Genomic Dissection of Peduncle Morphology in Barley through Nested Association Mapping
Abstract
1. Introduction
2. Results and Discussion
2.1. Descriptive Statistics
2.2. Correlations
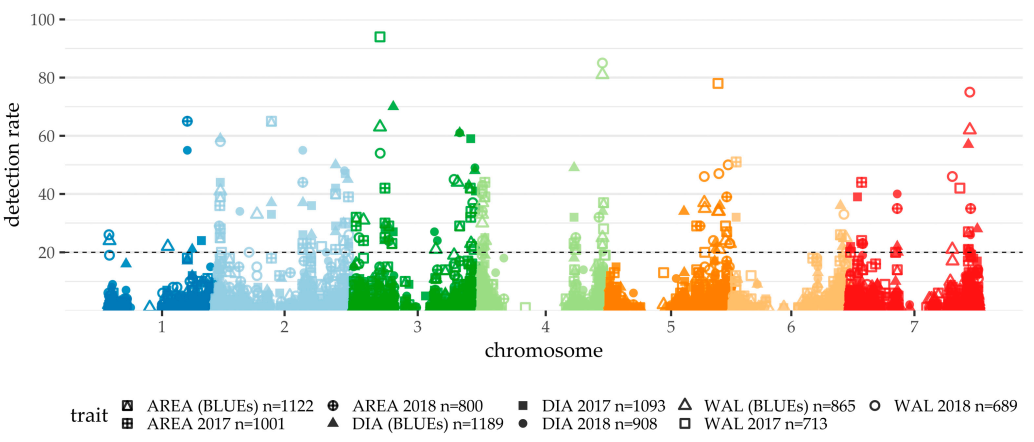
2.3. GWAS
2.3.1. QPed.shared.2H-1
2.3.2. QPed.shared.2H-2
2.3.3. QPed.shared.2H-3
2.3.4. QPed.shared.3H-1
2.3.5. QPed.shared.3H-2
2.3.6. QPed.shared.3H-3
2.3.7. QPed.shared.4H-1
2.4. Impact of Plant Development on Peduncle Morphology
3. Materials and Methods
3.1. Plant Material and Field Trials
3.2. Phenotyping
3.3. Statistics and Significance Tests
3.4. Genome-Wide Association Study (GWAS)
3.5. Exome Capture Sequencing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Statistisches Bundesamt—Destatis. Wachstum und ernte-feldfrüchte—2019. In Land-und Forstwirtschaft, Fischerei; Statistisches Bundesamt: Wiesbaden, Germany, 2020; Volume Fachserie 3 Reihe 3.2.1, pp. 12–13. [Google Scholar]
- Simon, S.M. Szenarien Nachhaltiger Bioenergiepotenziale Bis 2030—Modellierung für Deutschland, Polen, Tschechien und Ungarn; Technische Universität München: München, Germany, 2006. [Google Scholar]
- Schutyser, W.; Renders, T.; Van den Bosch, S.; Koelewijn, S.-F.; Beckham, G.; Sels, B. Chemicals from lignin: An interplay of lignocellulose fractionation, depolymerisation, and upgrading. Chem. Soc. Rev. 2018, 47, 852–908. [Google Scholar] [CrossRef] [PubMed]
- Huber, G.W.; Iborra, S.; Corma, A. Synthesis of transportation fuels from biomass: Chemistry, catalysts, and engineering. Chem. Rev. 2006, 106, 4044–4098. [Google Scholar] [CrossRef] [PubMed]
- Giurca, A.; Späth, P. A forest-based bioeconomy for germany? Strengths, weaknesses and policy options for lignocellulosic biorefineries. J. Clean. Prod. 2017, 153, 51–62. [Google Scholar] [CrossRef]
- Hedden, P. The genes of the green revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef]
- Serrano-Mislata, A.; Bencivenga, S.; Bush, M.; Schiessl, K.; Boden, S.; Sablowski, R. Della genes restrict inflorescence meristem function independently of plant height. Nat. Plants 2017, 3, 749–754. [Google Scholar] [CrossRef]
- Schnurbusch, T. Wheat and barley biology: Towards new frontiers. J. Integr. Plant Biol. 2019, 61, 198–203. [Google Scholar] [CrossRef]
- McKim, S.M. How plants grow up. J. Integr. Plant Biol. 2019, 61, 257–277. [Google Scholar] [CrossRef]
- Gol, L.; Tomé, F.; Von Korff, M. Floral transitions in wheat and barley: Interactions between photoperiod, abiotic stresses, and nutrient status. J. Exp. Bot. 2017, 68, 1399–1410. [Google Scholar] [CrossRef]
