Detection and Verification of QTL for Salinity Tolerance at Germination and Seedling Stages Using Wild Barley Introgression Lines
Abstract
:1. Introduction
2. Results
2.1. Variations and Heritability
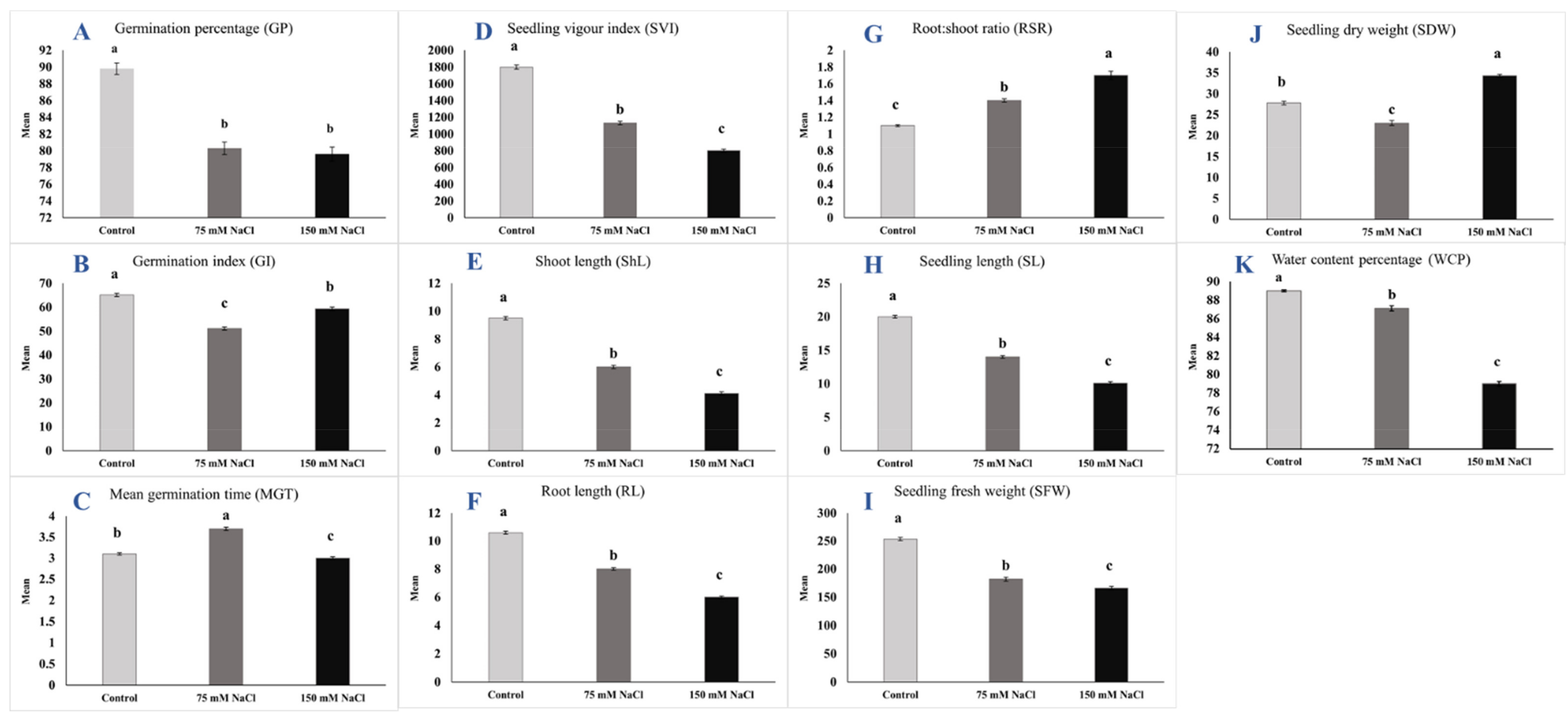
2.2. Trait Means and Reduction Percentage
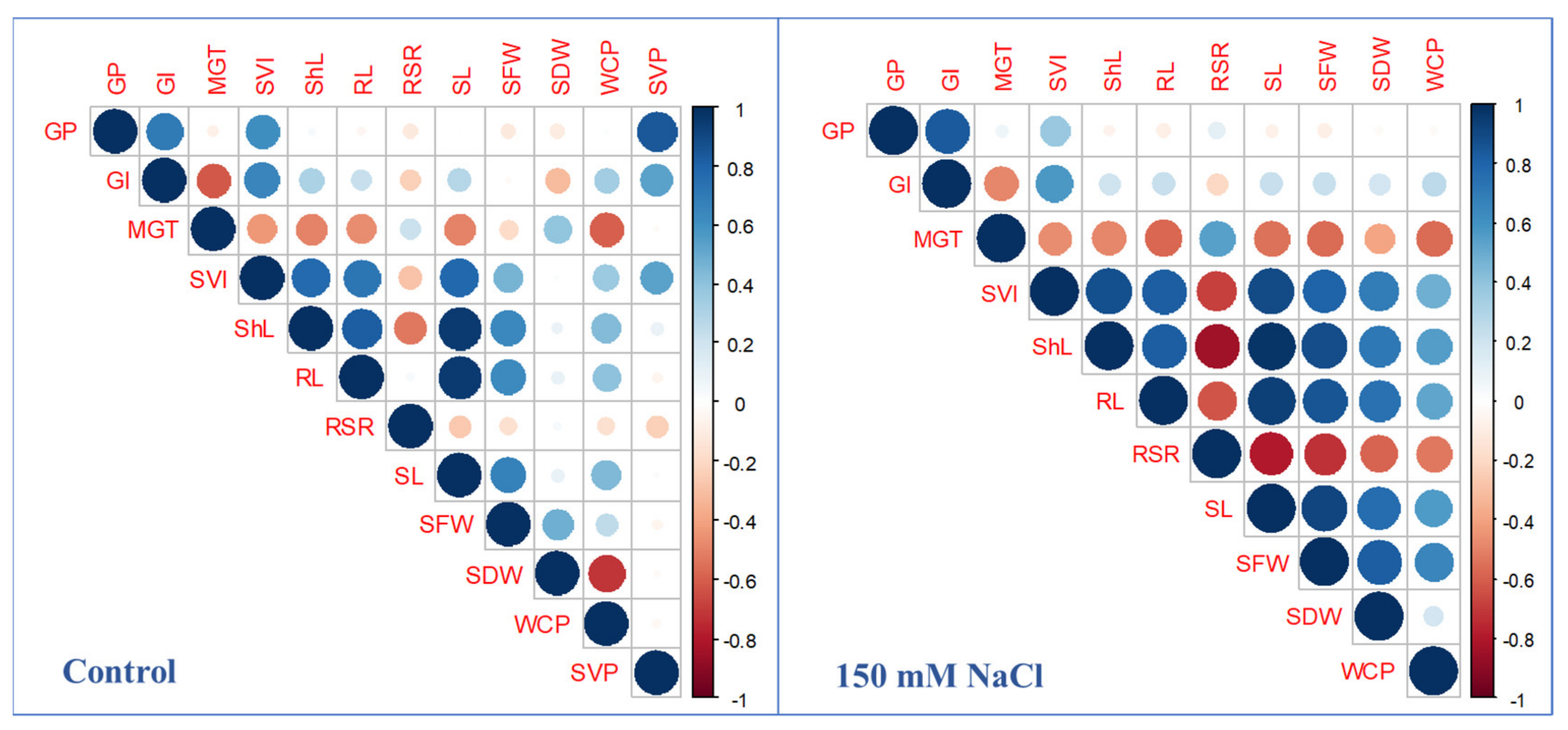
2.3. Phenotypic Correlation among Studied Traits
2.4. Salinity Tolerance Indices
2.5. QTL Identification for Germination and Seedling Related Traits
2.5.1. Seed Variability and Germination Percentages
2.5.2. Shoot and Root Lengths
2.5.3. Seedling Length and Seedling Vigor Index
2.5.4. Seedling Fresh and Dry Weights
2.5.5. Root/Shoot Ratio and Water Content Percentage
2.6. QTL Detection for Salinity Tolerance Indices
2.6.1. Germinability Tolerance Index (GTI)
2.6.2. Seedling Length Tolerance Index (SLTI)
2.6.3. Seedling Fresh (SFWTI) and Dry Weights Tolerance Indices (SDWTI)
2.6.4. Water Content Percentage Tolerance Index (WCPTI)
3. Discussion
3.1. S42ILs by Trait Associations
3.2. Germination Percentage Linked QTL
3.3. Seedling Growth-Related Traits Linked QTL
3.4. QTL Validation for Salt Tolerance
4. Materials and Methods
4.1. Plant Material and Genotyping
4.2. Seed Viability
4.3. Salinity Stress Treatments
4.4. Evaluation of Germination Parameters
- Germination percentage (GP; %):
- Germination index (GI) was calculated according to Ranal [66] as follows:where, N1, N2…N10, is the number of seeds germinated on the first, second and subsequent days until 10th day and the multipliers (i.e., 10, 9…etc.) are weights given to the days of the germination.GI = (10 × N1) + (9 × N2) +…+(1 × N10);
- Mean germination time (MGT) was calculated according to Mudaris [67] as follows:where, Ni is number of the new germinated seeds at time Ti.MGT = Σ(Ti × Ni)/ΣNi
4.5. Evaluation of Seedling Growth Parameters
4.6. Stress Tolerance Index (STI)
4.7. Statistical Analyses
4.8. QTL Detection
5. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ziemann, M.; Kamboj, A.; Hove, R.M.; Loveridge, S.; El-Osta, A.; Bhave, M. Analysis of the barley leaf transcriptome under salinity stress using mRNA-Seq. Acta Physiol. Plant. 2013, 35, 1915–1924. [Google Scholar] [CrossRef]
- Long, N.V.; Dolstra, O.; Malosetti, M.; Kilian, B.; Graner, A.; Visser, R.G.F.; van der Linden, C.G. Association mapping of salt tolerance in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2013, 126, 2335–2351. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassani, A.; Azapagic, A.; Shokri, N. Predicting long-term dynamics of soil salinity and sodicity on a global scale. Proc. Natl. Acad. Sci. USA 2021, 117, 33017–33027. [Google Scholar] [CrossRef] [PubMed]
- Pirasteh-Anosheh, H.; Ranjbar, G.; Pakniyat, H.; Emam, Y. Physiological mechanisms of salt stress tolerance in plants: An overview. Plant-Environ. Interact. Responses Approaches Mitigate Stress 2015, 141–160. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández, J.A.; Ferrer, M.A.; Jiménez, A.; Barceló, A.R.; Sevilla, F. Antioxidant systems and O2·-/H2O2 production in the apoplast of pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiol. 2001, 127, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, K. Alleviation of the Adverse Effects of Salinity Stress on Maize (Zea mays L.) by Exogenous Application of Glycine; Faculty of Sciences, University of Agriculture: Faisalabad, Pakistan, 2007. [Google Scholar]
