An Optimized Protein Extraction Method for Gel-Free Proteomic Analysis of Opuntia Ficus-Indica
Abstract
:1. Introduction
2. Results and Discussion
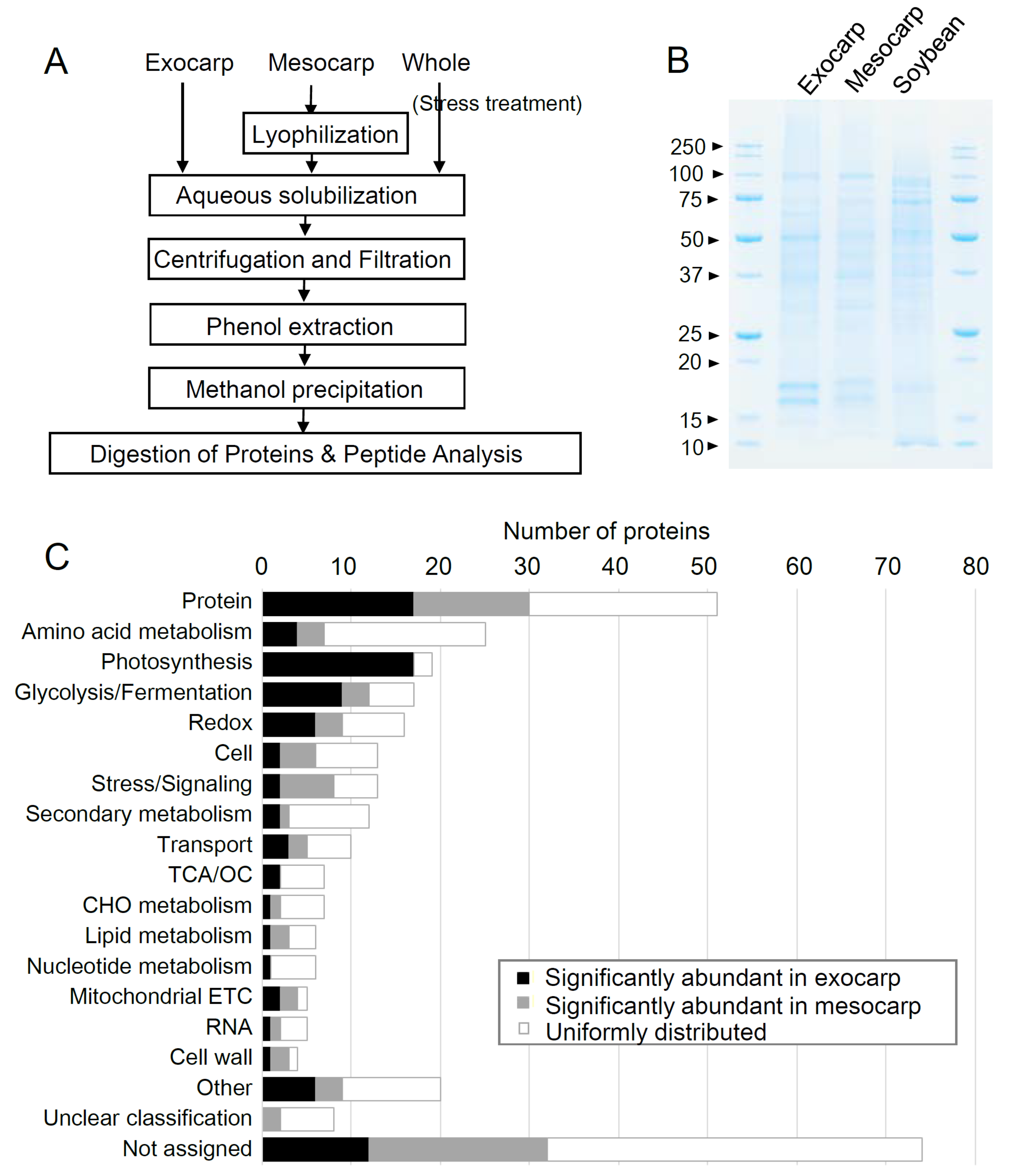
2.1. The Optimization of the Protein Extraction Method for Gel-Free Proteomic Analysis
2.2. Opuntia Ficus-Indica Salt Stress Response
3. Materials and Methods
3.1. Plant Materials
3.2. Protein Extraction, Enrichment, and Digestion for Mass Spectrometry Analysis
3.3. Nanoliquid Chromatography–Tandem Mass Spectrometry Analysis
3.4. Analysis of the Differential Abundance of Proteins Acquired Using Mass Spectrometry
3.5. Bioinformatic and Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Griffith, M.P. The origins of an important cactus crop, Opuntia ficus-indica (Cactaceae): New molecular evidence. Am. J. Bot. 2004, 91, 1915–1921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scalisi, A.; Morandi, B.; Inglese, P.; Bianco, R.L. Cladode growth dynamics in Opuntia ficus-indica under drought. Environ. Exp. Bot. 2016, 122, 158–167. [Google Scholar] [CrossRef]
- Falcão, H.M.; Oliveira, M.T.; Mergulhão, A.C.; Silva, M.V.; Santos, M.G. Ecophysiological performance of three Opuntia ficus-indica cultivars exposed to carmine cochineal under field conditions. Sci. Hort. 2013, 150, 419–424. [Google Scholar] [CrossRef]
- Gouws, C.A.; Georgousopoulou, E.N.; Mellor, D.D.; McKune, A.; Naumovski, N. Effects of the consumption of prickly pear cacti (Opuntia spp.) and its products on blood glucose levels and insulin: A systematic review. Medicina 2019, 55, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Héliès-Toussaint, C.; Fouché, E.; Naud, N.; Estrada, F.B.Y.; Díaz, M.D.S.S.; Salvayre, A.N.; De La Rosa, A.P.B.; Guéraud, F. Opuntia cladode powders inhibit adipogenesis in 3 T3-F442A adipocytes and a high-fat-diet rat model by modifying metabolic parameters and favouring faecal fat excretion. BMC Complement. Med. Ther. 2020, 20, 33. [Google Scholar] [CrossRef]
- Guevara-Figueroa, T.; Jiménez-Islas, H.; Reyes-Escogido, M.L.; Mortensen, A.G.; Laursen, B.B.; Lin, L.-W.; De León-Rodríguez, A.; Fomsgaard, I.S.; Barba de la Rosa, A.P. Proximate composition, phenolic acids, and flavonoids characterization of commercial and wild nopal (Opuntia spp.). J. Food Compos. Anal. 2010, 23, 525–532. [Google Scholar] [CrossRef]
- Al Juhaimi, F.; Ghafoor, K.; Uslu, N.; Mohamed Ahmed, I.A.; Babiker, E.E.; Özcan, M.M.; Fadimu, G.J. The effect of harvest times on bioactive properties and fatty acid compositions of prickly pear (Opuntia ficus-barbarica A. Berger) fruits. Food Chem. 2020, 303, 125387. [Google Scholar] [CrossRef]
