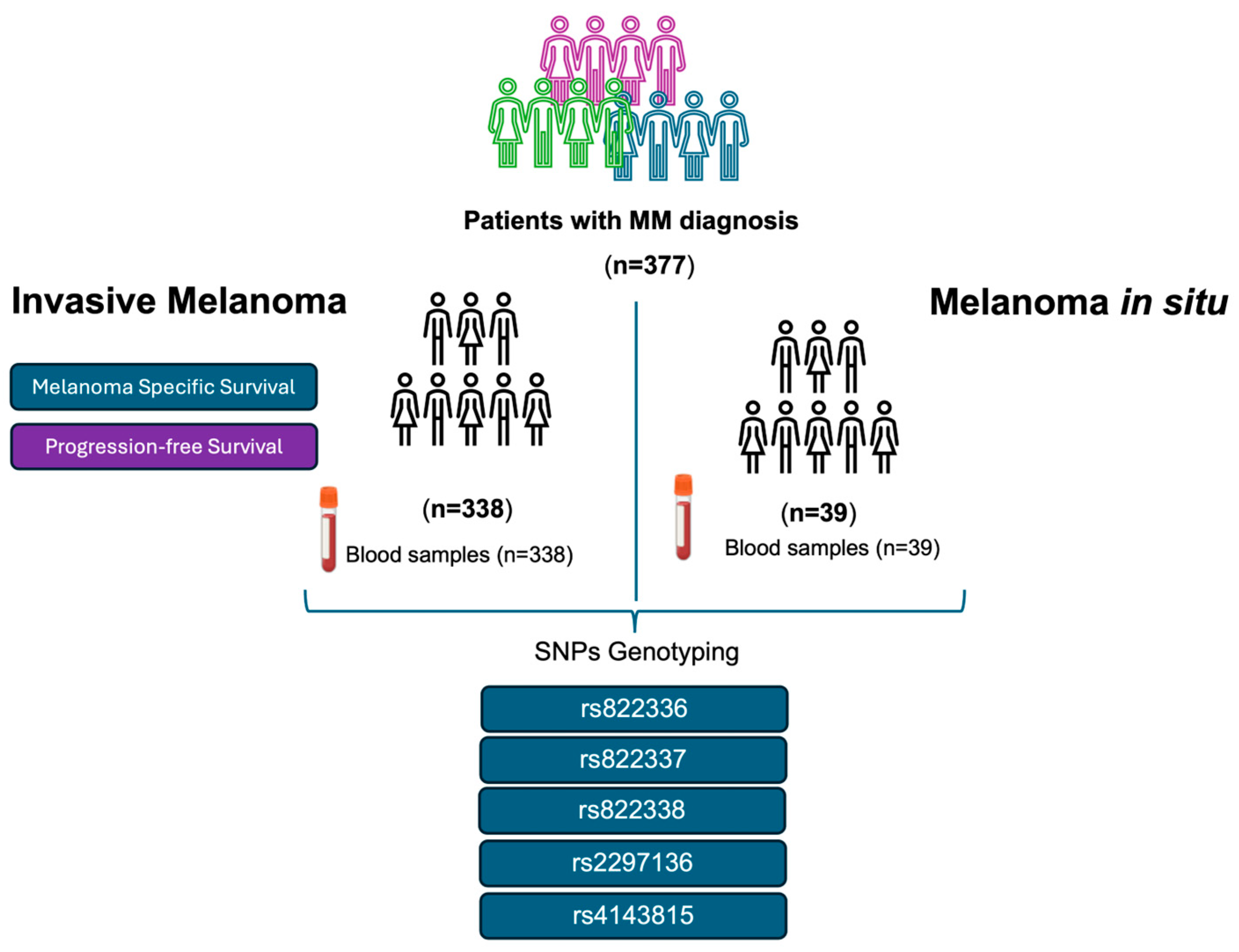
Following the systematic sampling method, we retrospectively screened blood samples from 377 patients of self-reported European origin with MM diagnosis who were followed by the Dermatology Department at “Complejo Hospitalario Universitario Nuestra Señora de Candelaria” (CHUNSC) (Santa Cruz de Tenerife, Spain) between the years 1975 and 2022 and who accepted to be included in this study. The samples corresponded to 338 patients with invasive melanoma and 39 with melanoma in situ (stage 0 of the AJCC TNM staging system). The following variables were analysed for the patients with invasive melanoma: sex, age at diagnosis, thickness (Breslow index), ulceration, and stage were analysed (Table 1). Figure 1 shows the patients included in this study and the workflow chart.

Table 1.
Clinicopathological characteristics of participating MM patients vs. melanoma progression and death due to melanoma.
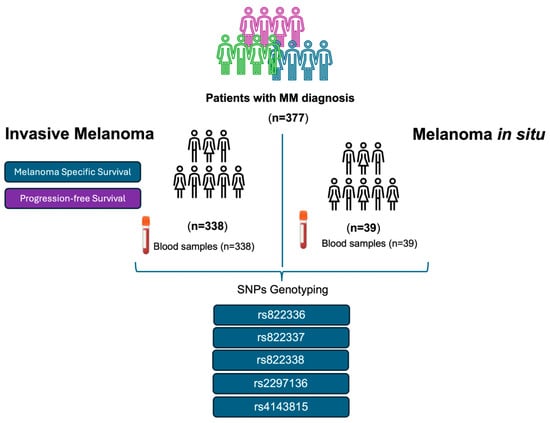
Figure 1.
Patients with invasive melanoma and melanoma in situ were included in this study. Flowchart of the methodology carried out.
This study was approved by the local Ethics Committee (C.P. MO—C.I. PI-57/17 and C.P. MO—C.I. PI-39/14) of HUNSC. Informed consent was obtained from all participants.
