Development and Application of Gene-Specific Markers for Tomato Yellow Leaf Curl Virus Resistance in Both Field and Artificial Infections
Abstract
1. Introduction
2. Results
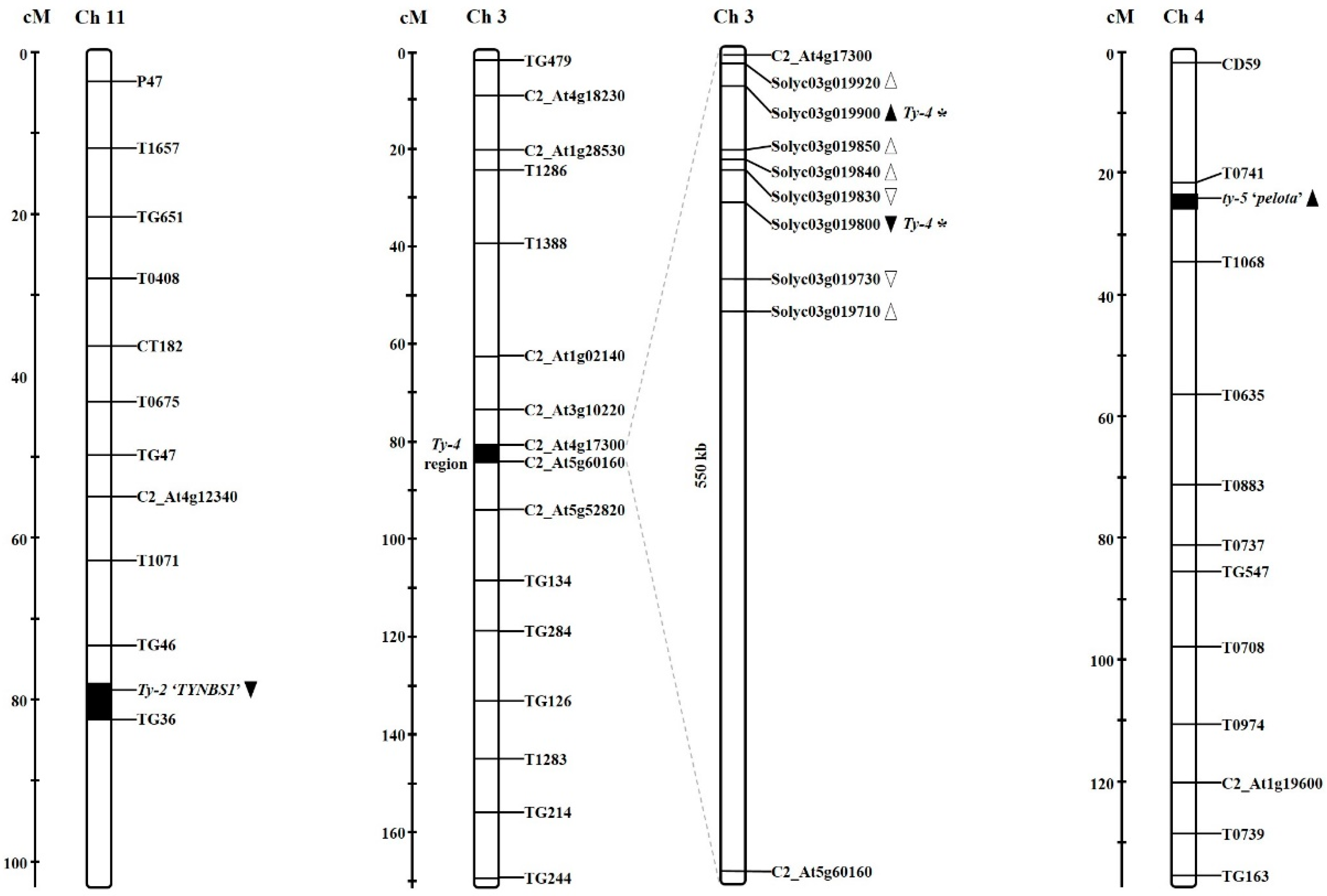
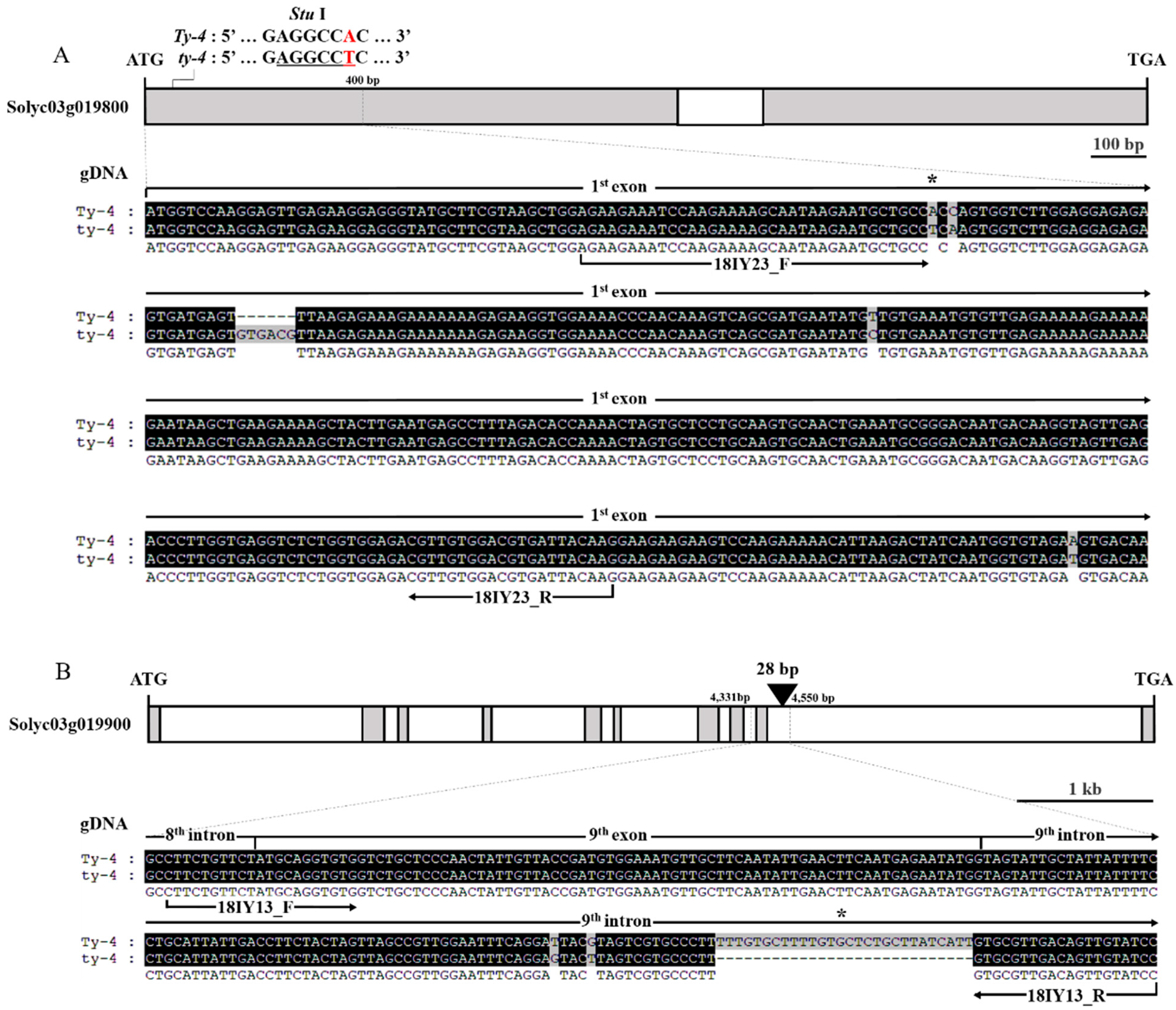
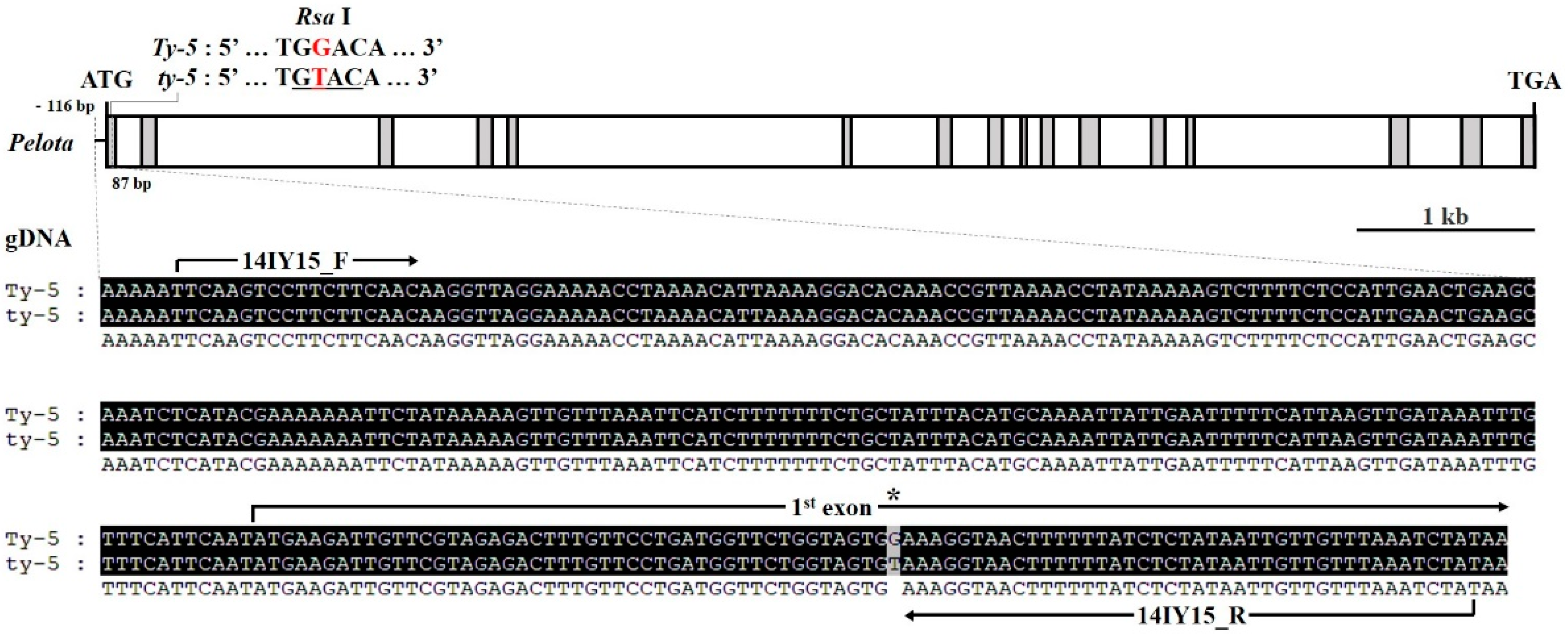
2.1. Development of Gene-Specific Markers for Ty-2, Ty-4, and ty-5 Resistances
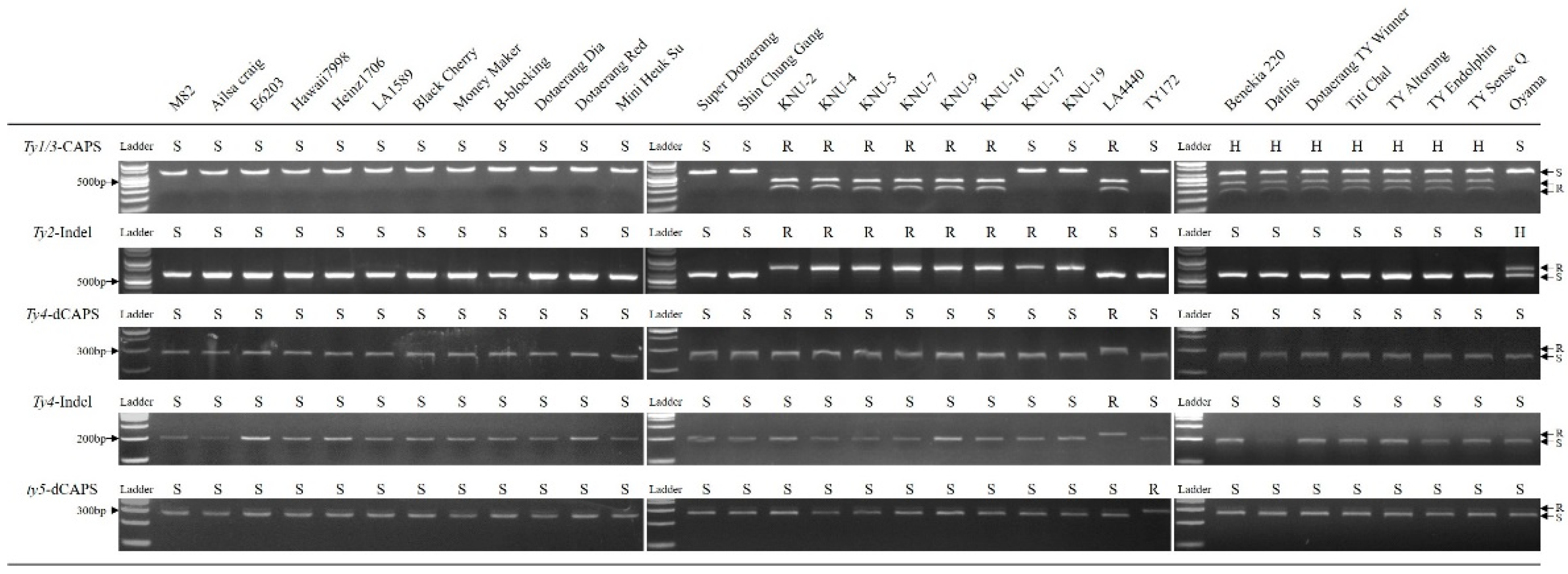
2.2. Application of Molecular Markers
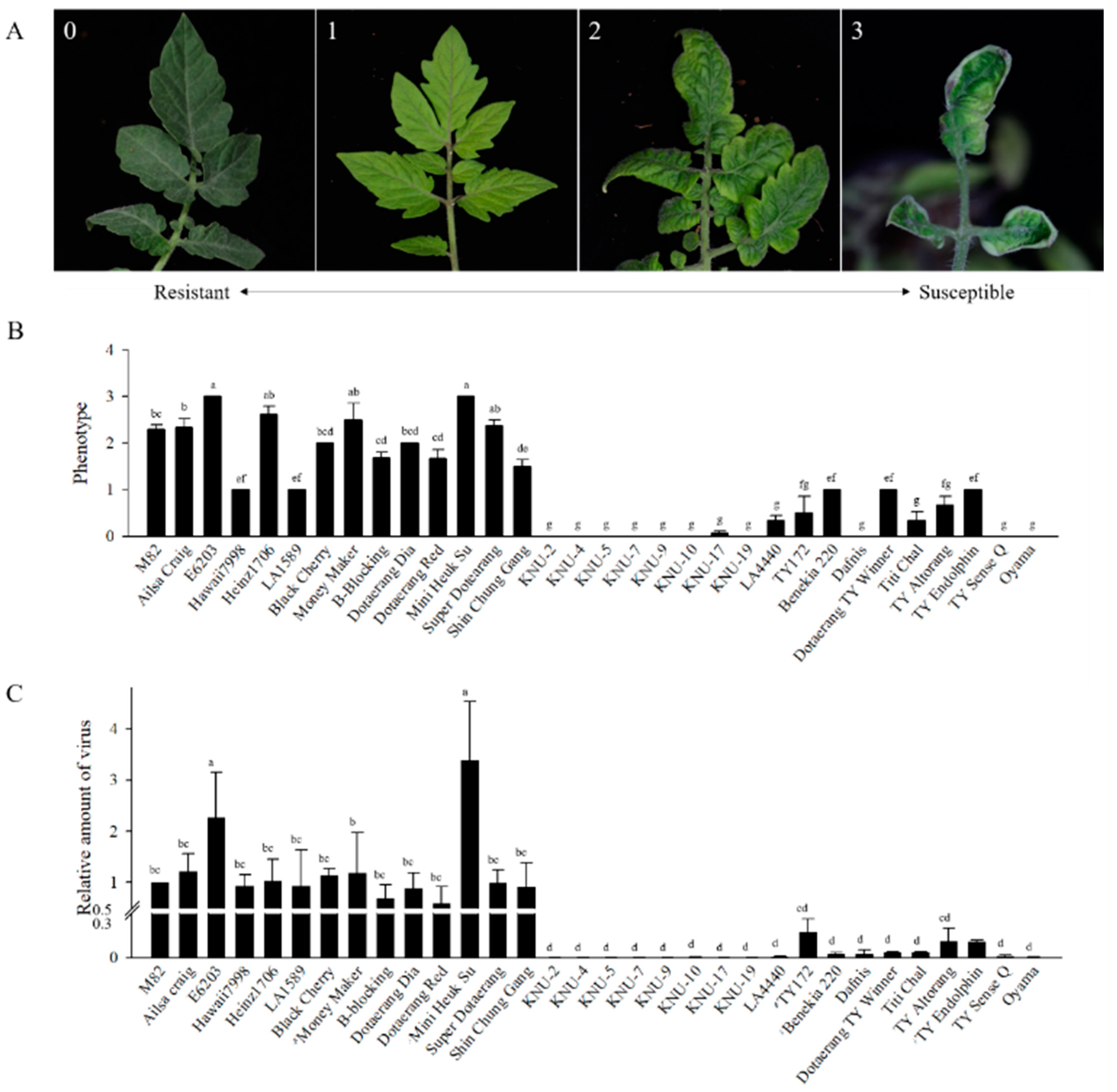
2.3. Disease Evaluation Using Natural Infection
2.4. Disease Evaluation Using Infectious Clones
3. Discussion
3.1. Genotyping of TYLCV Resistances with Molecular Markers
3.2. Comparison of Outcomes of Natural and Artificial Infection
3.3. Investigation of the Phenotypes of TYLCV Resistance and Their Combinations
4. Materials and Methods
4.1. Plant Materials and Marker Analysis
4.2. Virus Inoculation
4.3. Analysis of Disease Severity
4.4. Virus Accumulation Test
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Foolad, M.R. Genome mapping and molecular breeding of tomato. Int. J. Plant. Genom. 2007, 2007, 64358. [Google Scholar] [CrossRef]
- Rybicki, E.P. A Top Ten list for economically important plant viruses. Arch. Virol. 2015, 160, 17–20. [Google Scholar] [CrossRef]
- Moriones, E.; Navas-Castillo, J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 2000, 71, 123–134. [Google Scholar] [CrossRef]
- Abhary, M.; Patil, B.L.; Fauquet, C.M. Molecular Biodiversity, Taxonomy, and Nomenclature of Tomato Yellow Leaf Curl-like Viruses. In Tomato Yellow Leaf Curl Virus Disease; Czosnek, H., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 85–118. [Google Scholar] [CrossRef]
- Ning, W.; Shi, X.; Liu, B.; Pan, H.; Wei, W.; Zeng, Y.; Sun, X.; Xie, W.; Wang, S.; Wu, Q. Transmission of Tomato Yellow Leaf Curl Virus by Bemisia tabaci as Affected by Whitefly Sex and Biotype. Sci. Rep. 2015, 5, 10744. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Antignus, Y. Tomato Yellow Leaf Curl Virus, a Whitefly-Borne Geminivirus of Tomatoes. In Advances in Disease Vector Research; Harris, K.F., Ed.; Springer: New York, NY, USA, 1994; pp. 259–288. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef] [PubMed]