- Chen, W.Y.; Liu, Z.M.; Deng, G.B.; Pan, Z.F.; Liang, J.J.; Zeng, X.Q.; Tashi, N.M.; Long, H.; Yu, M.Q. Genetic relationship between lodging and lodging components in barley (hordeum vulgare) based on unconditional and conditional quantitative trait locus analyses. Genet. Mol. Res. 2014, 13, 1909–1925. [Google Scholar] [CrossRef]
- Bayer, M.M.; Rapazote-Flores, P.; Ganal, M.; Hedley, P.E.; Macaulay, M.; Plieske, J.; Ramsay, L.; Russell, J.; Shaw, P.D.; Thomas, W.; et al. Development and evaluation of a barley 50k iselect snp array. Front. Plant Sci. 2017, 8, 1792. [Google Scholar] [CrossRef]
- Maurer, A.; Draba, V.; Jiang, Y.; Schnaithmann, F.; Sharma, R.; Schumann, E.; Kilian, B.; Reif, J.C.; Pillen, K. Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genom. 2015, 16, 290. [Google Scholar] [CrossRef] [PubMed]
- Maurer, A.; Draba, V.; Pillen, K. Genomic dissection of plant development and its impact on thousand grain weight in barley through nested association mapping. J. Exp. Bot. 2016, 67, 2507–2518. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Sayre, K.; Kaul, J.N.; Narang, R.S. Growth and morphology of spring wheat (triticum aestivum. L.) culms and their association with lodging: Effects of genotypes, n levels and ethephon. Field Crop. Res. 2003, 84, 271–290. [Google Scholar] [CrossRef]
- Kashiwagi, T.; Togawa, E.; Hirotsu, N.; Ishimaru, K. Improvement of lodging resistance with qtls for stem diameter in rice (oryza sativa l.). Theor. Appl. Genet. 2008, 117, 749–757. [Google Scholar] [CrossRef]
- Gemmer, M.R.; Richter, C.; Jiang, Y.; Schmutzer, T.; Raorane, M.L.; Junker, B.; Pillen, K.; Maurer, A. Can metabolic prediction be an alternative to genomic prediction in barley? PLoS ONE 2020, 15, e0234052. [Google Scholar] [CrossRef]
- Monat, C.; Padmarasu, S.; Lux, T.; Wicker, T.; Gundlach, H.; Himmelbach, A.; Ens, J.; Li, C.; Muehlbauer, G.J.; Schulman, A.H.; et al. Tritex: Chromosome-scale sequence assembly of triticeae genomes with open-source tools. Genome Biol. 2019, 20, 284. [Google Scholar] [CrossRef]
- Wiegmann, M.; Maurer, A.; Pham, A.; March, T.J.; Al-Abdallat, A.; Thomas, W.T.B.; Bull, H.J.; Shahid, M.; Eglinton, J.; Baum, M.; et al. Barley yield formation under abiotic stress depends on the interplay between flowering time genes and environmental cues. Sci. Rep. 2019, 9, 6397. [Google Scholar] [CrossRef]