- Moustafa, E.S.A.; El-Sobky, E.S.E.A.; Farag, H.I.A.; Yasin, M.A.T.; Attia, A.; Rady, M.O.A.; Awad, M.F.; Mansour, E. Sowing date and genotype influence on yield and quality of dual-purpose barley in a salt-affected arid region. Agronomy 2021, 11, 717. [Google Scholar] [CrossRef]
- Mwando, E.; Angessa, T.T.; Han, Y.; Zhou, G.; Li, C. Quantitative Trait Loci Mapping for Vigour and Survival Traits of Barley Seedlings after Germinating under Salinity Stress. Agronomy 2021, 11, 103. [Google Scholar] [CrossRef]
- Gorzolka, K.; Kölling, J.; Nattkemper, T.W.; Niehaus, K. Spatio-Temporal metabolite profiling of the barley germination process by MALDI MS imaging. PLoS ONE 2016, 11, e0150208. [Google Scholar] [CrossRef]
- Wu, D. Genetic variation of HvCBF genes and their association with salinity tolerance in Tibetan annual wild barley. PLoS ONE 2011, 6, e22938. [Google Scholar] [CrossRef] [Green Version]
- Mwando, E.; Angessa, T.T.; Han, Y.; Li, C. Salinity tolerance in barley during germination—homologs and potential genes. J. Zhejiang Univ. Sci. B 2020, 21, 93–121. [Google Scholar] [CrossRef]
- Mwando, E.; Han, Y.; Angessa, T.T.; Zhou, G.; Hill, C.B.; Zhang, X.Q.; Li, C. Genome-Wide Association Study of Salinity Tolerance During Germination in Barley (Hordeum vulgare L.). Front. Plant Sci. 2020, 11, 118. [Google Scholar] [CrossRef] [Green Version]
- Askari, H.; Kazemitabar, S.K.; Zarrini, H.N.; Saberi, M.H. Salt tolerance assessment of barley (Hordeum vulgare L.) genotypes at germination stage by tolerance indices. Open Agric. 2016, 1, 37–44. [Google Scholar] [CrossRef]
- Zaare, M.; Jafary, J. Quantitative trait loci diversity for salt tolerance at the early growth stage of barley (Hordeum vulgare L.). Crop Breed J. 2013, 3, 69–77. [Google Scholar]
- Mano, Y.; Takeda, K. Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.). Euphytica 1997, 94, 263–272. [Google Scholar] [CrossRef]
- Hernandez, J.; Meints, B.; Hayes, P. Introgression Breeding in Barley: Perspectives and Case Studies. Front. Plant Sci. 2020, 11, 761. [Google Scholar] [CrossRef]
- Nevo, E.; Chen, G. Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ. 2010, 33, 670–685. [Google Scholar] [CrossRef]
- Zamir, D. Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2001, 2, 983–989. [Google Scholar] [CrossRef]
- Soleimani, B.; Sammler, R.; Backhaus, A.; Beschow, H.; Schumann, E.; Mock, H.P.; von Wirén, N.; Seiffert, U.; Pillen, K. Genetic regulation of growth and nutrient content under phosphorus deficiency in the wild barley introgression library S42IL. Plant Breed. 2017, 136, 892–907. [Google Scholar] [CrossRef]
- Hoffmann, A.; Maurer, A.; Pillen, K. Detection of nitrogen deficiency QTL in juvenile wild barley introgression lines growing in a hydroponic system. BMC Genet. 2012, 13, 88. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Schmalenbach, I.; von Korff, M.; Léon, J.; Kilian, B.; Rode, J.; Pillen, K. Association of barley photoperiod and vernalization genes with QTLs for flowering time and agronomic traits in a BC2DH population and a set of wild barley introgression lines. Theor. Appl. Genet. 2010, 120, 1559–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnaithmann, F.; Pillen, K. Detection of exotic QTLs controlling nitrogen stress tolerance among wild barley introgression lines. Euphytica 2013, 189, 67–88. [Google Scholar] [CrossRef]