- Polston, J.E.; De Barro, P.; Boykin, L.M. Transmission specificities of plant viruses with the newly identified species of the Bemisia tabaci species complex. Pest. Manag. Sci. 2014, 70, 1547–1552. [Google Scholar] [CrossRef]
- Schuster, D.J.; Mann, R.S.; Toapanta, M.; Cordero, R.; Thompson, S.; Cyman, S.; Shurtleff, A.; Morris, R.F., II. Monitoring neonicotinoid resistance in biotype B of Bemisia tabaci in Florida. Pest. Manag. Sci. 2010, 66, 186–195. [Google Scholar] [CrossRef]
- Palumbo, J.C.; Horowitz, A.; Prabhaker, N. Insecticidal control and resistance management for Bemisia tabaci. Crop. Prot. 2001, 20, 739–765. [Google Scholar] [CrossRef]
- Berlinger, M.; Mordechi, S.; Leeper, A. Application of screens to prevent whitefly penetration into greenhouses in the Mediterranean basin. Bull. OILB SROP 1991, 5, 105–110. [Google Scholar]
- Antignus, Y.; Lapidot, M.; Hadar, D.; Messika, Y.; Cohen, S. Ultraviolet-Absorbing Screens Serve as Optical Barriers to Protect Crops from Virus and Insect Pests. J. Econ. Entomol. 1998, 91, 1401–1405. [Google Scholar] [CrossRef]
- Díaz-Pendón, J.; Cañizares, M.; Moriones, E.; Bejarano, E.; Czosnek, H.; Navas-Castillo, J. Tomato yellow leaf curl viruses: Menage a trois between the virus complex, the plant and the whitefly vector. Mol. Plant. Pathol. 2010, 11, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Dhaliwal, M.; Jindal, S.; Sharma, A.; Prasanna, H. Tomato yellow leaf curl virus disease of tomato and its management through resistance breeding: A review. J. Hortic. Sci. Biotechnol. 2020, 95, 425–444. [Google Scholar] [CrossRef]