2.2. Genotyping of PDCDL1 Single-Nucleotide Polymorphisms
DNA extractions from whole blood were performed using the ‘IllustraTM blood genomicPrep Mini Spin’ kit (GE Healthcare Life Science, Chicago, IL, USA). DNA concentration was measured using the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). The study of PDCDL1 gene variants consisted of the selection of five single-nucleotide polymorphisms (SNPs) previously reported of interest in the literature: rs822336, rs822337, rs822338, rs2297136, and rs4143815 [18,20,21,22,23]. The genotype analysis of the selected SNPs was carried out by Helix BioS Company (Madrid, Spain). The DNA samples were genotyped for the cited SNPs, in duplicate, with qPCR using TaqMan probes in a QuantStudio™ 12K Flex Real-Time PCR System (Thermo Fisher Scientific Inc., Waltham, MA, USA), giving a total of 4170 trials. Negative controls were also included in the assay. An average overall quality of 98.96 (+/−0.17)% was obtained in the SNP calling.
2.3. Statistical Analysis
Hardy–Weinberg equilibrium (HWE) was determined using the quick exact test for HWE by Wigginton, Cutler, and Abecasis from the SNPassoc package 1.9-2-1. Likewise, for each SNP, the frequency of the minor allele (MAF) for this study and the SNP calling rate (%) were estimated.
Different models of genetic inheritance were evaluated: co-dominant, dominant, and recessive. Initially, the genotypes for each SNP were coded under the co-dominant genetic model. The differences between the genotypes of the SNPs involved in patients with MM and their clinical characteristics were evaluated using contingency tables based on the Chi-square test corrected by Yeats in the case of qualitative variables. For age and Breslow index, the ANOVA test and the Kruskal–Wallis test were used, respectively, evaluating in pairs and correcting for multiple comparisons. The cut-off point for the significance level was set at 0.05, and bilateral contrasts were performed.
Melanoma-specific survival (MSS) and progression-free survival (PFS) were estimated using the Kaplan–Meier (KM) method. Differences between curves were evaluated using the log-rank test, analysing both clinical and genotypic variables. A Cox regression for proportional hazards was used to assess possible associations between clinical and genotypic characteristics and the MSS and PFS endpoints. The hazard ratio (HR) was estimated with its 95% confidence interval (95% CI) in a univariate and multivariate analysis. Significance levels were established at a p < 0.05 value. HR was represented by “forest plot” graphs.
Analysis was performed using R statistical software version 3.6.2 (http://www.r-project.org/, accessed on 30 October 2022). Survival analysis and graphs were carried out with the Survival 3.4-0 (https://CRAN.R-project.org/package=survival, accessed on 30 October 2022) and Survminer 0.4.9 software (https://CRAN.R-project.org/package=survminer, accessed on 30 October 2022). The forestploter 0.2.0 software (https://CRAN.R-project.org/package=forestploter, accessed on 30 October 2022) was used to create the forest plots and the treatment and analysis of the genetic variants with the SNPassoc 1.9-2-1 (https://CRAN.R-project.org/package=SNPassoc, accessed on 30 October 2022) and SNPStats v0.95 (https://www.snpstats.net/, accessed on 30 October 2022) software.
3. Results
Thirty-nine patients (10.3%) showed in situ MM. Their mean age at diagnosis was 51.0 (±18) years. Twenty-nine were female (74.4%). This group of patients was not included in the prognostic analysis, as they were cured after excision and appropriate widening of surgical margins. The mean age at diagnosis for invasive MM patients was 53.1 (±16.3) years, and 142 (42%) were men. Table 1 shows the main clinical features of the 338 cases with invasive MM by specific death and progression of MM.
3.1. Clinical Features and Survival Analyses
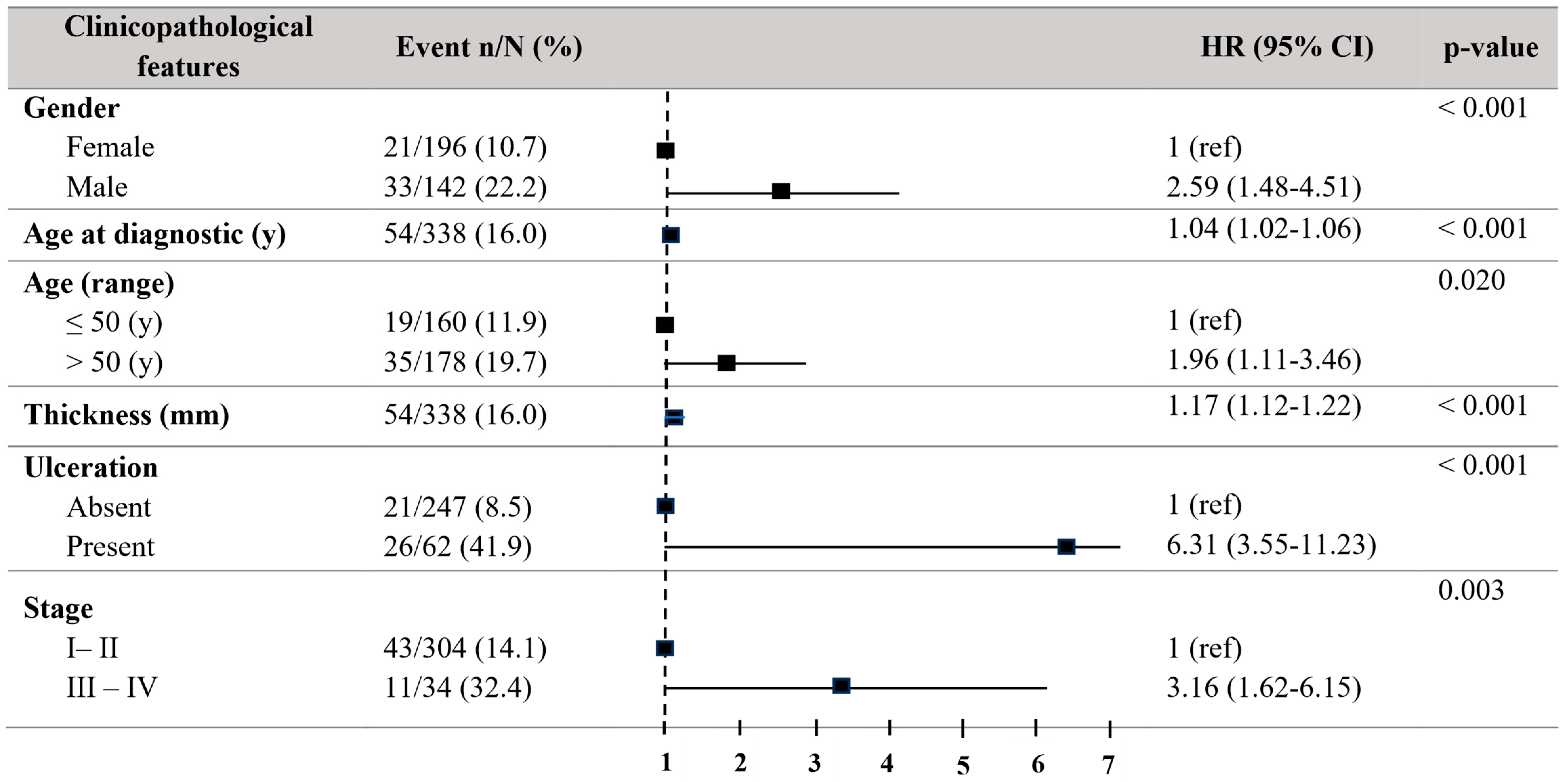
Regarding melanoma-specific survival (MSS), patients older than 50 years showed a significant decrease in MSS (HR = 1.96, 95% CI = 1.11–3.46; p = 0.021) in the univariate Cox regression analyses. Males revealed a decreased MSS (HR = 2.59, 95% CI = 1.48–4.51; p < 0.001). The tumour thickness (HR = 1.17, 95% CI = 1.12–1.22; p < 0.001) and the presence of ulceration (HR = 6.31, 95% CI = 3.55–11.23; p < 0.001) were in relationship to decreased MSS. Stages III–IV exhibited a significantly reduced MSS (HR = 3.16, 95% CI = 1.62–6.15; p = 0.003) (Figure 2).