- Frimpong, F.; Windt, C.W.; van Dusschoten, D.; Naz, A.A.; Frei, M.; Fiorani, F. A Wild Allele of Pyrroline-5-Carboxylate Synthase1 Leads to Proline Accumulation in Spikes and Leaves of Barley Contributing to Improved Performance under Reduced Water Availability. Front. Plant Sci. 2021, 12, 180. [Google Scholar] [CrossRef] [PubMed]
- Naz, A.A.; Ehl, A.; Pillen, K.; Léon, J. Validation for root-related quantitative trait locus effects of wild origin in the cultivated background of barley (Hordeum vulgare L.). Plant Breed. 2012, 131, 392–398. [Google Scholar] [CrossRef]
- Riaz, M.; Arif, M.S.; Ashraf, M.A.; Mahmood, R.; Yasmeen, T.; Shakoor, M.B.; Shahzad, S.M.; Ali, M.; Saleem, I.; Arif, M.; et al. A Comprehensive Review on Rice Responses and Tolerance to Salt Stress; Elsevier Inc.: Amsterdam, The Netherlands, 2018; ISBN 9780128143322. [Google Scholar]
- Tuan, P.A.; Sun, M.; Nguyen, T.N.; Park, S.; Ayele, B.T. Molecular Mechanisms of Seed Germination; Elsevier Inc.: Amsterdam, The Netherlands, 2018; ISBN 9780128115251. [Google Scholar]
- Lamichhane, J.R.; Messéan, A.; Ricci, P. Research and innovation priorities as defined by the Ecophyto plan to address current crop protection transformation challenges in France. Adv. Agron. 2019, 154, 81–152. [Google Scholar] [CrossRef]
- Zhu, J.; Fan, Y.; Li, C.; Shabala, S.; Zhao, C.; Hong, Y.; Lv, C.; Guo, B.; Xu, R.; Zhou, M. Candidate genes for salinity tolerance in barley revealed by RNA-seq analysis of near-isogenic lines. Plant Growth Regul. 2020, 92, 571–582. [Google Scholar] [CrossRef]
- Rajeswari, S.; Sood, N.; Swarup, T.G.; Subramanian, R. Assessing salt-stress tolerance in barley. Univ. Sci. 2019, 24, 91–109. [Google Scholar] [CrossRef] [Green Version]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef]
- Flowers, T.J. Improving crop salt tolerance. J. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef]
- Schmalenbach, I.; Léon, J.; Pillen, K. Identification and verification of QTLs for agronomic traits using wild barley introgression lines. Theor. Appl. Genet. 2009, 118, 483–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naz, A.A.; Arifuzzaman, M.; Muzammil, S.; Pillen, K.; Léon, J. Wild barley introgression lines revealed novel QTL alleles for root and related shoot traits in the cultivated barley ( Hordeum vulgare L.). BMC Genet. 2014, 15, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayed, M.A.; Schumann, H.; Pillen, K.; Naz, A.A.; Léon, J. AB-QTL analysis reveals new alleles associated to proline accumulation and leaf wilting under drought stress conditions in barley (Hordeum vulgare L.). BMC Genet. 2012, 13, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honsdorf, N.; March, T.J.; Berger, B.; Tester, M.; Pillen, K. High-throughput phenotyping to detect drought tolerance QTL in wild barley introgression lines. PLoS ONE 2014, 9, e97047. [Google Scholar] [CrossRef] [Green Version]
- Zahn, S.; Koblenz, B.; Christen, O.; Pillen, K.; Maurer, A. Evaluation of wild barley introgression lines for agronomic traits related to nitrogen fertilization. Euphytica 2020, 216, 39. [Google Scholar] [CrossRef] [Green Version]
- Honsdorf, N.; March, T.J.; Pillen, K. QTL controlling grain filling under terminal drought stress in a set of wild barley introgression lines. PLoS ONE 2017, 12, e0185983. [Google Scholar] [CrossRef] [Green Version]
- Sayed, M.A.; Hamada, A.; Lèon, J.; Naz, A.A. Genetic mapping reveals novel exotic QTL alleles for seminal root architecture in barley advanced backcross double haploid population. Euphytica 2017, 213, 54. [Google Scholar] [CrossRef]