- Zamir, D.; Ekstein-Michelson, I.; Zakay, Y.; Navot, N.; Zeidan, M.; Sarfatti, M.; Eshed, Y.; Harel, E.; Pleban, T.; Van-Oss, H. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, TY-1. Theor. Appl. Genet. 1994, 88, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Schuster, D.J.; Scott, J.W. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol. Breed. 2007, 20, 271–284. [Google Scholar] [CrossRef]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.; Scott, J.W.; Edwards, J.D.; Bai, Y. The Tomato Yellow Leaf Curl Virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA-dependent RNA polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef]
- Butterbach, P.; Verlaan, M.G.; Dullemans, A.; Lohuis, D.; Visser, R.G.; Bai, Y.; Kormelink, R. Tomato yellow leaf curl virus resistance by Ty-1 involves increased cytosine methylation of viral genomes and is compromised by cucumber mosaic virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 12942–12947. [Google Scholar] [CrossRef]
- Hanson, P.M.; Bernacchi, D.; Green, S.; Tanksley, S.D.; Muniyappa, V.; Padmaja, A.S.; Chen, H.-M.; Kuo, G.; Fang, D.; Chen, J.-T. Mapping a wild tomato introgression associated with tomato yellow leaf curl virus resistance in a cultivated tomato line. J. Am. Soc. Hortic. Sci. 2000, 125, 15–20. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Ohnishi, J.; Saito, A.; Ohyama, A.; Nunome, T.; Miyatake, K.; Fukuoka, H. An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 Begomovirus resistance locus of tomato. Theor. Appl. Genet. 2018, 131, 1345–1362. [Google Scholar] [CrossRef]
- Ji, Y.; Scott, J.W.; Schuster, D.J.; Maxwell, D.P. Molecular Mapping of Ty-4, a New Tomato Yellow Leaf Curl Virus Resistance Locus on Chromosome 3 of Tomato. J. Am. Soc. Hortic. Sci. 2009, 134, 281–288. [Google Scholar] [CrossRef]
- Kadirvel, P.; De la Peña, R.; Schafleitner, R.; Huang, S.; Geethanjali, S.; Kenyon, L.; Tsai, W.; Hanson, P. Mapping of QTLs in tomato line FLA456 associated with resistance to a virus causing tomato yellow leaf curl disease. Euphytica 2012, 190, 297–308. [Google Scholar] [CrossRef]