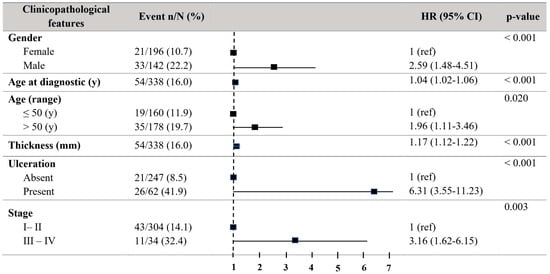
Figure 2.
Forest plot with the clinicopathological characteristics of melanoma patients. Univariate analysis of melanoma-specific survival (MSS). Hazard ratio (HR) (CI 95%). Patients with HR > 1 present a high risk of death. Statistically significant p-values < 0.05.
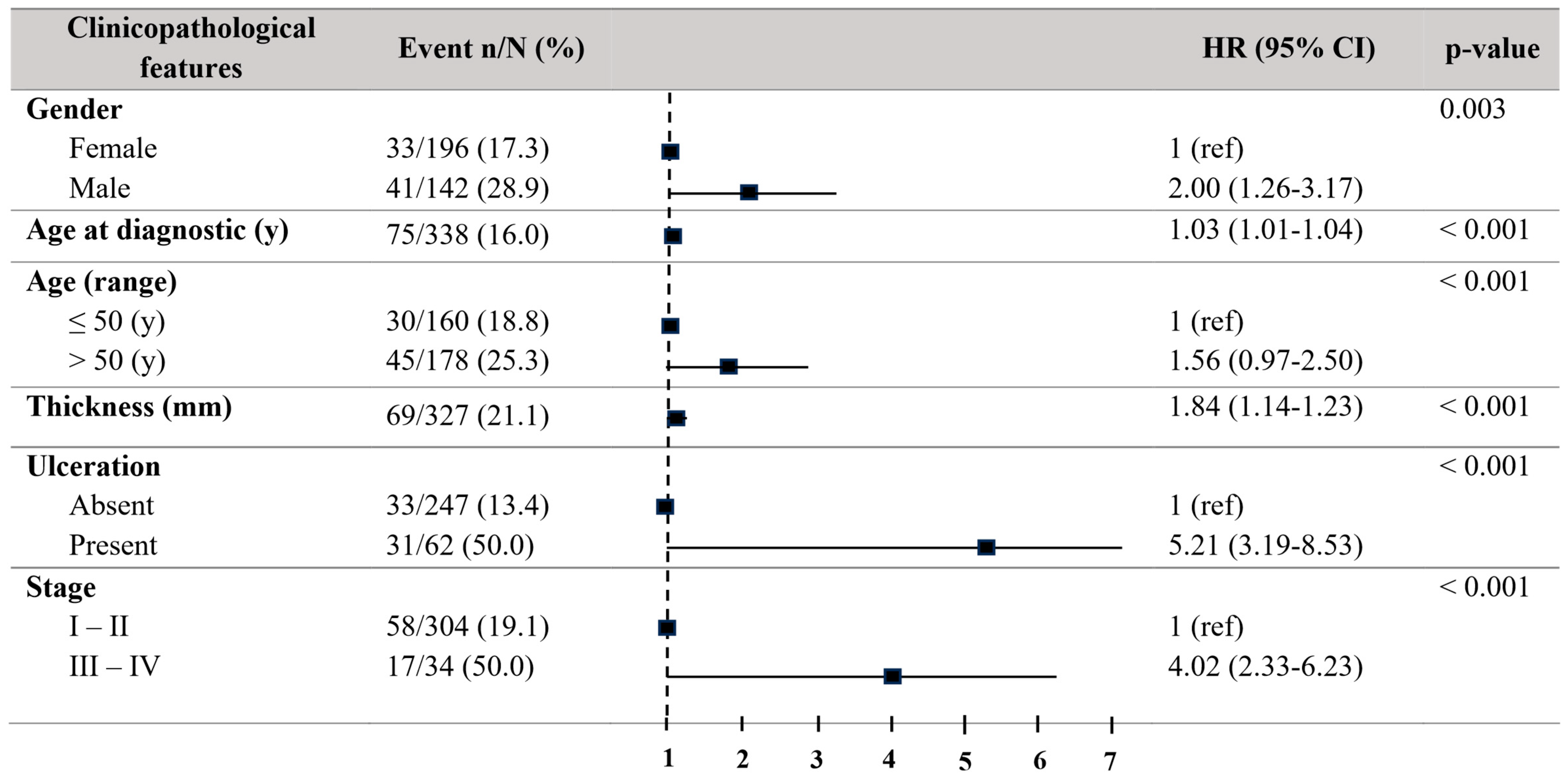
Concerning progression-free survival (PFS), patients older than 50 years also showed a decreased PFS (HR = 2.00, 95% CI = 1.26–3.17; p = 0.003). Males revealed a decreased PFS (HR = 2.00, 95% CI = 1.26–3.17; p = 0.003). The tumour thickness (HR01.84, 95% CI = 1.14–1.23; p < 0.001) and the existence of ulceration (HR = 5.21, 95% CI = 3.19–9.53; p < 0.001) disclosed a relationship to a reduced PFS. Stages III–IV also exhibited a significantly reduced PFS (HR = 4.02, 95% CI = 2.33–6.93; p < 0.001) (Figure 3).