- Hessini, K.; Ferchichi, S.; Ben Youssef, S.; Werner, K.H.; Cruz, C.; Gandour, M. How does salinity duration affect growth and productivity of cultivated barley? Agron. J. 2015, 107, 174–180. [Google Scholar] [CrossRef]
- Angessa, T.T.; Zhang, X.Q.; Zhou, G.; Broughton, S.; Zhang, W.; Li, C. Early growth stages salinity stress tolerance in CM72 x Gairdner doubled haploid barley population. PLoS ONE 2017, 12, e0179715. [Google Scholar] [CrossRef] [Green Version]
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Effect of salinity stress on plants and its tolerance strategies: A review. Environ. Sci. Pollut. Res. 2015, 22, 4056–4075. [Google Scholar] [CrossRef]
- Sayar, R.; Bchini, H.; Mosbahi, M.; Ezzine, M. Effects of salt and drought stresses on germination, emergence and seedling growth of Durum wheat (Triticum durum Desf.). Afr. J. Agric. Res. 2010, 5, 2008–2016. [Google Scholar]
- Zhang, H.; Irving, L.J.; McGill, C.; Matthew, C.; Zhou, D.; Kemp, P. The effects of salinity and osmotic stress on barley germination rate: Sodium as an osmotic regulator. Ann. Bot. 2010, 106, 1027–1035. [Google Scholar] [CrossRef]
- Baǧci, S.A.; Ekiz, H.; Yilmaz, A. Determination of the salt tolerance of some barley genotypes and the characteristics affecting tolerance. Turk. J. Agric. For. 2003, 27, 253–260. [Google Scholar] [CrossRef]
- Schmalenbach, I.; Körber, N.; Pillen, K. Selecting a set of wild barley introgression lines and verification of QTL effects for resistance to powdery mildew and leaf rust. Theor. Appl. Genet. 2008, 117, 1093–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmalenbach, I.K.P. Detection and verification of malting quality QTLs using wild barley introgression lines. Theor. Appl. Genet. 2009, 118, 1411–1427. [Google Scholar] [CrossRef] [Green Version]
- Arifuzzaman, M.; Sayed, M.A.; Muzammil, S.; Pillen, K.; Schumann, H.; Naz, A.A.; Léon, J. Detection and validation of novel QTL for shoot and root traits in barley (Hordeum vulgare L.). Mol. Breed. 2014, 34, 1373–1387. [Google Scholar] [CrossRef]
- Muzammil, S. Genetic Dissection of Shoot Traits and Proline Content under Control and Drought Conditions in Barley. Ph.D. Thesis, University of Bonn, Bonn, Germany, 2018. [Google Scholar]
- Xu, R. A single locus is responsible for salinity tolerance in a chinese landrace barley (Hordeum vulgare L.). PLoS ONE 2012, 7, e43079. [Google Scholar] [CrossRef]
- Thabet, S.G.; Moursi, Y.S.; Sallam, A.; Karam, M.A.; Alqudah, A.M. Genetic associations uncover candidate SNP markers and genes associated with salt tolerance during seedling developmental phase in barley. Environ. Exp. Bot. 2021, 188, 104499. [Google Scholar] [CrossRef]
- Sayed, M.A.; Nassar, S.M.; Moustafa, E.S.; Said, M.T.; Börner, A.; Hamada, A. Genetic Mapping Reveals Novel Exotic and Elite QTL Alleles for Salinity Tolerance in Barley. Agronomy 2021, 11, 1774. [Google Scholar] [CrossRef]
- Ma, Y.; Shabala, S.; Li, C.; Liu, C.; Zhang, W.; Zhou, M. Quantitative trait loci for salinity tolerance identified under drained and waterlogged conditions and their association with flowering time in barley (Hordeum vulgare. L). PLoS ONE 2015, 10, e0134822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saade, S.; Brien, C.; Pailles, Y.; Berger, B.; Shahid, M.; Russell, J.; Waugh, R.; Negrão, S.; Tester, M. Dissecting new genetic components of salinity tolerance in two-row spring barley at the vegetative and reproductive stages. PLoS ONE 2020, 15, e0236037. [Google Scholar] [CrossRef]