- Lapidot, M.; Karniel, U.; Gelbart, D.; Fogel, D.; Evenor, D.; Kutsher, Y.; Makhbash, Z.; Nahon, S.; Shlomo, H.; Chen, L. A novel route controlling begomovirus resistance by the messenger RNA surveillance factor pelota. PLoS Genet. 2015, 11, e1005538. [Google Scholar] [CrossRef] [PubMed]
- Gill, U.; Scott, J.W.; Shekasteband, R.; Ogundiwin, E.; Schuit, C.; Francis, D.M.; Sim, S.-C.; Smith, H.; Hutton, S.F. Ty-6, a major begomovirus resistance gene on chromosome 10, is effective against Tomato yellow leaf curl virus and Tomato mottle virus. Theor. Appl. Genet. 2019, 132, 1543–1554. [Google Scholar] [CrossRef] [PubMed]
- Vidavski, F.; Czosnek, H.; Gazit, S.; Levy, D.; Lapidot, M. Pyramiding of genes conferring resistance to Tomato yellow leaf curl virus from different wild tomato species. Plant Breed. 2008, 127, 625–631. [Google Scholar] [CrossRef]
- García-Cano, E.; Resende, R.; Boiteux, L.; Giordano, L.; Fernández-Muñoz, R.; Moriones, E. Phenotypic expression, stability, and inheritance of a recessive resistance to monopartite begomoviruses associated with tomato yellow leaf curl disease in tomato. Phytopathology 2008, 98, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, J.; Yamaguchi, H.; Saito, A. Analysis of the Mild strain of tomato yellow leaf curl virus, which overcomes Ty-2 gene-mediated resistance in tomato line H24. Arch. Virol. 2016, 161, 2207–2217. [Google Scholar] [CrossRef]
- Jung, J.; Kim, H.J.; Lee, J.M.; Oh, C.S.; Lee, H.-J.; Yeam, I. Gene-based molecular marker system for multiple disease resistances in tomato against Tomato yellow leaf curl virus, late blight, and verticillium wilt. Euphytica 2015, 205, 599–613. [Google Scholar] [CrossRef]
- Kim, M. Development and application of Tomato leaf curl virus (TYLCV)-Resistance Molecular Markers. Master’s Thesis, Andong National University, Andong, Korea, 2015. [Google Scholar]
- Friedmann, M.; Lapidot, M.; Cohen, S.; Pilowsky, M. A Novel Source of Resistance to Tomato Yellow Leaf Curl Virus Exhibiting a Symptomless Reaction to Viral Infection. J. Am. Soc. Hortic. Sci. 1998, 123, 1004–1007. [Google Scholar] [CrossRef]