- Silva-Ortega, C.O.; Ochoa-Alfaro, A.E.; Reyes-Agüero, J.A.; Aguado-Santacruz, G.A.; Jiménez-Bremont, J.F. Salt stress increases the expression of P5CS gene and induces proline accumulation in cactus pear. Plant Physiol. Biochem. 2008, 46, 82–92. [Google Scholar] [CrossRef]
- Hashiguchi, A.; Sakata, K.; Komatsu, S. Proteome Analysis of Early-Stage Soybean Seedlings under Flooding Stress. J. Proteome Res. 2009, 8, 2058–2069. [Google Scholar] [CrossRef]
- Barkla, B.J.; Vera-Estrella, R.; Raymond, C. Single-cell-type quantitative proteomic and ionomic analysis of epidermal bladder cells from the halophyte model plant Mesembryanthemum crystallinum to identify salt-responsive proteins. BMC Plant Biol. 2016, 16, 110. [Google Scholar] [CrossRef] [Green Version]
- Vadivel, A.-K.-A. Gel-based proteomics in plants: Time to move on from the tradition. Front. Plant Sci. 2015, 6, 369. [Google Scholar]
- Shakeel, S.N.; Aman, S.; Haq, N.U.; Heckathorn, S.A.; Luthe, D. Proteomic and transcriptomic analyses of Agave americana in response to heat stress. Plant Mol. Biol. Rep. 2013, 31, 840–851. [Google Scholar] [CrossRef]
- Cabello-Ruiz, E.D.; Torres-de la Cruz, V.M.; Rivas-Morales, C.; Molina-Salinas, G.M.; Núñez-González, M.A.; Verde-Star, M.J.; Leos-Rivas, C. Proteomic analysis of a bioactive Aloe vera extract. Curr. Proteome 2019, 16, 181–187. [Google Scholar] [CrossRef]
- Lledías, F.; Hernández, F.; Rivas, V.; García-Mendoza, A.; Cassab, G.I.; Nieto-Sotelo, J. A Rapid and Reliable Method for Total Protein Extraction from Succulent Plants for Proteomic Analysis. Protein J. 2017, 36, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Pichereaux, C.; Hernández-Domínguez, E.-E.; Santos-Diaz, M.D.S.; Reyes-Agüero, A.; Astello-García, M.; Guéraud, F.; Salvayre, A.N.; Schiltz, O.; Rossignol, M.; De La Rosa, A.P.B. Comparative shotgun proteomic analysis of wild and domesticated Opuntia spp. species shows a metabolic adaptation through domestication. J. Proteom. 2016, 143, 353–364. [Google Scholar] [CrossRef]
- Tran, N.T.; Oguchi, T.; Akatsuka, N.; Matsunaga, E.; Kawaoka, A.; Yamada, A.; Ozeki, Y.; Watanabe, K.N.; Kikuchi, A. Development and evaluation of novel salt-tolerant Eucalyptus trees by molecular breeding using an RNA-Binding-Protein gene derived from common ice plant (Mesembryanthemum crystallinum L.). Plant Biotechnol. J. 2019, 17, 801–811. [Google Scholar] [CrossRef]
- Kubota, S.; Hisamatsu, T.; Koshioka, M. Estimation of malic acid metabolism by measuring pH of hot water extracts of Phalaenopsis leaves. Sci. Hortic. 1997, 71, 251–255. [Google Scholar] [CrossRef]
