Pathwalker: A New Individual-Based Movement Model for Conservation Science and Connectivity Modelling
Abstract
1. Introduction
- This being human is a guest house.
- Every morning a new arrival.
- A joy, a depression, a meanness,
- some momentary awareness comes
- as an unexpected visitor.
- Welcome and entertain them all!...
- The dark thought, the shame, the malice,
- meet them at the door laughing,
- and invite them in.
- Be grateful for whoever comes,
- because each has been sent
- as a guide from beyond.
1.1. Landscape Connectivity
1.1.1. Resistance Surfaces
1.1.2. Modelling Connectivity
1.1.3. Individual-Based Models
1.2. Pathwalker
2. Methods: The Pathwalker Model
2.1. Input Layers
2.2. Model Parameters
2.2.1. Movement Mechanisms
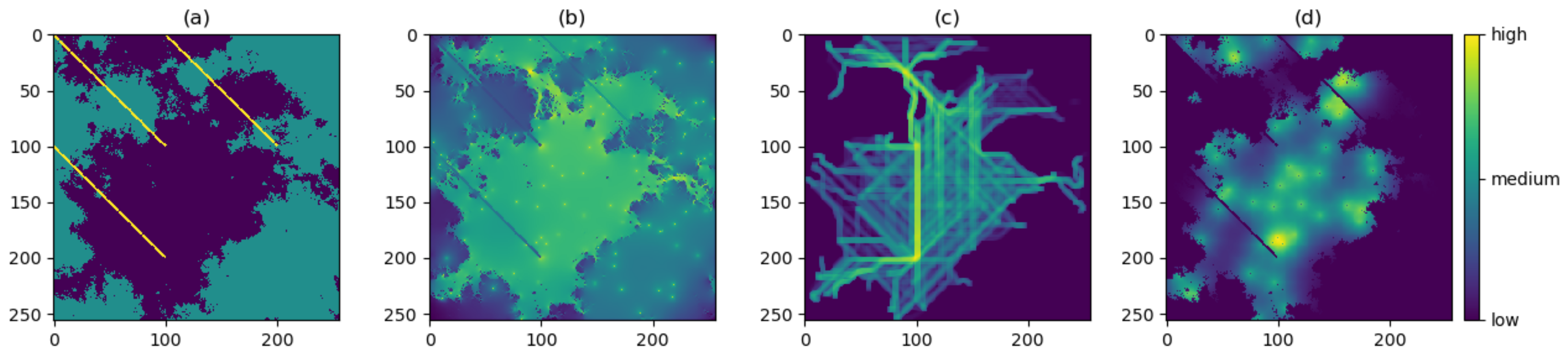
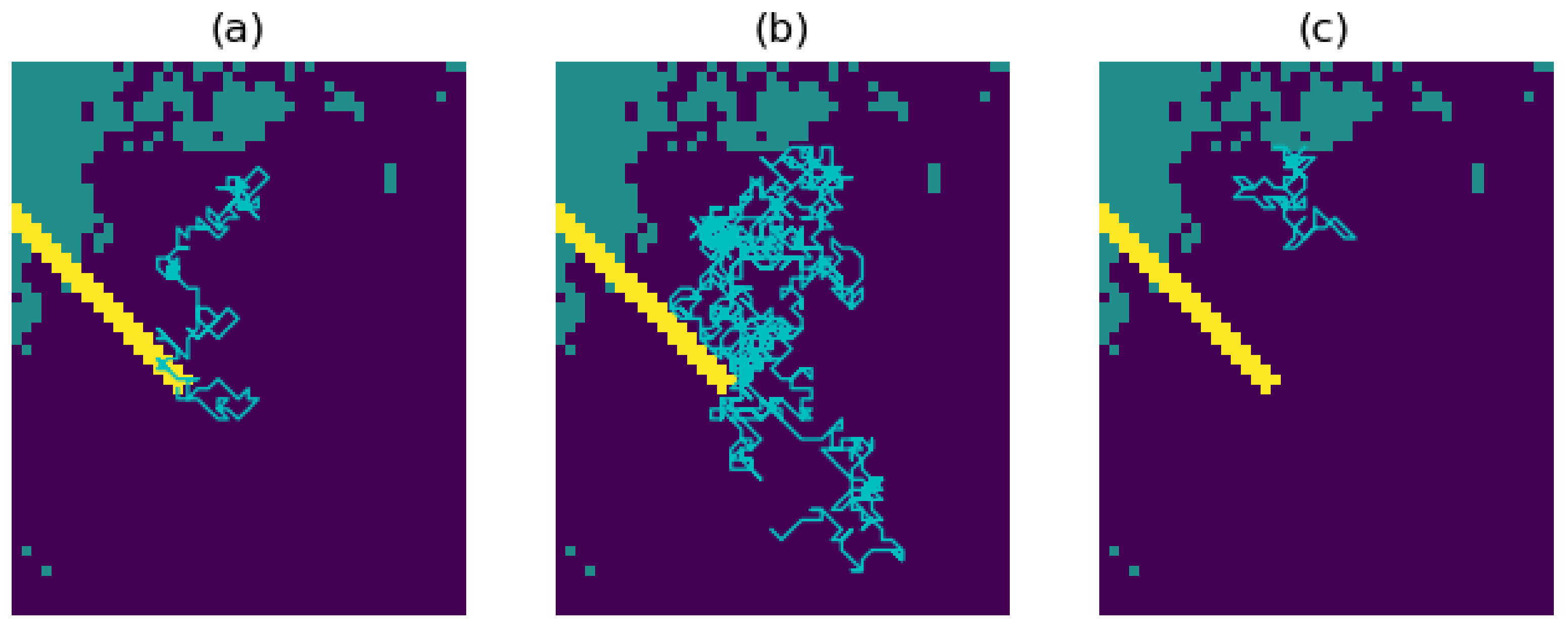
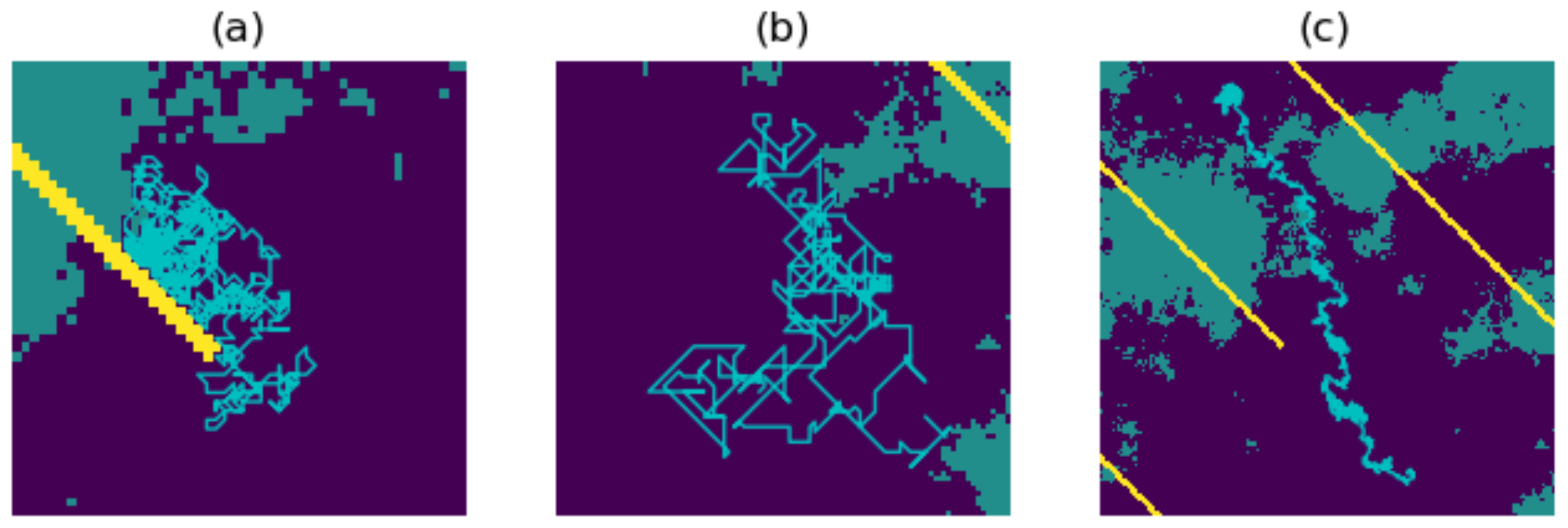
- Energy. We first specify a value for the ‘total energy’ parameter, which represents the maximum allowed cumulative energetic cost of movement. The walker then follows an unbiased random walk on the resistance surface, and thus at each step chooses any one of the nine pixels with equal probability. For the pixels traversed in the walk, a cumulative sum of the resistance values is computed. The walk ends once this sum reaches or exceeds the chosen total energy value, or once the maximum number of steps has been reached.
- Attraction. The walker now follows a resistance-biased random walk. The probability of choosing any one of the nine pixels is given by the inverse of that pixel’s resistance value (where the inverse resistance values are scaled so that the nine inverse values give a probability distribution; in other words, these nine inverse values sum to 1). Thus, the walker will be more likely to move to pixels of lower resistance value and vice versa. The four diagonally adjacent pixels are given a weighting of 1/ to account for the increased distance when moving diagonally. This mechanism does not require any additional parameters to be specified, and the walk ends once the maximum number of steps has been reached.
- Risk. If risk is the only chosen factor in the movement, then we first specify a chosen risk surface; this may be proportional to our resistance surface (the default setting), or a different surface may be chosen but must be scaled so that the values of the risk surface lie between 0 (no risk) and 1 (highest risk). The walker then follows an unbiased random walk on the risk surface. At each step, the probability that the walk ends is given by the value of that pixel in the risk surface. The maximum length of the walk capped at the chosen maximum number of steps.