- Iqbal, M.T. Physiological, Biochemical and Molecular Responses of Different Barley Varieties to Drought and Salinity. Ph.D. Thesis, University of Bonn, Bonn, Germany, 2018. [Google Scholar]
- Zhou, G.; Johnson, P.; Ryan, P.R.; Delhaize, E.; Zhou, M. Quantitative trait loci for salinity tolerance in barley (Hordeum vulgare L.). Mol. Breed. 2012, 29, 427–436. [Google Scholar] [CrossRef]
- Tarawneh, R.F.A.; Gutachter, J.; Pillen, K.; Doz Andreas Börner, P.; Khlestkina, E. Mapping and Identifying Genes for Drought Tolerance in Barley (Hordeum vulgare L.). Ph.D. Thesis, Martin-Luther-Universität Halle-Wittenberg, Halle, Germany, 2020. [Google Scholar]
- Foolad, M. Comparison of salt tolerance during seed germination and vegetative growth in tomato by QTL mapping. Genome 1999, 42, 727–734. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Xu, B.; Athman, A.; Conn, S.J.; Jordans, C.; Byrt, C.S.; Hare, R.A.; Tyerman, S.D.; Tester, M.; et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012, 30, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, M.W.; Xie, M.; Liu, X.; Ni, M.; Shao, G.; Song, C.; Kay-Yuen Yim, A.; Tao, Y.; Wong, F.L.; et al. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 2014, 5, 4340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Korff, M.; Wang, H.; Léon, J.; Pillen, K. Development of candidate introgression lines using an exotic barley accession (Hordeum vulgare ssp. spontaneum) as donor. Theor. Appl. Genet. 2004, 109, 1736–1745. [Google Scholar] [CrossRef]
- Schmalenbach, I.; March, T.J.; Pillen, K.; Bringezu, T.; Waugh, R. High-resolution genotyping of wild barley introgression lines and fine-mapping of the threshability locus thresh-1 using the illumina goldengate assay. G3 Genes Genomes Genet. 2011, 1, 187–196. [Google Scholar] [CrossRef] [Green Version]
- Comadran, J. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 2012, 44, 1388–1392. [Google Scholar] [CrossRef]
- Ista International rules for seed testing. In 12. ISTA (1993) International Rules for Seed Testing; ISTA: Zurich, Switzerland, 1993; Volume 12, pp. 345–376.
- Ranal, M.A.; de Santana, D.G.; Ferreira, W.R.; Mendes-Rodrigues, C. Calculando medidas de germinação e organizando planilhas eletrônicas. Rev. Bras. Bot. 2009, 32, 849–855. [Google Scholar] [CrossRef] [Green Version]
- Mudaris, M.A. Notes on Various Parameter s Recording the Speed of Seed Germination. J. Agric. Trop. Subtrop. 1984, 99, 147–154. [Google Scholar]
- Abdul-Baki, A.A.; Anderson, J.D. Vigor Determination in Soybean Seed by Multiple Criteria 1. Crop Sci. 1973, 13, 630–633. [Google Scholar] [CrossRef]
- Fernandez, G.C.J. Effective selection criteria for assessing plant stress tolerance. In Proceedings of the International Symposium on Adaptation of Food Crops to Temperature and Water Stress, Taiwan, 13–18 August 1992; pp. 257–270. [Google Scholar] [CrossRef]
- SAS Institute (2011). The SAS System for Windows. Release 9.2. SAS Inst., Cary, NC. 2011. Available online: http://www.sciepub.com/reference/166089 (accessed on 4 July 2021).
- Padi, F.K. Genotype × environment interaction and yield stability in a cowpea-based cropping system. Euphytica 2007, 158, 11–25. [Google Scholar] [CrossRef]

| Trait | Control | 75 mM NaCL | 150 mM NaCL | Combined ANOVA | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MS | C.V.% | R2 | Hb | MS | C.V.% | R2 | Hb | MS | C.V.% | R2 | Hb | G | T | G × T | |