- Ji, Y.; Scott, J.W.; Schuster, D.J. Toward fine mapping of the Tomato yellow leaf curl virus resistance gene Ty-2 on chromosome 11 of tomato. HortScience 2009, 44, 614–618. [Google Scholar] [CrossRef]
- Mejía, L.; Teni, R.; Vidavski, F.; Czosnek, H.; Lapidot, M.; Nakhla, M.; Maxwell, D. Evaluation of tomato germplasm and selection of breeding lines for resistance to begomoviruses in Guatemala. In Proceedings of the International Symposium on Tomato Diseases, Orlando, FL, USA, 21–24 June 2004. [Google Scholar]
- Anbinder, I.; Reuveni, M.; Azari, R.; Paran, I.; Nahon, S.; Shlomo, H.; Chen, L.; Lapidot, M.; Levin, I. Molecular dissection of Tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor. Appl. Genet. 2009, 119, 519–530. [Google Scholar] [CrossRef]
- Costa, A. Whitefly-transmitted plant diseases. Annu. Rev. Phytopathol. 1976, 14, 429–449. [Google Scholar] [CrossRef]
- Ber, R.; Navot, N.; Zamir, D.; Antignus, Y.; Cohen, S.; Czosnek, H. Infection of tomato by the tomato yellow leaf curl virus: Susceptibility to infection, symptom development, and accumulation of viral DNA. Arch. Virol. 1990, 112, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Pérez-de-Castro, A.; Díez, M.J.; Hutton, S.F.; Visser, R.G.; Wolters, A.-M.A.; Bai, Y.; Li, J. Resistance to Tomato Yellow Leaf Curl Virus in Tomato Germplasm. Front. Plant. Sci. 2018, 9, 1198. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, M.; Legg, J.P.; Wintermantel, W.M.; Polston, J.E. Management of whitefly-transmitted viruses in open-field production systems. Adv. Virus Res. 2014, 90, 147–206. [Google Scholar] [CrossRef] [PubMed]
- Picó, B.; Díez, M.J.; Nuez, F. Evaluation of whitefly-mediated inoculation techniques to screen Lycopersicon esculentum and wild relatives for resistance to tomato yellow leaf curl virus. Euphytica 1998, 101, 259–271. [Google Scholar] [CrossRef]