2.2.2. Spatial Scale of Movement Choice
- The window size determines the spatial scale at which the movement responds to resistance values. For example, if the chosen window size is 7-by-7, then the movement will be affected by resistance values in a 7-by-7 pixel neighbourhood of each of the nine pixels. The default scale, a 1-by-1 window, is equivalent to not incorporating spatial scaling.
- The scaling function determines the way in which the walker responds to landscape resistance at a chosen spatial scale n. There are three choices for the scaling function: focal mean, focal maximum, and focal minimum. With the focal mean, the resistance value of a pixel is replaced by the mean average of the resistance values of all pixels in a n-by-n neighbourhood of that pixel. With the focal maximum, the value of a pixel is replaced by the maximum pixel value in that neighbourhood; with the focal minimum, it is replaced by the minimum value. The window size and scaling function also act in this way on the risk surface (if used).
2.2.3. Directionality: Autocorrelation and Destination Bias
- Autocorrelation. This parameter C takes values between 0 and 1, and determines the degree to which the walker is inclined to continue in the present direction of travel. The default value of C is 0, an uncorrelated random walk. If we increase the value of C, then our walk becomes more correlated, with the extreme case resulting in a straight line (in other words, a path in which the walker continues in the same direction with probability 1). For example, if we choose , then the nine movement probabilities are scaled to sum to instead of summing to 1, and there will now be an added probability of 0.3 for continuing in the same direction as the previous step.
- Destination bias. This parameter D takes values between 0 and 1, and determines the degree to which the walk will be biased towards a destination point X on the resistance surface. It works by giving additional preference to moving to the pixel closest to the direction of X. The default value is 0, in which there is no bias towards the destination. As we increase the value of D, the walk becomes more biased towards X, with the extreme case resulting in a path which is a straight line towards X. For example, if , then the nine movement probabilities are scaled to sum to instead of summing to 1, and there will now be an added probability of 0.2 for moving to the pixel closest to the direction of X.
2.2.4. Additional Parameters
2.3. Output Layers
2.4. Summary of Pathwalker Setup
3. Case Study
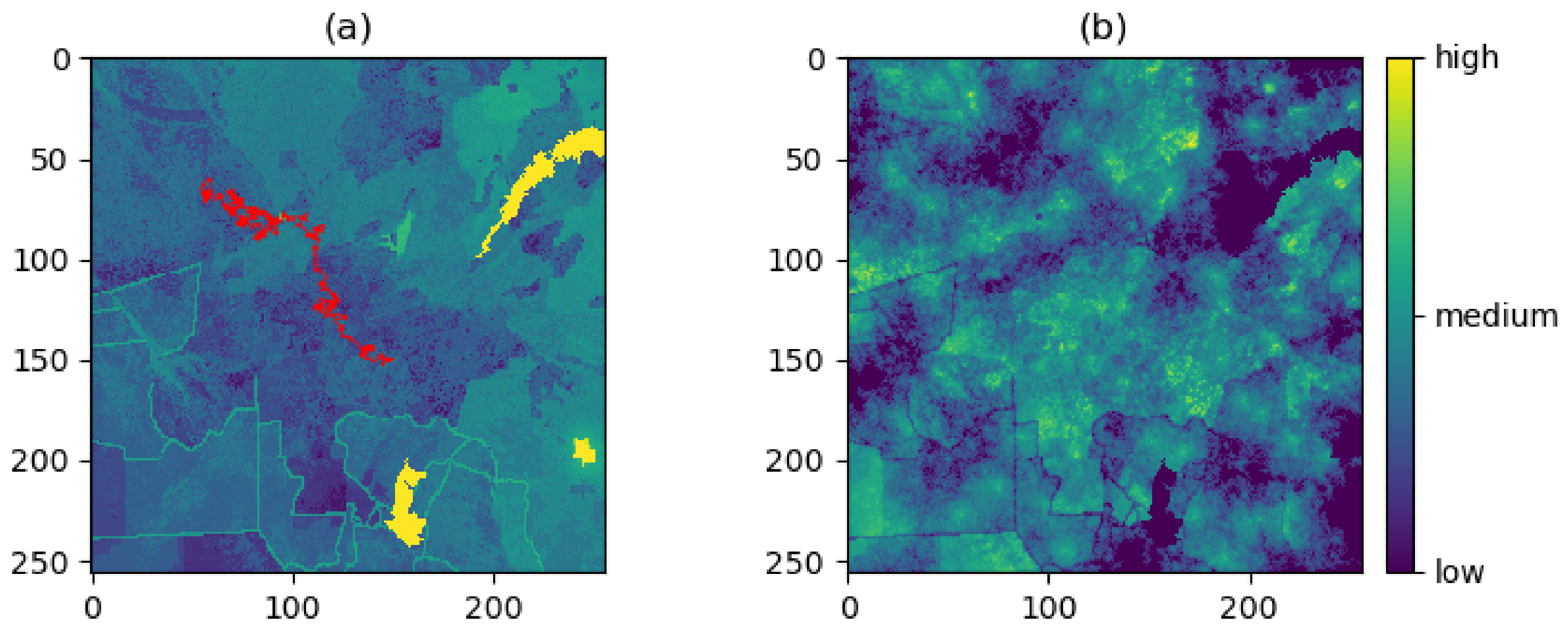
3.1. Producing the Density Surfaces
3.2. Comparison of Density Surfaces
4. Results
5. Discussion
5.1. Relation to Popular Connectivity Models
5.2. Limitations, Further Developments, and the Wider Context
5.3. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abram, D. The Spell of the Sensuous: Perception and Language in a More-Than-Human World; Vintage: New York City, NY, USA, 1996. [Google Scholar]
- Ingold, T. Being Alive: Essays on Movement, Knowledge and Description; Routledge: Abingdon-on-Thames, UK, 2011. [Google Scholar] [CrossRef]