| GP | 172.49 ** | 5.5 | 0.78 | 85.98 | 196.75 ** | 7.2 | 0.74 | 82.98 | 244.55 ** | 8.1 | 0.74 | 83.02 | 346.6 ** | 148.8 ** | 133.6 ** |
| GI | 200.3 ** | 5.0 | 0.90 | 94.71 | 145.96 ** | 10.3 | 0.72 | 81.14 | 174.22 ** | 10.4 | 0.69 | 78.01 | 277.7 ** | 7583 ** | 121.4 |
| MGT | 0.47 ** | 5.5 | 0.88 | 93.62 | 0.54 ** | 9.9 | 0.66 | 75.02 | 0.34 ** | 9.2 | 0.68 | 77.36 | 0.44 ** | 20.5 ** | 0.46 ** |
| SVI | 190,565 ** | 10.9 | 0.71 | 80.00 | 166,431 ** | 11.9 | 0.82 | 89.05 | 134,821 ** | 13.4 | 0.85 | 91.38 | 163,643 ** | 39,506,781 ** | 164,087 ** |
| ShL | 4.44 ** | 11.2 | 0.66 | 74.76 | 6.44 ** | 13.1 | 0.83 | 90.24 | 6.79 ** | 16.8 | 0.87 | 92.98 | 5.1 ** | 1120.1 ** | 6.3 ** |
| RL | 4.15 ** | 10.5 | 0.62 | 70.40 | 2.66 ** | 9.9 | 0.67 | 76.18 | 2.98 ** | 10.5 | 0.79 | 86.85 | 3.02 ** | 815.1 ** | 3.4 ** |
| RSR | 0.02 ** | 11.0 | 0.42 | 30.92 | 0.24 ** | 13.1 | 0.78 | 86.41 | 1.03 ** | 21.5 | 0.80 | 87.63 | 0.47 ** | 10.6 ** | 0.41 ** |
| SL | 15.65 ** | 9.4 | 0.68 | 77.17 | 15.16 ** | 10.1 | 0.79 | 86.75 | 17.21 ** | 11.2 | 0.87 | 92.61 | 13.5 ** | 3839.5 ** | 17.3 ** |
| SFW | 2166.4 ** | 12.0 | 0.53 | 57.39 | 3972.9 ** | 16.9 | 0.67 | 76.37 | 2749.5 ** | 14.3 | 0.70 | 79.39 | 2875.9 ** | 328,179 ** | 3006.5 ** |
| SDW | 48.32 ** | 18.0 | 0.49 | 48.59 | 102.15 ** | 21.4 | 0.67 | 76.27 | 77.80 ** | 17.7 | 0.51 | 52.65 | 63.5 ** | 4981.3 ** | 82.4 ** |
| WCP | 5.66 ** | 1.4 | 0.63 | 71.09 | 26.47 ** | 2.4 | 0.75 | 83.37 | 18.00 ** | 3.2 | 0.57 | 63.69 | 14.5 ** | 4294.6 ** | 17.8 ** |
| SVP | 123.50 * | 6.5 | 0.66 | 74.99 | |||||||||||
| Trait | Control | 75 mM NaCl | 150 mM NaCl | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean ± S.E. | Range | Mean ± S.E. | Range | R% a | Mean ± S.E. | Range | R% b | R% c | |
| GP | 89.8 ± 0.69 | 60–100 | 80.3 ± 0.75 | 60–100 | −10.6 | 79.6 ± 0.84 | 53.3–100 | −11.4 | −0.9 |
| GI | 65.1 ± 0.69 | 45–85 | 51.1 ± 0.65 | 33–68 | −21.5 | 59.3 ± 0.73 | 37–79 | −8.9 | 16.0 |
| MGT | 3.1 ± 0.03 | 2.3–4.2 | 3.7 ± 0.04 | 2.45–5.08 | 19.4 | 3.3 ± 0.03 | 2–4.36 | −3.2 | −18.9 |
| SVI | 1799.1 ± 24.01 | 1026–2470 | 1132 ± 20.9 | 553.3–1800 | −37.1 | 801.6 ± 18.4 | 271.1–1466.6 | −55.4 | −29.2 |
| ShL | 9.5 ± 0.12 | 5–13.7 | 6 ± 0.12 | 2.2–10.47 | −36.8 | 4.1 ± 0.12 | 0.6–8 | −56.8 | −31.7 |
| RL | 10.6 ± 0.11 | 6.5–15.33 | 8 ± 0.09 | 4.7–11.1 | −24.5 | 6 ± 0.09 | 2.83–8.33 | −43.4 | −25.0 |
| RSR | 1.1 ± 0.01 | 0.8–1.67 | 1.4 ± 0.02 | 0.91–3 | 27.3 | 1.7 ± 0.05 | 0.91–5.56 | 54.5 | 21.4 |
| SL | 20 ± 0.22 | 12.2–29 | 14 ± 0.20 | 8.3–20.33 | −30 | 10.1 ± 0.21 | 3.83–15.6 | −49.5 | −27.9 |
| SFW | 253 ± 2.95 | 165.7–351.7 | 181.8 ± 3.55 | 58.4–272 | −28.2 | 166.1 ± 2.89 | 35.3–241.7 | −34.4 | −8.6 |
| SDW | 27.8 ± 0.46 | 12.8–43.5 | 23 ± 0.57 | 7.6–40.5 | −17.3 | 34.3 ± 0.34 | 11.93–48.97 | 23.4 | 49.1 |
| WCP | 89 ± 0.13 | 86.4–94.9 | 87.1 ± 0.27 | 80.2–92.7 | −2.1 | 79 ± 0.25 | 61.68–87.29 | −11.2 | −9.3 |
| SVP | 85 ± 0.63 | 65–100 | |||||||
| Control-75 mM NaCl | Control-150 mM NaCl | |||||
|---|---|---|---|---|---|---|
| MS | R2 | Hb | MS | R2 | Hb | |
| GTI | 0.0288 ** | 0.67 | 75.68 | 0.0327 ** | 0.68 | 77.30 |
| SLTI | 0.0886 ** | 0.78 | 85.88 | 0.0683 ** | 0.81 | 88.17 |
| SFWTI | 0.0856 ** | 0.64 | 72.86 | 0.0682 ** | 0.61 | 69.06 |
| SDWTI | 0.2550 ** | 0.64 | 72.54 | 0.3020 ** | 0.49 | 48.21 |
| WCTI | 0.0046 ** | 0.75 | 83.49 | 0.0030 ** | 0.62 | 69.60 |
| Mean ± S.E. | C.V.% | Range | Mean ± S.E. | C.V.% | Range | |
| GTI | 0.90 ± 0.01 | 9.3 | 0.66–1.25 | 0.89 ± 0.01 | 9.7 | 0.62–1.25 |