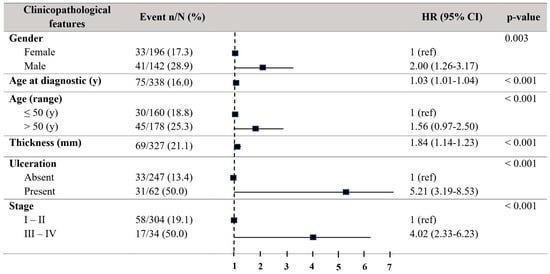
Figure 3.
Forest plot with the clinicopathological characteristics of melanoma patients. Univariate analysis of progression-free survival (PFS). Hazard ratio (HR) (IC 95%). Patients with HR > 1 present a high risk of death. Statistically significant p-values < 0.05.
3.2. PDCDL1 Polymorphisms and Survival Analyses
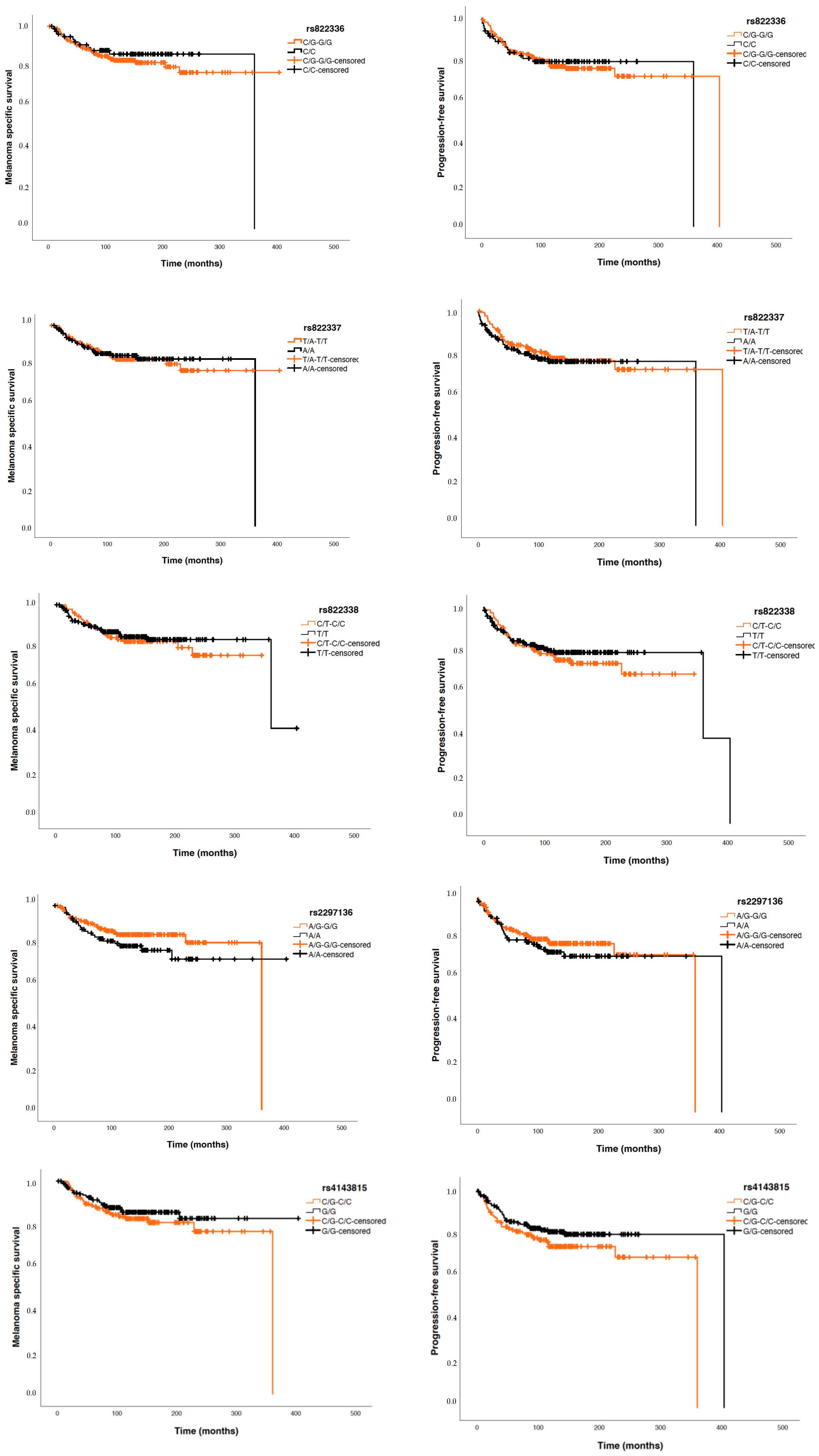
Considering patients with invasive tumours, Cox regression studies did not show significant differences between the five analysed PDCDL1 polymorphisms and PFS nor MSS (Figure 4).