- Fahrig, L. Effects of habitat fragmentation on biodiversity. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 487–515. [Google Scholar] [CrossRef]
- Cushman, S.A. Effects of habitat loss and fragmentation on amphibians: A review and prospectus. Biol. Conserv. 2006, 128, 231–240. [Google Scholar] [CrossRef]
- Cushman, S.A.; Elliot, N.B.; Macdonald, D.W.; Loveridge, A.J. A multi-scale assessment of population connectivity in African lions (Panthera leo) in response to landscape change. Landsc. Ecol. 2016, 31, 1337–1353. [Google Scholar] [CrossRef]
- Lorimer, J. Wildlife in the Anthropocene: Conservation after Nature; University of Minnesota Press: Minneapolis, MN, USA, 2015. [Google Scholar]
- Abram, D. Becoming Animal: An Earthly Cosmology; Vintage: New York City, NY, USA, 2010. [Google Scholar]
- Ingold, T. Point, line and counterpoint: From environment to fluid space. In Neurobiology of “Umwelt”; Springer: Berlin/Heidelberg, Germany, 2009; pp. 141–155. [Google Scholar]
- Levin, S.A. The problem of pattern and scale in ecology: The Robert H. MacArthur award lecture. Ecology 1992, 73, 1943–1967. [Google Scholar] [CrossRef]
- Gibbs, J.P. Amphibian movements in response to forest edges, roads, and streambeds in southern New England. J. Wildl. Manag. 1998, 62, 584–589. [Google Scholar] [CrossRef]
- Ingold, T. The Perception of the Environment: Essays on Livelihood, Dwelling and Skill; Routledge: Abingdon-on-Thames, UK, 2000. [Google Scholar]
- Tischendorf, L.; Fahrig, L. On the usage and measurement of landscape connectivity. Oikos 2000, 90, 7–19. [Google Scholar] [CrossRef]
- Rudnick, D.; Ryan, S.J.; Beier, P.; Cushman, S.A.; Dieffenbach, F.; Epps, C.; Gerber, L.R.; Hartter, J.N.; Jenness, J.S.; Kintsch, J.; et al. The role of landscape connectivity in planning and implementing conservation and restoration priorities. Issues Ecol. 2012, 16, 1–23. [Google Scholar]
- Hilty, J.A.; Lidicker, W.Z., Jr.; Merenlender, A.M. Corridor Ecology: The Science and Practice of Linking Landscapes for Biodiversity Conservation; Island Press: Washington, DC, USA, 2012. [Google Scholar]
- Cushman, S.A.; McRae, B.H.; Adriaensen, F.; Beier, P.; Shirley, M.; Zeller, K. Biological corridors and connectivity. In Key Topics in Conservation Biology 2; Macdonald, D.W., Willis, K.J., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2013; Chapter 21; pp. 384–404. [Google Scholar]
- Zeller, K.A.; Jennings, M.K.; Vickers, T.W.; Ernest, H.B.; Cushman, S.A.; Boyce, W.M. Are all data types and connectivity models created equal? Validating common connectivity approaches with dispersal data. Divers. Distrib. 2018, 24, 868–879. [Google Scholar] [CrossRef]
- Zeller, K.A.; McGarigal, K.; Whiteley, A.R. Estimating landscape resistance to movement: A review. Landsc. Ecol. 2012, 27, 777–797. [Google Scholar] [CrossRef]
- Elliot, N.B.; Cushman, S.A.; Macdonald, D.W.; Loveridge, A.J. The devil is in the dispersers: Predictions of landscape connectivity change with demography. J. Appl. Ecol. 2014, 51, 1169–1178. [Google Scholar] [CrossRef]
- McRae, B.H.; Dickson, B.G.; Keitt, T.H.; Shah, V.B. Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology 2008, 89, 2712–2724. [Google Scholar] [CrossRef] [PubMed]