- Rodríguez-López, M.; Garzo, E.; Bonani, J.; Fereres, A.; Fernández-Muñoz, R.; Moriones, E. Whitefly resistance traits derived from the wild tomato Solanum pimpinellifolium affect the preference and feeding behavior of Bemisia tabaci and reduce the spread of Tomato yellow leaf curl virus. Phytopathology 2011, 101, 1191–1201. [Google Scholar] [CrossRef]
- Wang, K.; Tsai, J.H. Temperature Effect on Development and Reproduction of Silverleaf Whitefly (Homoptera: Aleyrodidae). Ann. Entomol. Soc. Am. 1996, 89, 375–384. [Google Scholar] [CrossRef]
- Rochester, D.E.; Kositratana, W.; Beachy, R.N. Systemic movement and symptom production following agroinoculation with a single DNA of tomato yellow leaf curl geminivirus (Thailand). Virology 1990, 178, 520–526. [Google Scholar] [CrossRef]
- Vidavsky, F.; Leviatov, S.; Milo, J.; Rabinowitch, H.; Kedar, N.; Czosnek, H. Response of tolerant breeding lines of tomato, Lycopersicon esculentum, originating from three different sources (L. peruvianum, L. pimpinellifolium and L. chilense) to early controlled inoculation by tomato yellow leaf curl virus (TYLCV). Plant Breed. 1998, 117, 165–169. [Google Scholar] [CrossRef]
- Kil, E.-J.; Kim, S.; Lee, Y.-J.; Byun, H.-S.; Park, J.; Seo, H.; Kim, C.-S.; Shim, J.-K.; Lee, J.-H.; Kim, J.-K. Tomato yellow leaf curl virus (TYLCV-IL): A seed-transmissible geminivirus in tomatoes. Sci. Rep. 2016, 6, 19013. [Google Scholar] [CrossRef]
- Lee, J.M.; Jahn, M.M.; Yeam, I. Allelic relationships at the pvr1 locus in Capsicumannuum. Euphytica 2013, 194, 417–424. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, J.; Stall, R.; Vallejos, C. Genomic localization of tomato genes that control a hypersensitive reaction to Xanthomonas campestris pv. vesicatoria (Doidge) dye. Genetics 1995, 141, 675–682. [Google Scholar] [PubMed]
- Murray, M.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, M.; Jeong, E.S.; Lee, J.M.; Choi, D. Harnessing anthocyanin-rich fruit: A visible reporter for tracing virus-induced gene silencing in pepper fruit. Plant Methods 2017, 13, 3. [Google Scholar] [CrossRef]