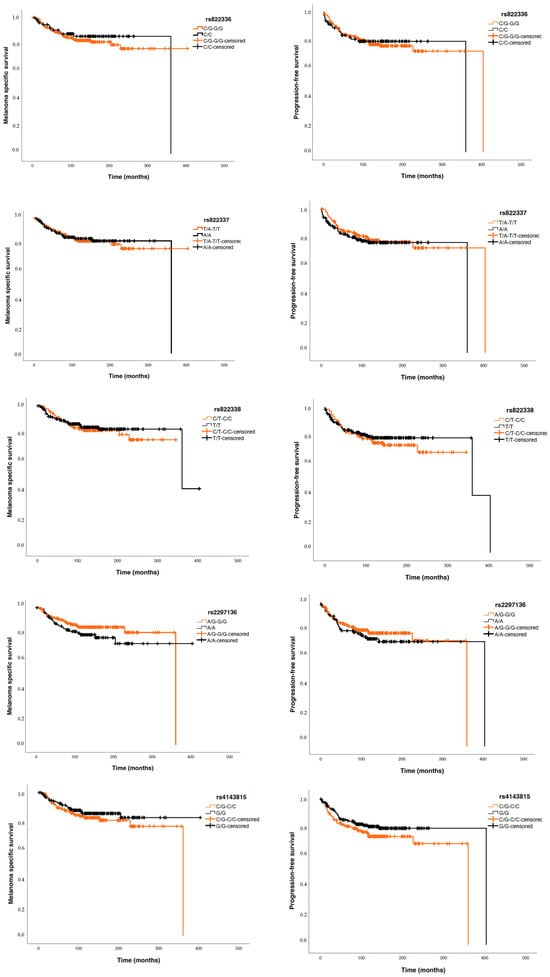
Figure 4.
Kaplan–Meier curves for melanoma-specific survival (MSS) and progression-free survival (PFS) based on the five ana-lysed SNPs.
3.3. PDCDL1 Polymorphisms and Tumour Invasiveness
The genetic variants analysis revealed an association between four of the five studied PDCDL1 SNPs and the invasiveness of the tumour. We found that patients who presented, under a dominant model of inheritance, the rs822336 CG or GG genotypes (OR = 3.01, 95% CI = 1.53–5.92; p = 0.0017), the TA or TT genotypes in rs822337 (OR = 2.45, 95% CI = 1.22–4.93; p = 0.0098), and the CT or CC genotype of rs822338 (OR = 2.23, 95% CI = 1.05–4.73; p = 0.028) were at an increased risk of developing invasive melanomas compared to those that presented an in situ melanoma (Table 2). On the other hand, those MM cases that harboured the AG or GG genotype in rs2297136 were at a lower risk of developing invasive tumours (OR = 0.29, 95% CI = 0.11–0.75; p = 0.0038) (Table 2).

Table 2.
Association of Single-Nucleotide Polymorphisms (SNPs) in PDCDL1 with tumour invasion.
The genetic analysis at the haplotype level resulted in similar findings. Patients harbouring the haplotype GTCAC, which contains the rs822336 rs822337, rs822338, and rs4143815 minor alleles and the rs2297136 frequent A allele, were at an increased risk of developing an invasive melanoma (OR = 2.95, 95% CI = 1.08–8.10, p = 0.036) (Table 3) compared to those who did not harbour it.

Table 3.
Distribution of the PDCDL1 haplotypes and their association with tumour invasiveness (in situ vs. invasive).
Regarding patients with invasive MM, a significant association was found between the SNP rs2297136 and the Breslow index. Patients carrying the homozygous AA genotype have thicker tumours (1.4 mm; IQR25–75: 0.6–2.6 mm) than those harbouring the AG (1.0 mm; IQR25–75: 0.6–2 mm) or GG (0.8 mm; IQR25–75: 0.5–1.7 mm) (p = 0.030). No association between other clinicopathological features and the studied gene variants was found. (Table 4).

Table 4.
Association analysis between the five polymorphisms of the PDCDL1 gene studied and the main clinical characteristics of patients with melanoma.
4. Discussion
Melanoma is one of the most aggressive forms of skin cancer, and its development is influenced by various genetic factors. However, the exact contribution of genetic variants to melanoma susceptibility and clinical characteristics such as primary tumour thickness remains poorly understood. For example, relevant studies have found that the variants rs12913832 in HERC2, rs3798577 in ESR1 [25], and rs183471242 on chromosome 11 are linked to both MM risk and tumour thickness, suggesting that germline genetic variation plays a crucial role in disease development and progression [26]. Up to now, data regarding the relationship between PDCDL1 SNPs and MM features have been scarce. This study provides novel descriptive and experimental analyses of the relationship between five PDCDL1 single-nucleotide variants and relevant features of the disease. The activation of the PD-1/PD-L1 axis is vital for tumour cells to evade the immune system. This is why the study of genetic variants in genes involved in immune control may be used to predict the course of the disease and the response to cancer treatment [23,27].
To our knowledge, this is the first study that analyses PDCDL1 gene variants and MM features. In our research, a significant association was found between four PDCDL1 SNPs and the invasive features of MM. Patients with invasive MM exhibited higher frequencies of the G allele of rs822336, the T allele of rs822337, and the C allele of rs822338 compared to patients with in situ MM. These results suggest that carrying these alleles is associated with an increased risk of tumour invasiveness. Overlapping findings have been reported for the rs822336 and rs822337 polymorphisms in non-small-cell lung cancer: individuals carrying the G and T alleles, respectively, showed poorer overall survival [28].
The polymorphism rs22997136 has been the focus of previous research. The study by Boutros et al. (2023) investigated its role in predicting the tumour response and development of immune-related adverse effects in patients with advanced MM with anti-PD-1 treatment without finding any significant results [29]. In the current study, the patients carrying the G allele of rs2297136 showed a significant association with in situ MM. Consistent with this finding, in the group of cases with invasive MM, individuals carrying the homozygous genotype A/A in rs2297136 had significantly thicker tumours compared to those with the A/G or G/G genotypes. To our knowledge, the relationship between this SNP and tumour thickness has not been reported in previous studies. However, other gene variants such as rs12203592 in IRF4 have already been associated with Breslow thickness and worse MSS [30] in patients of European origin.
The study of haplotypes, a combination of alleles of different loci on the same chromosome, has been valuable in identifying individuals at risk for developing a disease or its progression. An important finding in this study lies in the genetic association between the GTCAC haplotype (rs822336, rs822337, rs822338, rs2297136, rs4143815) in PDCDL1 and the increased risk of developing invasive MM: those patients carry this haplotype at higher frequencies than patients with in situ melanomas. Few studies focus on the study of haplotypes in PDCDL1. The first one analysed an Iranian population with basal cell carcinoma (BCC) [31]. The second focused on rheumatoid arthritis in the Chinese population [32]. Given the scarcity of data on the subject, the findings established here are relevant for conducting further studies.