- Adriaensen, F.; Chardon, J.; De Blust, G.; Swinnen, E.; Villalba, S.; Gulinck, H.; Matthysen, E. The application of ‘least-cost’modelling as a functional landscape model. Landsc. Urban Plan. 2003, 64, 233–247. [Google Scholar] [CrossRef]
- Cushman, S.A.; McKelvey, K.S.; Schwartz, M.K. Use of empirically derived source–destination models to map regional conservation corridors. Conserv. Biol. 2009, 23, 368–376. [Google Scholar] [CrossRef]
- Landguth, E.; Hand, B.; Glassy, J.; Cushman, S.; Sawaya, M. UNICOR: A species connectivity and corridor network simulator. Ecography 2012, 35, 9–14. [Google Scholar] [CrossRef]
- Moilanen, A. On the limitations of graph-theoretic connectivity in spatial ecology and conservation. J. Appl. Ecol. 2011, 48, 1543–1547. [Google Scholar] [CrossRef]
- Compton, B.W.; McGarigal, K.; Cushman, S.A.; Gamble, L.R. A resistant-kernel model of connectivity for amphibians that breed in vernal pools. Conserv. Biol. 2007, 21, 788–799. [Google Scholar] [CrossRef] [PubMed]
- Dray, S.; Royer-Carenzi, M.; Calenge, C. The exploratory analysis of autocorrelation in animal-movement studies. Ecol. Res. 2010, 25, 673–681. [Google Scholar] [CrossRef]
- Cushman, S.A.; Chase, M.; Griffin, C. Elephants in space and time. Oikos 2005, 109, 331–341. [Google Scholar] [CrossRef]
- Wiens, J.A. Spatial scaling in ecology. Funct. Ecol. 1989, 3, 385–397. [Google Scholar] [CrossRef]
- Osipova, L.; Okello, M.; Njumbi, S.; Ngene, S.; Western, D.; Hayward, M.; Balkenhol, N. Using step-selection functions to model landscape connectivity for African elephants: Accounting for variability across individuals and seasons. Anim. Conserv. 2019, 22, 35–48. [Google Scholar] [CrossRef]
- Kaszta, Ż.; Cushman, S.A.; Slotow, R. Temporal Non-stationarity of Path-Selection Movement Models and Connectivity: An Example of African Elephants in Kruger National Park. Front. Ecol. Evol. 2021, 9, 207. [Google Scholar] [CrossRef]
- Gorini, L.; Linnell, J.D.; May, R.; Panzacchi, M.; Boitani, L.; Odden, M.; Nilsen, E.B. Habitat heterogeneity and mammalian predator–prey interactions. Mammal Rev. 2012, 42, 55–77. [Google Scholar] [CrossRef]
- DeAngelis, D.L.; Mooij, W.M. Individual-based modeling of ecological and evolutionary processes. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 147–168. [Google Scholar] [CrossRef]
- Kareiva, P.; Shigesada, N. Analyzing insect movement as a correlated random walk. Oecologia 1983, 56, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Schumaker, N.H. Using landscape indices to predict habitat connectivity. Ecology 1996, 77, 1210–1225. [Google Scholar] [CrossRef]
- Hargrove, W.W.; Hoffman, F.M.; Efroymson, R.A. A practical map-analysis tool for detecting potential dispersal corridors. Landsc. Ecol. 2005, 20, 361–373. [Google Scholar] [CrossRef]
- Schumaker, N.H.; Brookes, A. HexSim: A modeling environment for ecology and conservation. Landsc. Ecol. 2018, 33, 197–211. [Google Scholar] [CrossRef]
- Bocedi, G.; Palmer, S.C.; Malchow, A.K.; Zurell, D.; Watts, K.; Travis, J.M. RangeShifter 2.0: An extended and enhanced platform for modelling spatial eco-evolutionary dynamics and species’ responses to environmental changes. Ecography 2021, 44, 1453–1462. [Google Scholar] [CrossRef]