- Turner, A.; Beales, J.; Faure, S.; Dunford, R.P.; Laurie, D.A. The pseudo-response regulator ppd-h1 provides adaptation to photoperiod in barley. Science 2005, 310, 1031–1034. [Google Scholar] [CrossRef]
- Campoli, C.; Shtaya, M.; Davis, S.J.; Von Korff, M. Expression conservation within the circadian clock of a monocot: Natural variation at barley ppd-h1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 2012, 12, 97. [Google Scholar] [CrossRef]
- Comadran, J.; Kilian, B.; Russell, J.; Ramsay, L.; Stein, N.; Ganal, M.; Shaw, P.; Bayer, M.; Thomas, W.; Marshall, D.; et al. Natural variation in a homolog of antirrhinum centroradialis contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 2012, 44, 1388–1392. [Google Scholar] [CrossRef]
- Badr, A.; M, K.; Sch, R.; Rabey, H.E.; Effgen, S.; Ibrahim, H.H.; Pozzi, C.; Rohde, W.; Salamini, F. On the origin and domestication history of barley (hordeum vulgare). Mol. Biol. Evol. 2000, 17, 499–510. [Google Scholar] [CrossRef]
- Bi, X.; Van Esse, W.; Mulki, M.A.; Kirschner, G.; Zhong, J.; Simon, R.; Von Korff, M. Centroradialis interacts with flowering locus t -like genes to control floret development and grain number. Plant Physiol. 2019, 180, 1013–1030. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Wei, Y.; Xu, R.; Lin, S.; Luan, H.; Lv, C.; Zhang, X.; Song, X.; Xu, R. Genome-wide analysis of apetala2/ethylene-responsive factor (ap2/erf) gene family in barley (hordeum vulgare l.). PLoS ONE 2016, 11, e0161322. [Google Scholar] [CrossRef]
- Patil, V.; McDermott, H.I.; McAllister, T.; Cummins, M.; Silva, J.C.; Mollison, E.; Meikle, R.; Morris, J.; Hedley, P.E.; Waugh, R.; et al. Apetala2 control of barley internode elongation. Development 2019, 146, dev170373. [Google Scholar] [CrossRef]
- Wang, G.; Gao, Y.; Wang, J.; Yang, L.; Song, R.; Li, X.; Shi, J. Overexpression of two cambium-abundant chinese fir (cunninghamia lanceolata) α-expansin genes clexpa1 and clexpa2 affect growth and development in transgenic tobacco and increase the amount of cellulose in stem cell walls. Plant Biotechnol. J. 2011, 9, 486–502. [Google Scholar] [CrossRef]
- Jia, Q.; Li, C.; Shang, Y.; Zhu, J.; Hua, W.; Wang, J.; Yang, J.; Zhang, G. Molecular characterization and functional analysis of barley semi-dwarf mutant riso no. 9265. BMC Genom. 2015, 16, 927. [Google Scholar] [CrossRef]
- Kuczyńska, A.; Wyka, T. The effect of the denso dwarfing gene on morpho-anatomical characters in barley recombinant inbred lines. Breed. Sci. 2011, 61, 275–280. [Google Scholar] [CrossRef]