Addressing polymorphisms to predict the progression of diseases is challenging. Regarding cancer, results vary significantly based on the specific molecular profiles of each tumour, the tumour microenvironment, and the presence of genetic heterogeneity. Research suggests that the herein-studied gene variants may be related to the prognosis of other types of cancer. Harbouring the rs822336 CC genotype was reported as a favourable prognostic factor in gastric cancer in a Chinese population [33]. The SNPs rs822336 (G/C) and rs822337 (T/A) have been linked to worse survival in triple-negative breast cancer patients in a European population [20]. Additionally, rs4143815 C/G was reported to be associated with increased susceptibility to developing gastric cancer, bladder cancer, and hepatocellular carcinoma [18]. However, the current study did not observe significant associations between the analysed PDCDL1 SNPs with MSS or PFS.
Importantly, genetic variants may influence the response to therapy. Therefore, the relationship between polymorphisms and disease prognosis is not always straightforward. For example, Gong et al. [34] found that Asian individuals with advanced non-small-cell lung cancer carrying the AA genotype of the rs2297136 SNP who received anti-PD-1 therapy had worse overall survival and PFS than those patients with the AG/GG genotype [34]. However, other investigations have provided contradictory results for the rs2297136 AA genotype and survival in ovarian [24] and gastric cancer [33]. These controversies may be due to differences in the tumour microenvironment, genetic regulation, and epigenetic modifications. Furthermore, population diversity is a crucial factor in the study of polymorphisms, as allele frequencies vary significantly across ethnic groups due to evolutionary history and local adaptations [35].
The rs2297136 is an A-to-G mutation in the 3′-UTR of PDCDL1. It is shown that rs2297136 could affect PD-L1 expression by modulating the miRNA–mRNA interaction [36,37]. An increase in the expression of PD-L1 in tumour cells or the tumour microenvironment, such as in tumour-associated macrophages, may allow melanoma to evade the immune response and facilitate an immunosuppressive environment that favours tumour growth, greater invasiveness and, therefore, a thicker tumour. The expression of PD-L1 is not necessarily associated with tumour development, as it may be induced as part of inflammatory processes that do not involve neoplasia or tumour development [38]. A meta-analysis including thirteen published articles with 1062 enrolled patients, conducted by Yang J. et al. [39], concluded that PD-L1 expression may not predict a worse prognosis in melanoma patients. Instead, elevated PD-L1 expression was found to be associated with the absence of lymph node metastasis [39].
As strengths of our study, we highlight the presence of a well-characterized cohort of patients with MM who underwent prolonged follow-up. The main clinical parameters were recorded for nearly every patient, and they are consistent with the data in the literature. Thus, it should be considered as a representative cohort [3,40]. Gender, Breslow index, ulceration, and stage at diagnosis were related to MSS and PFS [18,19,22]. However, our research also shows limitations. First, a larger series would provide more reliable results. Second, further genetic analyses adjusting for potential population structure would enhance the precision of our findings. Lastly, we recognise the importance of confirming the expression of PD-L1 in the tumour tissue. This data would allow us to complement the present findings, so further research will focus on it.
5. Conclusions
This study supports that SNPs in the PDCDL1 gene are associated with invasive features of MM. In particular, the G allele of rs822336, the T allele of rs822337, and the C allele of rs822338 are associated with a higher risk of tumour invasiveness. In addition, the A/A genotype of rs2297136 is associated with thicker tumours. No relationship with MSS or PFS was found. Further studies on PDCDL1 polymorphisms, considering diverse genetic and epigenetic factors, are necessary to understand their impact on cancer prognosis and treatment. Further large-scale epidemiologic studies are needed to confirm present findings.
Author Contributions
Conceptualization, R.F.-d.-M., E.C.-L. and O.G.-P.; methodology, O.G.-P., A.D.-d.-B., L.M.-V., E.C.-L. and R.F.-d.-M.; validation, L.M.-V. and E.C.-L.; formal analysis, R.F.-d.-M., E.C.-L. and O.G.-P.; investigation, E.C.-L., O.G.-P. and A.D.-d.-B.; resources, R.F.-d.-M.; data, R.F.-d.-M., E.C.-L. and O.G.-P.; curation, R.F.-d.-M., E.C.-L. and O.G.-P.; writing original draft preparation, R.F.-d.-M., E.C.-L. and O.G.-P.; visualization, R.F.-d.-M. and E.C.-L.; supervision, R.F.-d.-M. and E.C.-L.; project administration, R.F.-d.-M. and E.C.-L.; funding acquisition, R.F.-d.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Fundación Canaria Instituto de Investigación Sanitaria de Canarias, PIFIISC21/15, Cabildo Insular de Tenerife 2023–2028, PI-CC202302222, Cabildo.23, Consorcio Centro de Investigación Biomédica (CIBER) de Enfermedades Infecciosas (CIBERINFEC) (CB21/13/00100); Instituto de Salud Carlos III, 28006 Madrid, Spain.
Institutional Review Board Statement
This study was conducted with the Declaration of Helsinki and approved by the Ethical Committee Board of Hospital Universitario N/S de Candelaria, Santa Cruz de Tenerife (protocol code: C.P. MO—C.I. PI-57/17, 25 July 2017 and C.P. MO—C.I. PI-39/14, 23 January 2015).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data are available upon reasonable request. All data relevant to this study are included in the article.