- Kasrawi, M.A. Inheritance of resistance to tomato yellow leaf curl virus (TYLCV) in Lycopersicon pimpinellifolium. Plant Dis. 1989, 73, 435–437. [Google Scholar] [CrossRef]
- Powell, M.E.; Cuthbertson, A.G.; Bell, H.A.; Boonham, N.; Morris, J.; Northing, P. First record of the Q Biotype of the sweetpotato whitefly, Bemisia tabaci, intercepted in the UK. Eur. J. Plant Pathol. 2012, 133, 797–801. [Google Scholar] [CrossRef]


| No. | Plant Information | |||
|---|---|---|---|---|
| Name | Type | Species | Source b | |
| 1 | M82 a | Accession | S. lycopersicum | TGRC |
| 2 | Ailsa Craig | S. lycopersicum | ||
| 3 | E6203 a | S. lycopersicum | ||
| 4 | Hawaii7998 a | S. lycopersicum | ||
| 5 | Heinz 1706 | S. lycopersicum | ||
| 6 | LA1589 | S. pimpinellifolium | ||
| 7 | Black Cherry | S. lycopersicum | ||
| 8 | Money Maker | S. lycopersicum | ||
| 9 | LA4440 | S. chilense | ||
| 10 | TY172 a | Accession | S. peruvianum | Volcani center |
| 11 | KNU-2 a | S. lycopersicum | AVRDC | |
| 12 | KNU-4 a | S. lycopersicum | ||
| 13 | KNU-5 a | S. lycopersicum | ||
| 14 | KNU-7 a | S. lycopersicum | ||
| 15 | KNU-9 a | S. lycopersicum | ||
| 16 | KNU-10 a | S. lycopersicum | ||
| 17 | KNU-17 a | S. lycopersicum | ||
| 18 | KNU-19 a | S. lycopersicum | ||
| 19 | Miniheuksu | Commercial cultivar | S. lycopersicum | Asia seed |
| 20 | Benekia220 a | S. lycopersicum | Bunong seed | |
| 21 | TYEndolphin | S. lycopersicum | Bunong seed | |
| 22 | Shinchunggang | S. lycopersicum | Farm Hannong | |
| 23 | Super Dotaerang | S. lycopersicum | Koregon | |
| 24 | TitiChal a | S. lycopersicum | Nongwoo bio | |
| 25 | TYAltorang a | S. lycopersicum | Nongwoo bio | |
| 26 | TYSenseQ a | S. lycopersicum | Nongwoo bio | |
| 27 | Dotaerang Red | S. lycopersicum | Sakata seed | |
| 28 | Oyama a | S. lycopersicum | Sakata seed | |
| 29 | Dafnis | S. lycopersicum | Syngenta | |
| 30 | B Blocking | S. lycopersicum | Takii seed | |
| 31 | Dotaerang Dia | S. lycopersicum | Takii seed | |
| 32 | Dotaerang TY Winner a | S. lycopersicum | Takii seed | |