- Herzig, P.; Maurer, A.; Draba, V.; Sharma, R.; Draicchio, F.; Bull, H.; Milne, L.; Thomas, W.T.B.; Flavell, A.J.; Pillen, K. Contrasting genetic regulation of plant development in wild barley grown in two european environments revealed by nested association mapping. J. Exp. Bot. 2018, 69, 1517–1531. [Google Scholar] [CrossRef]
- Sharma, R.; Draicchio, F.; Bull, H.; Herzig, P.; Maurer, A.; Pillen, K.; Thomas, W.T.B.; Flavell, A.J. Genome-wide association of yield traits in a nested association mapping population of barley reveals new gene diversity for future breeding. J. Exp. Bot. 2018, 69, 3811–3822. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, X.; Ding, Y.; Fei, Z.; Jiao, C.; Fan, M.; Yao, B.; Xin, P.; Chu, J.; Wei, Q. Cellular and molecular characterization of a thick-walled variant reveal a pivotal role of shoot apical meristem in transverse development of bamboo culm. J Exp Bot 2019, 70, 3911–3926. [Google Scholar] [CrossRef]
- Yong, W.; Link, B.; O’Malley, R.; Tewari, J.; Hunter, C.T.; Lu, C.-A.; Li, X.; Bleecker, A.B.; Koch, K.E.; McCann, M.C.; et al. Genomics of plant cell wall biogenesis. Planta 2005, 221, 747–751. [Google Scholar] [CrossRef] [PubMed]
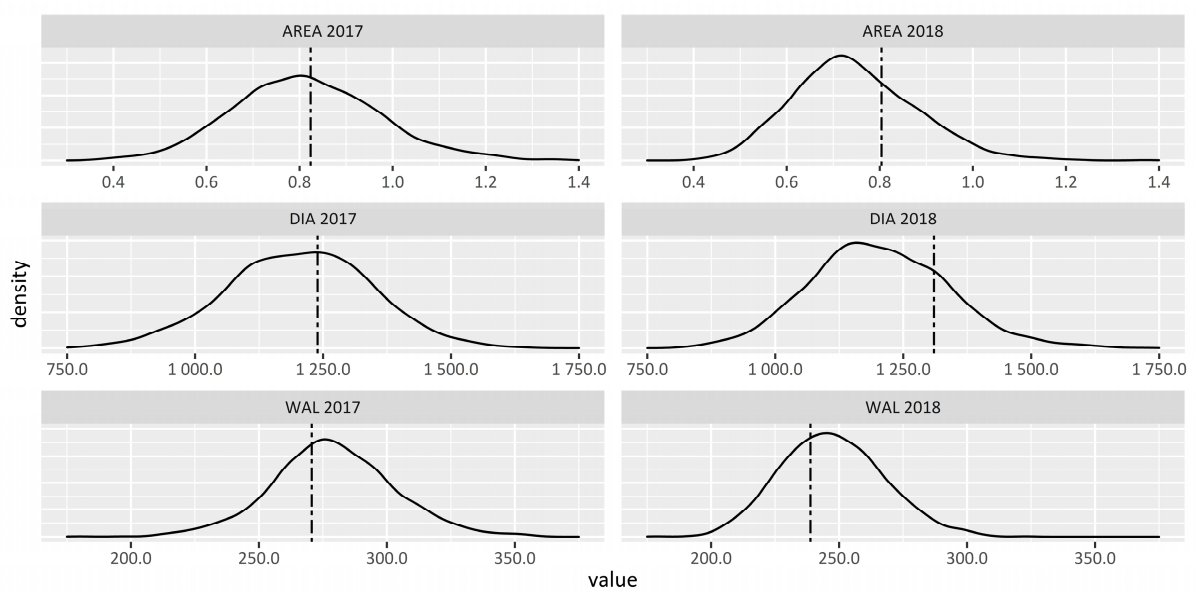



| Trait a | N b | Mean c | SD d | Min e | Max f | CV g | Barke h |
|---|---|---|---|---|---|---|---|
| DIA17 | 1232 | 1202.82 | 141.08 | 776.00 | 1640.91 | 0.12 | 1239.35 |
| DIA18 | 1343 | 1204.25 | 137.89 | 833.50 | 1931.10 | 0.11 | 1309.98 |
| DIA | 1411 | 1204.33 | 126.87 | 841.76 | 1876.26 | 0.11 | 1274.66 |
| WAL17 | 1232 | 279.01 | 24.01 | 179.96 | 385.95 | 0.09 | 270.66 |
| WAL18 | 1343 | 247.32 | 19.98 | 178.74 | 322.83 | 0.08 | 238.83 |
| WAL | 1411 | 263.16 | 18.84 | 179.35 | 338.69 | 0.07 | 254.74 |
| AREA17 | 1232 | 8.14 × 105 | 1.57 × 105 | 3.78 × 105 | 1.35 × 106 | 0.19 | 8.24 × 105 |
| AREA18 | 1343 | 7.46 × 105 | 1.32 × 105 | 4.12 × 105 | 1.37 × 106 | 0.18 | 8.04 × 105 |
| AREA | 1411 | 7.81 × 105 | 1.29 × 105 | 4.04 × 105 | 1.29 × 106 | 0.16 | 8.16 × 105 |
| Trait | Vg a | Ve b | Vr c | H2 d |
|---|---|---|---|---|
| DIA | 57.8 | 0.0 | 42.2 | 73.3 |
| WAL | 17.4 | 51.1 | 31.4 | 52.6 |
| AREA | 45.2 | 10.4 | 44.4 | 67.0 |
| Trait | Prediction Ability | ⌀Number of Sig. SNPs |
|---|---|---|
| DIA | 0.47 | 36.81 |
| DIA17 | 0.45 | 31.67 |
| DIA18 | 0.28 | 25.75 |
| WAL | 0.20 | 27.15 |
| WAL17 | 0.10 | 21.41 |
| WAL18 | 0.21 | 24.42 |
| AREA17 | 0.33 | 28.46 |
| AREA18 | 0.28 | 23.35 |
| AREA | 0.40 | 32.70 |