Acknowledgments
We thank Hilaria González for her technical assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Welch, H.G.; Mazer, B.L.; Adamson, A.S. The Rapid Rise in Cutaneous Melanoma Diagnoses. N. Engl. J. Med. 2021, 384, 72–79. [Google Scholar] [CrossRef]
- Ríos, L.; Nagore, E.; López, J.; Redondo, P.; Martí, R.; Fernández-De-Misa, R.; Soler, B. Melanoma characteristics at diagnosis from the Spanish National Cutaneous Melanoma Registry: 15 years of experience. Actas Dermosifiliogr. 2013, 104, 789–799, Erratum in Ann. Surg. Oncol. 2018, 25 (Suppl. S3), 993–994. [Google Scholar] [CrossRef] [PubMed]
- Gershenwald, J.E.; Scolyer, R.A. Melanoma Staging: American Joint Committee on Cancer (AJCC) 8th Edition and Beyond. Ann. Surg. Oncol. 2018, 25, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Tejera-Vaquerizo, A.; Solís-García, E.; Ríos-Martín, J.J.; Moreno-Ramírez, D. Factores pronósticos en el melanoma cutáneo primario no incluidos en la clasificación de la American Joint Committee on Cancer (AJCC) [Primary cutaneous melanoma: Prognostic factors not included in the classification of the American Joint Committee on Cancer]. Actas Dermosifiliogr. 2011, 102, 255–263. [Google Scholar] [CrossRef]
- Balch, C.M.; Gershenwald, J.E.; Soong, S.-J.; Thompson, J.F.; Atkins, M.B.; Byrd, D.R.; Buzaid, A.C.; Cochran, A.J.; Coit, D.G.; Ding, S.; et al. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009, 27, 6199–6206. [Google Scholar] [CrossRef] [PubMed]
- Waseh, S.; Lee, J.B. Advances in melanoma: Epidemiology, diagnosis, and prognosis. Front. Med. 2023, 10, 1268479. [Google Scholar] [CrossRef]
- Zitvogel, L.; Kroemer, G. Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology 2012, 1, 1223–1225. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Barber, D.L.; Wherry, E.J.; Masopust, D.; Zhu, B.; Allison, J.P.; Sharpe, A.H.; Freeman, G.J.; Ahmed, R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006, 439, 682–687. [Google Scholar] [CrossRef]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 pathway: Current researches in cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar]
- Wang, X.; Teng, F.; Kong, L.; Yu, J. PD-L1 expression in human cancers and its association with clinical outcomes. OncoTargets Ther. 2016, 9, 5023–5039. [Google Scholar] [CrossRef]
- Hino, R.; Kabashima, K.; Kato, Y.; Yagi, H.; Nakamura, M.; Honjo, T.; Okazaki, T.; Tokura, Y. Tumor cell expression of programmed cell death-1 ligand 1 is a prognostic factor for malignant melanoma. Cancer 2010, 116, 1757–1766. [Google Scholar] [CrossRef] [PubMed]
- Hamanishi, J.; Mandai, M.; Iwasaki, M.; Okazaki, T.; Tanaka, Y.; Yamaguchi, K.; Higuchi, T.; Yagi, H.; Takakura, K.; Minato, N.; et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 3360–3365. [Google Scholar] [CrossRef] [PubMed]
- Kythreotou, A.; Siddique, A.; Mauri, F.A.; Bower, M.; Pinato, D.J. PD-L1. J. Clin. Pathol. 2018, 71, 189–194. [Google Scholar] [CrossRef]
- Daud, A.I.; Wolchok, J.D.; Robert, C.; Hwu, W.-J.; Weber, J.S.; Ribas, A.; Hodi, F.S.; Joshua, A.M.; Kefford, R.; Hersey, P.; et al. Programmed Death-Ligand 1 Expression and Response to the Anti-Programmed Death 1 Antibody Pembrolizumab in Melanoma. J. Clin. Oncol. 2016, 34, 4102–4109. [Google Scholar] [CrossRef]
- National Library of Medicine. NCBI Gene Resource CD274 Molecule [Homo sapiens (Human)]-Gene-NCBI. 2017. Available online: https://www.ncbi.nlm.nih.gov/gene (accessed on 29 June 2017).
- Sakran, M.I.; Alalawy, A.I.; Alharbi, A.A.; El-Hefnawy, M.E.; Alzahrani, S.M.; Alfuraydi, A.; Alzuaibr, F.M.; Zidan, N.S.; Elsaid, A.M.; Toraih, E.A.; et al. The blockage signal for PD-L1/CD274 gene variants and their potential impact on lung carcinoma susceptibility. Int. Immunopharmacol. 2023, 125 Pt A, 111180. [Google Scholar] [CrossRef]
- Zou, J.; Wu, D.; Li, T.; Wang, X.; Liu, Y.; Tan, S. Association of PD-L1 gene rs4143815 C>G polymorphism and human cancer susceptibility: A systematic review and meta-analysis. Pathol. Res. Pract. 2019, 215, 229–234. [Google Scholar] [CrossRef]
- Zhou, R.-M.; Li, Y.; Liu, J.-H.; Wang, N.; Huang, X.; Cao, S.-R.; Shan, B.-E. Programmed death-1 ligand-1 gene rs2890658 polymorphism associated with the risk of esophageal squamous cell carcinoma in smokers. Cancer Biomark. 2017, 21, 65–71. [Google Scholar] [CrossRef]
- Makrantonakis, A.-E.; Zografos, E.; Gazouli, M.; Dimitrakakis, K.; Toutouzas, K.G.; Zografos, C.G.; Kalapanida, D.; Tsiakou, A.; Samelis, G.; Zagouri, F. PD-L1 Gene Polymorphisms rs822336 G>C and rs822337 T>A: Promising Prognostic Markers in Triple Negative Breast Cancer Patients. Medicina 2022, 58, 1399. [Google Scholar] [CrossRef]
- Nomizo, T.; Ozasa, H.; Tsuji, T.; Funazo, T.; Yasuda, Y.; Yoshida, H.; Yagi, Y.; Sakamori, Y.; Nagai, H.; Hirai, T.; et al. Clinical Impact of Single Nucleotide Polymorphism in PD-L1 on Response to Nivolumab for Advanced Non-Small-Cell Lung Cancer Patients. Sci. Rep. 2017, 7, 45124. [Google Scholar] [CrossRef]