- Nanjo, Y.; Škultéty, L.; Ashraf, Y.; Komatsu, S. Comparative Proteomic Analysis of Early-Stage Soybean Seedlings Responses to Flooding by Using Gel and Gel-Free Techniques. J. Proteome Res. 2010, 9, 3989–4002. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for compre-hensive analysis of (prote) omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Xu, X.; Tian, J.; Zhang, L.; Komatsu, S. Proteomic analysis of Lonicera japonica Thunb. immature flower buds using combinatorial peptide ligand libraries and polyethylene glycol fractionation. J. Proteome Res. 2016, 15, 166–181. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Hong, Z.; Su, W.; Li, J. A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2009, 106, 13612–13617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginestra, G.; Parker, M.L.; Bennett, R.N.; Robertson, J.; Mandalari, G.; Narbad, A.; Lo Curto, R.B.; Bisignano, G.; Faulds, C.B.; Waldron, K.W. Anatomical, chemical, and biochemical characterization of cladodes from prickly pear (Opuntia ficus-indica (L.) Mill.). J. Agric. Food Chem. 2009, 57, 10323–10330. [Google Scholar] [CrossRef]
- DePaoli, H.C.; Borland, A.M.; Tuskan, G.A.; Cushman, J.C.; Yang, X. Synthetic biology as it relates to CAM photosynthesis: Challenges and opportunities. J. Exp. Bot. 2014, 65, 3381–3393. [Google Scholar] [CrossRef] [Green Version]
- Ceusters, N.; Luca, S.; Feil, R.; Claes, J.E.; Lunn, J.E.; Ende, W.V.D.; Ceusters, J. Hierarchical clustering reveals unique features in the diel dynamics of metabolites in the CAM orchid Phalaenopsis. J. Exp. Bot. 2019, 70, 3269–3281. [Google Scholar] [CrossRef]
- Mallona, I.; Egea-Cortines, M.; Weiss, J. Conserved and Divergent Rhythms of Crassulacean Acid Metabolism-Related and Core Clock Gene Expression in the Cactus Opuntia ficus-indica. Plant Physiol. 2011, 156, 1978–1989. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashiguchi, A.; Yamaguchi, H.; Hitachi, K.; Watanabe, K. An Optimized Protein Extraction Method for Gel-Free Proteomic Analysis of Opuntia Ficus-Indica. Plants 2021, 10, 115. https://doi.org/10.3390/plants10010115
Hashiguchi A, Yamaguchi H, Hitachi K, Watanabe K. An Optimized Protein Extraction Method for Gel-Free Proteomic Analysis of Opuntia Ficus-Indica. Plants. 2021; 10(1):115. https://doi.org/10.3390/plants10010115
Chicago/Turabian StyleHashiguchi, Akiko, Hisateru Yamaguchi, Keisuke Hitachi, and Kazuo Watanabe. 2021. "An Optimized Protein Extraction Method for Gel-Free Proteomic Analysis of Opuntia Ficus-Indica" Plants 10, no. 1: 115. https://doi.org/10.3390/plants10010115
APA StyleHashiguchi, A., Yamaguchi, H., Hitachi, K., & Watanabe, K. (2021). An Optimized Protein Extraction Method for Gel-Free Proteomic Analysis of Opuntia Ficus-Indica. Plants, 10(1), 115. https://doi.org/10.3390/plants10010115