| SLTI | 0.72 ± 0.016 | 15.6 | 0.37–1.56 | 0.52 ± 0.01 | 17.4 | 0.17–1.16 |
| SFWTI | 0.73 ± 0.017 | 20.8 | 0.19–1.39 | 0.68 ± 0.015 | 21.5 | 0.12–1.27 |
| SDWTI | 0.87 ± 0.029 | 26.5 | 0.29–2.33 | 1.31 ± 0.036 | 30.2 | 0.34–30.47 |
| WCTI | 0.98 ± 0.004 | 2.8 | 0.89–1.06 | 0.89 ± 0.003 | 3.5 | 0.69–1.01 |
| Trait | QTL | Int. Lines | Chr. | Interval (cM) | Line Main Effect | Control | 75 mM NaCl | 150 mM NaCl | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | RP% | Mean | RP% | Mean | RP% | Mean | RP% | |||||
| GP | Scarlett | 81.5 SC | 93.3 SC | 73.3 SC | 77.8 SC | |||||||
| QGP.S42IL.4H.a | S42IL-116 | 4H | 1.1–40 | 91.9 | 12.7 ** | 95.6 | 2.4 | 97.8 | 33.3 ** | 82.2 | 5.7 | |
| QGP.S42IL.6H.c | S42IL-132 | 6H | 94.9–108.3 | 91.9 | 12.7 ** | 88.9 | −4.8 | 91.1 | 24.2 ** | 95.6 | 22.9 ** | |
| ShL | Scarlett | 7.2 SC | 9.9 SC | 6.2 SC | 5.7 SC | |||||||
| QShl.S42IL.2H.a | S42IL-109 | 2H | 33.9–62.7 | 7.7 | −22.8 | 9.6 | 53.3 ** | 7.7 | 35.5 ** | |||
| QShl.S42IL.2H.c | S42IL-110 | 2H | 89.5–97.8 | 9 | −9.6 | 9.4 | 50.8 ** | 5.4 | −5 | |||
| RL | Scarlett | 8.1 SC | 11.3 SC | 7.2 SC | 5.8 SC | |||||||
| QRl.S42IL.2H.a | S42IL-109 | 2H | 33.9–62.7 | 8.3 | −26.8 * | 9.67 | 32.6 ** | 7.3 | 26.4 | |||
| QRl.S42IL.5H.a | S42IL-126 | 5H | 76.2–120.3 | 10.6 | −6.6 | 7.22 | −0.91 | 8.1 | 40.9 ** | |||
| QRl.S42IL.5H.b | S42IL-176 | 5H | 81.3–140.1 | 9.4 | −16.6 | 9.86 | 35.2 ** | 7.5 | 30.6 ** | |||
| SL | Scarlett | 15.4 SC | 21.3 SC | 13.5 SC | 11.5 SC | |||||||
| QSl.S42IL.2H.b | S42IL-109 | 2H | 33.9–62.7 | 16 | −24.9 | 19.2 | 42.1 ** | 15 | 30.9 ** | |||
| QSl.S42IL.2H.d | S42IL-110 | 2H | 89.5–97.8 | 19.4 | −8.8 | 18.3 | 35.1 ** | 11.3 | −1.2 | |||
| QSl.S42IL.3H.c | S42IL-115 | 3H | 120.7–155 | 20.6 | −3 | 17.4 | 28.4 * | 9.68 | −15.5 | |||
| QSl.S42IL.4H.a | S42IL-116 | 4H | 1.1–40 | 19.3 | −9.2 | 17.6 | 29.9 * | 8.5 | −25.8 * | |||
| QSl.S42IL.6H | S42IL-132 | 6H | 94.9–108.3 | 19.8 | −7 | 14.8 | 9.4 | 15.1 | 31.4 ** | |||
| SVI | Scarlett | 1289 SC | 1986 SC | 992 SC | 891 SC | |||||||
| QSvi.S42IL.2H.b | S42IL-109 | 2H | 33.9–62.7 | 1569 | −21 | 1537 | 55.0 ** | 1300 | 46.0 ** | |||
| QSvi.S42IL.4H.a | S42IL-116 | 4H | 1.1–40 | 1842 | −7.3 | 1718 | 73.2 ** | 700 | −21.4 | |||
| QSvi.S42IL.6H.a | S42IL-148 | 6H | 0.3–11.3 | 1004 | −22 ** | 1356 | −31.7 ** | 934 | −5.8 | 723 | −18.8 | |
| QSvi.S42IL.6H.b | S42IL-132 | 6H | 94.9–108.3 | 1514 | 17.4 * | 1758 | −11.5 | 1348 | 36.0 * | 1438 | 61.5 ** | |
| SFW | Scarlett | 211.5 SC | 278.7 SC | 184 SC | 172 SC | |||||||
| QSfw.S42IL.6H | S42IL-132 | 6H | 94.9–108.3 | 269.3 | −3.4 | 196.8 | 7 | 241.9 | 40.6 ** | |||
| SDW | Scarlett | 15.4 SC | 27 | 26.3 SC | 33.4 SC | |||||||
| QSdw.S42IL.2H | S42IL-107 | 2H | 12.5–41.2 | 21 | −22.4 | 11.5 | −56.0 ** | 40.6 | 21.8 | |||
| RSR | Scarlett | 1.1 SC | 1.1 SC | 1.2 SC | 1.0 SC | |||||||
| QRsr.S42IL.5H.a | S42IL-125 | 5H | 51.5–81.3 | 1.7 | 53.8 ** | 1.1 | 0 | 2.59 | 121.1 ** | 1.39 | 36.8 | |
| QRsr.S42IL.5H.b | S42IL-127 | 5H | 138.5–162.5 | 2.3 | 108 ** | 1.1 | −7.3 | 1.77 | 51.6 ** | 4.1 | 304.6 ** | |
| WCP | Scarlett | 85.5 SC | 90.3 SC | 85.8 SC | 80.6 SC | |||||||
| QWc.S42IL.2H.a | S42IL-109 | 2H | 33.9–62.7 | 87.3 | −3.3 | 92.1 | 7.3 ** | 82.5 | 2.3 | |||
| QTL | Int. Lines | Chr. | Interval | Line Main Effect | Control-75 mM NaCl | Control-150 mM NaCl | |||
|---|---|---|---|---|---|---|---|---|---|
| Mean | RP% | Mean | RP% | Mean | RP% | ||||
| Germination percentage tolerance index | |||||||||