| Locus | Primer Name | Sequence (5′-3′) | Annealing Temp (°C) | Product Size (bp) | Type (Enzyme) | Reference | |
|---|---|---|---|---|---|---|---|
| Ty-1/3 | 14IY218 | F | ATG AAG ACA AAA ACT GCT TC | 55 | R : 383, 226 | CAPS (SspI) | Jung et al., 2015 |
| R | TCA GGG TTT CAC TTC TAT GAA T | S : 609 | |||||
| Ty-2 | 20IY10 | F | GTT CTA TCA CAA GAC TTG CCA | 55 | R : 738 | Indel | In this assay |
| R | TGC ATT CAC CAT TGA TGT ATA AGA | S : 600 | |||||
| Ty-4 | 18IY23 | F | AGA AGA AAT CCA AGA AAA GCA ATA AGA ATG AGG CC | 55 | R : 304 | dCAPS (StuI) | In this assay |
| R | CTT GTA ATC ACG TCC ACA ACG | S : 269, 35 | |||||
| 18IY13 | F | CTT CTG TTC TAT GCA GGT GTG | 55 | R : 228 | Indel | ||
| R | GGA TAC AAC TGT CAA CGC AC | S : 200 | |||||
| ty-5 | 14IY5 | F | TTC AAG TCC TTC TTC AAC | 55 | R : 300 | dCAPS (RsaI) | In this assay |
| R | ATA GAT TTA AAC AAC AAT TAT AGA GAT AAA AAA GTT ACC TGT | S : 260, 40 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.H.; Chung, D.J.; Lee, J.M.; Yeam, I. Development and Application of Gene-Specific Markers for Tomato Yellow Leaf Curl Virus Resistance in Both Field and Artificial Infections. Plants 2021, 10, 9. https://doi.org/10.3390/plants10010009
Lee JH, Chung DJ, Lee JM, Yeam I. Development and Application of Gene-Specific Markers for Tomato Yellow Leaf Curl Virus Resistance in Both Field and Artificial Infections. Plants. 2021; 10(1):9. https://doi.org/10.3390/plants10010009
Chicago/Turabian StyleLee, Jang Hee, Dae Jun Chung, Je Min Lee, and Inhwa Yeam. 2021. "Development and Application of Gene-Specific Markers for Tomato Yellow Leaf Curl Virus Resistance in Both Field and Artificial Infections" Plants 10, no. 1: 9. https://doi.org/10.3390/plants10010009
APA StyleLee, J. H., Chung, D. J., Lee, J. M., & Yeam, I. (2021). Development and Application of Gene-Specific Markers for Tomato Yellow Leaf Curl Virus Resistance in Both Field and Artificial Infections. Plants, 10(1), 9. https://doi.org/10.3390/plants10010009