- Lee, S.Y.; Jung, D.K.; Choi, J.E.; Jin, C.C.; Hong, M.J.; Do, S.K.; Kang, H.-G.; Lee, W.K.; Seok, Y.; Lee, E.B.; et al. Functional polymorphisms in PD-L1 gene are associated with the prognosis of patients with early stage non-small cell lung cancer. Gene 2017, 599, 28–35. [Google Scholar] [CrossRef]
- Yeo, M.K.; Choi, S.Y.; Seong, I.O.; Suh, K.S.; Kim, J.M.; Kim, K.H. Association of PD-L1 expression and PD-L1 gene polymorphism with poor prognosis in lung adenocarcinoma and squamous cell carcinoma. Hum. Pathol. 2017, 68, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, Y.; Si, W.; Hua, T.; Chen, J.; Kang, S. Genetic Variation of PD-L1 Gene Affects Its Expression and Is Related to Clinical Outcome in Epithelial Ovarian Cancer. Front. Oncol. 2022, 12, 763134. [Google Scholar] [CrossRef] [PubMed]
- Calbet-Llopart, N.; Combalia, M.; Kiroglu, A.; Potrony, M.; Tell-Martí, G.; Combalia, A.; Brugues, A.; Podlipnik, S.; Carrera, C.; Puig, S.; et al. Common genetic variants associated with melanoma risk or naevus count in patients with wildtype MC1R melanoma. Br. J. Dermatol. 2022, 187, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Mangantig, E.; MacGregor, S.; Iles, M.M.; Scolyer, A.R.; Cust, E.A.; Hayward, N.K.; Montgomery, G.W.; Duffy, D.L.; Thompson, J.F.; Henders, A.; et al. Germline variants are associated with increased primary melanoma tumor thickness at diagnosis. Hum. Mol. Genet. 2021, 29, 3578–3587. [Google Scholar] [CrossRef]
- Lin, X.; Kang, K.; Chen, P.; Zeng, Z.; Li, G.; Xiong, W.; Yi, M.; Xiang, B. Regulatory mechanisms of PD-1/PD-L1 in cancers. Mol. Cancer. 2024, 23, 108. [Google Scholar] [CrossRef]
- Kang, M.K.; Lee, S.Y.; Choi, J.E.; Do, S.K.; Cho, M.; Kim, J.; Park, J.Y. Prognostic implication of PD-L1 polymorphisms in non-small cell lung cancer treated with radiotherapy. Cancer Med. 2021, 10, 8071–8078. [Google Scholar] [CrossRef]
- Boutros, A.; Carosio, R.; Campanella, D.; Spagnolo, F.; Banelli, B.; Morabito, A.; Pistillo, M.; Croce, E.; Cecchi, F.; Pronzato, P.; et al. The predictive and prognostic role of single nucleotide gene variants of PD-1 and PD-L1 in patients with advanced melanoma treated with PD-1 inhibitors. Immunooncol. Technol. 2023, 20, 100408. [Google Scholar] [CrossRef]
- Davari, D.R.; Orlow, I.; Kanetsky, P.A.; Luo, L.; Busam, K.J.; Sharma, A.; Kricker, A.; Cust, A.E.; Anton-Culver, H.; Gruber, S.B.; et al. Association of Melanoma-Risk Variants with Primary Melanoma Tumor Prognostic Characteristics and Melanoma-Specific Survival in the GEM Study. Curr. Oncol. 2021, 28, 4756–4771. [Google Scholar] [CrossRef]
- Fathi, F.; Ebrahimi, M.; Eslami, A.; Hafezi, H.; Eskandari, N.; Motedayyen, H. Association of programmed death-1 gene polymorphisms with the risk of basal cell carcinoma. Int. J. Immunogenet. 2019, 46, 444–450. [Google Scholar] [CrossRef]
- Kong, E.K.P.; Prokunina-Olsson, L.; Wong, W.H.S.; Lau, C.S.; Chan, T.M.; Alarcón-Riquelme, M.; Lau, Y.-L. A new haplotype of PDCD1 is associated with rheumatoid arthritis in Hong Kong Chinese. Arthritis Rheum. 2005, 52, 1058–1062. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhao, T.; Jia, Z.; Cao, D.; Cao, X.; Pan, Y.; Zhao, D.; Zhang, B.; Jiang, J. Polymorphism of the programmed death-ligand 1 gene is associated with its protein expression and prognosis in gastric cancer. J. Gastroenterol. Hepatol. 2019, 34, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Qie, H.L.; Dong, S.Y.; Jiang, H.T. Implication of PD-L1 polymorphisms rs2297136 on clinical outcomes of patients with advanced NSCLC who received PD-1 blockades: A retrospective exploratory study. Oncol. Lett. 2024, 27, 144. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hindorff, L.A.; Bonham, V.L.; Brody, L.C.; Ginoza, M.E.C.; Hutter, C.M.; Manolio, T.A.; Green, E.D. Prioritizing diversity in human genomics research. Nat. Rev. Genet. 2018, 19, 175–185. [Google Scholar] [CrossRef]
- Umemoto, M.; Yokoyama, Y.; Sato, S.; Tsuchida, S.; Al-Mulla, F.; Saito, Y. Carbonyl reductase as a significant predictor of survival and lymph node metastasis in epithelial ovarian cancer. Br. J. Cancer 2001, 85, 1032–1036. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Zhu, J.; Chen, Y.; Zeng, Y.; Shen, D.; Zhang, N.; Ning, W.; Liu, Z.; Huang, J.-A. Variant SNPs at the microRNA complementary site in the B7-H1 3′-untranslated region increase the risk of non-small cell lung cancer. Mol. Med. Rep. 2017, 16, 2682–2690. [Google Scholar] [CrossRef]
- Gong, J.; Chehrazi-Raffle, A.; Reddi, S.; Salgia, R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations. J. Immunother. Cancer 2018, 6, 8. [Google Scholar] [CrossRef]
- Yang, J.; Dong, M.; Shui, Y.; Zhang, Y.; Zhang, Z.; Mi, Y.; Zuo, X.; Jiang, L.; Liu, K.; Liu, Z.; et al. A pooled analysis of the prognostic value of PD-L1 in melanoma: Evidence from 1062 patients. Cancer Cell Int. 2020, 20, 96. [Google Scholar] [CrossRef]
- Rastrelli, M.; Tropea, S.; Rossi, C.R.; Alaibac, M. Melanoma: Epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo 2014, 28, 1005–1011. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).