| QTL Region | DIA [µm] | WAL [µm] | AREA [µm2] | Candidate Gene | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| shared-QTL | chromosome | Position a | 2017 | 2018 | BLUEs | 2017 | 2018 | BLUEs | 2017 | 2018 | BLUEs | ||
| QPed.shared.2H-1 | 2H | 18.3–26.8 cM | 21,508,891–29,254,219 bp | −101.5 | - | −67.5 | 5.6 | 6.5 | 5.5 | −8.3 × 104 | - | −4.4 × 104 | PPD-H1 [20] |
| QPed.shared.2H-2 | 2H | 55.5–60.8 cM | 164,050,763–518,354,808 bp | −181.6 | −153.7 | −168.0 | −10.2 | −7.5 | −9.5 | −1.8 × 105 | −1.4 × 105 | −1.6 × 105 | HvCEN [22] |
| QPed.shared.2H-3 | 2H | 82.0 cM | 586,582,931–586,594,551 bp | 11.3 | - | 16.3 | - | - | - | −1.3 × 104 | - | 1.0 × 104 | |
| QPed.shared.3H-1 | 3H | 47.7–51.8 cM | 128,830,583–394,780,497 bp | 72.7 | 100.9 | 91.1 | 10.5 | 14.4 | 14.2 | 8.8 × 104 | 1.2 × 105 | 1.1 × 105 | |
| QPed.shared.3H-2 | 3H | 71.5 cM | 517,985,664 bp | - | 17.0 | 16.3 | - | - | - | - | - | 2.1 × 104 | |
| QPed.shared.3H-3 | 3H | 106.6–109.5 cM | 569,528,666–573,503,074 bp | −50.3 | - | - | −8.8 | - | −2.0 | −6.0 × 104 | - | −2.4 × 104 | HvGA20ox2 [23] |
| QPed.shared.4H-1 | 4H | 0.9–2.1 cM | 393,618–3,342,910 bp | - | 25.8 | - | 5.1 | 8.1 | 6.3 | - | 3.7 × 104 | 2.5 × 104 | |
| QTL | HEA a | WAL b | DIA b | AREA b |
|---|---|---|---|---|
| QFt.HEB25-1b a | −1.4 | - | - | - |
| QPed.shared.2H-1 | −9.5 | 5.5 | −67.5 | −4.4 × 104 |
| QPed.shared.2H-2 | −3.0 | −9.5 | −168.0 | −1.6 × 105 |
| QPed.shared.3H-1 | - | 14.2 | 91.1 | 1.1 × 105 |
| QPed.shared.3H-3 | −3.1 | −2.0 | −22.3 | −2.4 × 104 |
| QPed.shared.4H-1 | 3.2 | 6.3 | 25.2 | 2.5 × 104 |
| QFt.HEB25-4e a | 2.2 | - | - | - |
| QFt.HEB25-5d a | 3.8 | −1.3 | −22.7 | - |
| QFt.HEB25-7a a | 4.1 | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zahn, S.; Schmutzer, T.; Pillen, K.; Maurer, A. Genomic Dissection of Peduncle Morphology in Barley through Nested Association Mapping. Plants 2021, 10, 10. https://doi.org/10.3390/plants10010010
Zahn S, Schmutzer T, Pillen K, Maurer A. Genomic Dissection of Peduncle Morphology in Barley through Nested Association Mapping. Plants. 2021; 10(1):10. https://doi.org/10.3390/plants10010010
Chicago/Turabian StyleZahn, Sebastian, Thomas Schmutzer, Klaus Pillen, and Andreas Maurer. 2021. "Genomic Dissection of Peduncle Morphology in Barley through Nested Association Mapping" Plants 10, no. 1: 10. https://doi.org/10.3390/plants10010010
APA StyleZahn, S., Schmutzer, T., Pillen, K., & Maurer, A. (2021). Genomic Dissection of Peduncle Morphology in Barley through Nested Association Mapping. Plants, 10(1), 10. https://doi.org/10.3390/plants10010010