- Schumaker, N.H.; Brookes, A.; Dunk, J.R.; Woodbridge, B.; Heinrichs, J.A.; Lawler, J.J.; Carroll, C.; LaPlante, D. Mapping sources, sinks, and connectivity using a simulation model of northern spotted owls. Landsc. Ecol. 2014, 29, 579–592. [Google Scholar] [CrossRef]
- Henry, R.C.; Palmer, S.C.; Watts, K.; Mitchell, R.J.; Atkinson, N.; Travis, J.M. Tree loss impacts on ecological connectivity: Developing models for assessment. Ecol. Inform. 2017, 42, 90–99. [Google Scholar] [CrossRef]
- Jeltsch, F.; Bonte, D.; Pe’er, G.; Reineking, B.; Leimgruber, P.; Balkenhol, N.; Schröder, B.; Buchmann, C.M.; Mueller, T.; Blaum, N.; et al. Integrating movement ecology with biodiversity research-exploring new avenues to address spatiotemporal biodiversity dynamics. Mov. Ecol. 2013, 1, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Unnithan Kumar, S.; Cushman, S.A. Connectivity modelling in conservation science: A comparative evaluation. 2022; Under review. [Google Scholar]
- Landguth, E.L.; Cushman, S.A. CDPOP: A spatially explicit cost distance population genetics program. Mol. Ecol. Resour. 2010, 10, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Landguth, E.L.; Cushman, S.A.; Schwartz, M.K.; McKelvey, K.S.; Murphy, M.; Luikart, G. Quantifying the lag time to detect barriers in landscape genetics. Mol. Ecol. 2010, 19, 4179–4191. [Google Scholar] [CrossRef] [PubMed]
- Shirk, A.J.; Landguth, E.L.; Cushman, S.A. A comparison of regression methods for model selection in individual-based landscape genetic analysis. Mol. Ecol. Resour. 2018, 18, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberl, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Cushman, S.A.; Landguth, E.L. Spurious correlations and inference in landscape genetics. Mol. Ecol. 2010, 19, 3592–3602. [Google Scholar] [CrossRef]
- Cushman, S.A.; Shirk, A.J.; Landguth, E.L. Separating the effects of habitat area, fragmentation and matrix resistance on genetic differentiation in complex landscapes. Landsc. Ecol. 2012, 27, 369–380. [Google Scholar] [CrossRef]
- Stronen, A.V.; Schumaker, N.H.; Forbes, G.J.; Paquet, P.C.; Brook, R.K. Landscape resistance to dispersal: Simulating long-term effects of human disturbance on a small and isolated wolf population in southwestern Manitoba, Canada. Environ. Monit. Assess. 2012, 184, 6923–6934. [Google Scholar] [CrossRef][Green Version]
- Heinrichs, J.A.; Lawler, J.J.; Schumaker, N.H. Intrinsic and extrinsic drivers of source–sink dynamics. Ecol. Evol. 2016, 6, 892–904. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.S.; Marra, P.P.; Haig, S.M.; Bensch, S.; Holmes, R.T. The importance of understanding migratory connectivity and seasonal interactions. Trends Ecol. Evol. 2002, 17, 76–83. [Google Scholar] [CrossRef]
- Ingold, T. The temporality of the landscape. World Archaeol. 1993, 25, 152–174. [Google Scholar] [CrossRef]
- Cushman, S.; Raphael, M.; Ruggiero, L.; Shirk, A.; Wasserman, T.; O’Doherty, E. Limiting factors and landscape connectivity: The American marten in the Rocky Mountains. Landsc. Ecol. 2011, 26, 1137–1149. [Google Scholar] [CrossRef]
- Wan, H.Y.; McGarigal, K.; Ganey, J.L.; Lauret, V.; Timm, B.C.; Cushman, S.A. Meta-replication reveals nonstationarity in multi-scale habitat selection of Mexican Spotted Owl. Condor Ornithol. Appl. 2017, 119, 641–658. [Google Scholar] [CrossRef]
- Vergara, M.; Cushman, S.A.; Ruiz-González, A. Ecological differences and limiting factors in different regional contexts: Landscape genetics of the stone marten in the Iberian Peninsula. Landsc. Ecol. 2017, 32, 1269–1283. [Google Scholar] [CrossRef]
- Cushman, S.A. Space and time in ecology: Noise or fundamental driver? In Spatial Complexity, Informatics, and Wildlife Conservation; Springer: Berlin/Heidelberg, Germany, 2010; pp. 19–41. [Google Scholar]
- Zeller, K.A.; Lewsion, R.; Fletcher, R.J.; Tulbure, M.G.; Jennings, M.K. Understanding the importance of dynamic landscape connectivity. Land 2020, 9, 303. [Google Scholar] [CrossRef]
- Bennett, N.J.; Roth, R.; Klain, S.C.; Chan, K.; Christie, P.; Clark, D.A.; Cullman, G.; Curran, D.; Durbin, T.J.; Epstein, G.; et al. Conservation social science: Understanding and integrating human dimensions to improve conservation. Biol. Conserv. 2017, 205, 93–108. [Google Scholar] [CrossRef]
- Kaszta, Ż.; Cushman, S.A.; Hearn, A.J.; Burnham, D.; Macdonald, E.A.; Goossens, B.; Nathan, S.K.; Macdonald, D.W. Integrating Sunda clouded leopard (Neofelis diardi) conservation into development and restoration planning in Sabah (Borneo). Biol. Conserv. 2019, 235, 63–76. [Google Scholar] [CrossRef]
- Benson, E.S. Movement Ecology and the Minimal Animal. LA+ 2016, 30–33. [Google Scholar]
- Pooley, S.; Barua, M.; Beinart, W.; Dickman, A.; Holmes, G.; Lorimer, J.; Loveridge, A.J.; Macdonald, D.W.; Marvin, G.; Redpath, S.; et al. An interdisciplinary review of current and future approaches to improving human–predator relations. Conserv. Biol. 2017, 31, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Peterson, M.N.; Birckhead, J.L.; Leong, K.; Peterson, M.J.; Peterson, T.R. Rearticulating the myth of human–wildlife conflict. Conserv. Lett. 2010, 3, 74–82. [Google Scholar] [CrossRef]
- Benson, E.S. Minimal animal: Surveillance, simulation, and stochasticity in wildlife biology. Antennae 2014, 30, 39–53. [Google Scholar]
- Barua, M. Bio-geo-graphy: Landscape, dwelling, and the political ecology of human-elephant relations. Environ. Plan. D Soc. Space 2014, 32, 915–934. [Google Scholar] [CrossRef]
- Salmón, E. Kincentric ecology: Indigenous perceptions of the human–nature relationship. Ecol. Appl. 2000, 10, 1327–1332. [Google Scholar]
- Nathan, R.; Getz, W.M.; Revilla, E.; Holyoak, M.; Kadmon, R.; Saltz, D.; Smouse, P.E. A movement ecology paradigm for unifying organismal movement research. Proc. Natl. Acad. Sci. USA 2008, 105, 19052–19059. [Google Scholar] [CrossRef]
- Cooke, S.J.; Blumstein, D.T.; Buchholz, R.; Caro, T.; Fernandez-Juricic, E.; Franklin, C.E.; Metcalfe, J.; O’Connor, C.M.; St. Clair, C.C.; Sutherland, W.J.; et al. Physiology, behavior, and conservation. Physiol. Biochem. Zool. 2014, 87, 1–14. [Google Scholar] [CrossRef]
- Lorimer, J. Nonhuman charisma. Environ. Plan. D Soc. Space 2007, 25, 911–932. [Google Scholar] [CrossRef]
- Ingold, T. The optimal forager and economic man. In Nature and Society; Routledge: Abingdon-on-Thames, UK, 2003; pp. 35–54. [Google Scholar]
- Sekar, N.; Shiller, D. Engage with animal welfare in conservation. Science 2020, 369, 629–630. [Google Scholar] [CrossRef]
- Paquet, P.C.; Darimont, C.T. Wildlife conservation and animal welfare: Two sides of the same coin. Anim. Welf. 2010, 19, 177–190. [Google Scholar]
- Wallach, A.D.; Bekoff, M.; Batavia, C.; Nelson, M.P.; Ramp, D. Summoning compassion to address the challenges of conservation. Conserv. Biol. 2018, 32, 1255–1265. [Google Scholar] [CrossRef]
- Parreñas, J.S. Decolonizing Extinction; Duke University Press: Durham, NC, USA, 2018. [Google Scholar]
- Cooke, S.J.; O’Connor, C.M. Making conservation physiology relevant to policy makers and conservation practitioners. Conserv. Lett. 2010, 3, 159–166. [Google Scholar] [CrossRef]
- Hodgetts, T.; Lorimer, J. Methodologies for animals’ geographies: Cultures, communication and genomics. Cult. Geogr. 2015, 22, 285–295. [Google Scholar] [CrossRef]
- Hodgetts, T. Connectivity as a multiple: In, with and as “nature”. Area 2018, 50, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Bunnefeld, N.; Nicholson, E.; Milner-Gulland, E.J. Decision-Making in Conservation and Natural Resource Management: Models for Interdisciplinary Approaches; Cambridge University Press: Cambridge, UK, 2017; Volume 22. [Google Scholar]
- Unnithan Kumar, S.; Maini, P.K.; Chiaverini, L.; Hearn, A.J.; Macdonald, D.W.; Kaszta, Ż.; Cushman, S.A. Smoothing and the environmental manifold. Ecol. Inform. 2021, 66, 101472. [Google Scholar] [CrossRef]
- Kimmerer, R. Braiding Sweetgrass: Indigenous Wisdom, Scientific Knowledge and the Teachings of Plants; Milkweed Editions: Minneapolis, MN, USA, 2013. [Google Scholar]
- Berkes, F. Sacred Ecology; Routledge: Abingdon-on-Thames, UK, 2017. [Google Scholar] [CrossRef]
- Ingold, T. Rethinking the animate, re-animating thought. Ethnos 2006, 71, 9–20. [Google Scholar] [CrossRef]



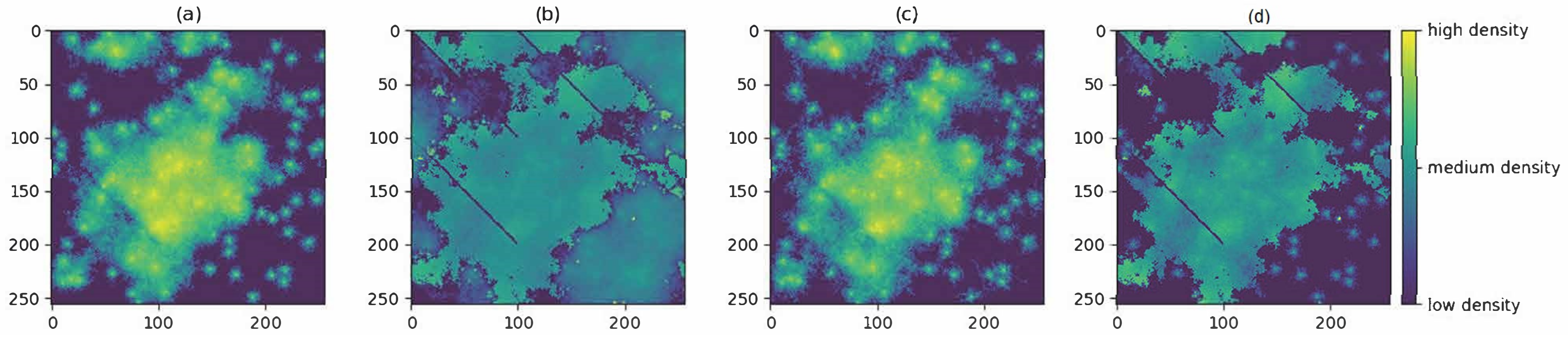
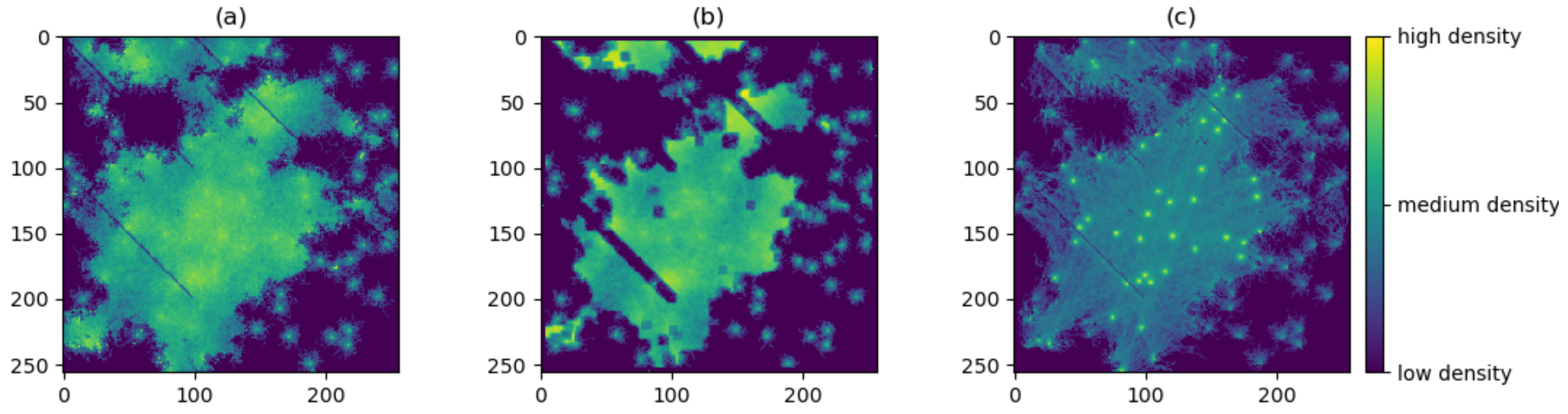
| Simulated surface | ||||
| Empirical surface | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Unnithan Kumar, S.; Kaszta, Ż.; Cushman, S.A. Pathwalker: A New Individual-Based Movement Model for Conservation Science and Connectivity Modelling. ISPRS Int. J. Geo-Inf. 2022, 11, 329. https://doi.org/10.3390/ijgi11060329
Unnithan Kumar S, Kaszta Ż, Cushman SA. Pathwalker: A New Individual-Based Movement Model for Conservation Science and Connectivity Modelling. ISPRS International Journal of Geo-Information. 2022; 11(6):329. https://doi.org/10.3390/ijgi11060329
Chicago/Turabian StyleUnnithan Kumar, Siddharth, Żaneta Kaszta, and Samuel A. Cushman. 2022. "Pathwalker: A New Individual-Based Movement Model for Conservation Science and Connectivity Modelling" ISPRS International Journal of Geo-Information 11, no. 6: 329. https://doi.org/10.3390/ijgi11060329
APA StyleUnnithan Kumar, S., Kaszta, Ż., & Cushman, S. A. (2022). Pathwalker: A New Individual-Based Movement Model for Conservation Science and Connectivity Modelling. ISPRS International Journal of Geo-Information, 11(6), 329. https://doi.org/10.3390/ijgi11060329