| Scarlett | 0.81 | 0.79 | 0.84 | ||||||
| QGpti.S42IL.4H.a | S42IL-116 | 4H | 1.1–40 | 1.03 | 30.0 ** | ||||
| QGpti.S42IL.4H.b | S42IL-117 | 4H | 17.8–49.9 | 1.03 | 30.0 ** | ||||
| QGpti.S42IL.4H.c | S42IL-121 | 4H | 51.9–81.2 | 1.03 | 24.7 * | 1.09 | 30.7 ** | ||
| QGpti.S42IL.5H.a | S42IL-126 | 5H | 76.2–120.3 | 1.06 | 30.6 ** | 1.07 | 35.2 ** | ||
| QGpti.S42IL.5H.b | S42IL-176 | 5H | 81.3–140.1 | 0.99 | 22.4 * | 1.01 | 28.4 ** | ||
| QGpti.S42IL.5H.c | S42IL-127 | 5H | 138.5–162.5 | 1.0 | 23.1 * | 1.08 | 29.2 ** | ||
| QGpti.S42IL.6H.a | S42IL-148 | 6H | 0.3–11.3 | 1.10 | 39.4 ** | ||||
| QGpti.S42IL.6H.b | S42IL-149 | 6H | 30–51 | 1.01 | 24.7 * | 1.06 | 34.1 ** | ||
| QGpti.S42IL.6H.c | S42IL-132 | 6H | 94.9–108.3 | 1.05 | 29.3 ** | 1.03 | 30.0 ** | 1.08 | 28.7 ** |
| QGpti.S42IL.7H.a | S42IL-133 | 7H | 12.7–37.6 | 1.03 | 30.2 ** | ||||
| QGpti.S42IL.7H.b | S42IL-135 | 7H | 67.8–118.5 | 1.05 | 29.5 ** | 1.11 | 32.0 ** | ||
| Seedling length tolerance index | |||||||||
| Scarlett | 0.58 | 0.64 | 0.54 | ||||||
| QSlti.S42IL.2H.a | S42IL-107 | 2H | 12.5–41.2 | 0.85 | 46.3 ** | 0.98 | 54.2 ** | 0.00 | |
| QSlti.S42IL.2H.b | S42IL-109 | 2H | 33.9–62.7 | 1.23 | 93.9 ** | 0.96 | 78.5 ** | ||
| QSlti.S42IL.2H.c | S42IL-153 | 2H | 60.7–68.6 | 0.22 | −59.4 ** | ||||
| QSlti.S42IL.2H.d | S42IL-110 | 2H | 89.5–97.8 | 0.95 | 48.9 ** | ||||
| QSlti.S42IL.3H.a | S42IL-111 | 3H | 43.1–55.2 | 0.96 | 51.6 ** | ||||
| QSlti.S42IL.3H.b | S42IL-114 | 3H | 75.9–144.9 | 1.25 | 96.3 ** | ||||
| QSlti.S42IL.5H.a | S42IL-176 | 5H | 81.3–140.1 | 0.90 | 53.9 ** | 0.98 | 53.4 ** | 0.83 | 54.7 ** |
| QSlti.S42IL.5H.b | S42IL-127 | 5H | 138.5–162.5 | 0.22 | −58.4 ** | ||||
| QSlti.S42IL.6H | S42IL-132 | 6H | 94.9–108.3 | 0.76 | 41.5 ** | ||||
| QSlti.S42IL.7H | S42IL-139 | 7H | 129.5–141.1 | 0.27 | −50.0 ** | ||||
| Seedling fresh weight tolerance index | |||||||||
| Scarlett | 0.63 | 0.66 | 0.62 | ||||||
| QSfwti.S42IL.2H.a | S42IL-107 | 2H | 12.5–41.2 | 1.00 | 61.8 ** | ||||
| QSfwti.S42IL.2H.b | S42IL-109 | 2H | 33.9–62.7 | 1.1 | 72.7 ** | 1.15 | 73.7 ** | 1.06 | 71.7 ** |
| Seedling dry weight tolerance index | |||||||||
| Scarlett | 1.1 | ||||||||
| QSdwti.S42IL.7H.a | S42IL-133 | 7H | 12.7–37.6 | 1.8 | 64.2 * | ||||
| QSdwti.S42IL.7H.b | S42IL-137 | 7H | 86–127.5 | 1.7 | 59.1 * | ||||
| Water content percentage tolerance index | |||||||||
| Scarlett | 0.92 | 0.95 | 0.89 | ||||||
| QWcti.S42IL.2H.a | S42IL-108 | 2H | 12.5–59.1 | 1.03 | 8.2 ** | ||||
| QWcti.S42IL.2H.b | S42IL-109 | 2H | 33.9–62.7 | 0.99 | 8.4 ** | 1.05 | 10.9 ** | ||
| QWcti.S42IL.2H.c | S42IL-153 | 2H | 60.7–68.6 | 0.78 | −12.4 ** | ||||
| QWcti.S42IL.3H.a | S42IL-111 | 3H | 43.1–55.2 | 1.03 | 8.7 ** | ||||
| QWcti.S42IL.3H.b | S42IL-114 | 3H | 75.9–144.9 | 1.04 | 9.9 ** | ||||
| QWcti.S42IL.4H.a | S42IL-116 | 4H | 1.1–40 | 1.03 | 8.4 ** | ||||
| QWcti.S42IL.4H.b | S42IL-146 | 4H | 55.7–81.2 | 1.03 | 8.8 ** | ||||
| QWcti.S42IL.5H | S42IL-147 | 5H | 73.3–81.3 | 1.05 | 10.0 ** | ||||
| QWcti.S42IL.6H.a | S42IL-148 | 6H | 0.3–11.3 | 0.99 | 7.6 * | 1.04 | 8.9 ** | ||
| QWcti.S42IL.6H.b | S42IL-129 | 6H | 47.5–79.6 | 1.03 | 7.9 ** | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sayed, M.A.; Tarawneh, R.; Youssef, H.M.; Pillen, K.; Börner, A. Detection and Verification of QTL for Salinity Tolerance at Germination and Seedling Stages Using Wild Barley Introgression Lines. Plants 2021, 10, 2246. https://doi.org/10.3390/plants10112246
Sayed MA, Tarawneh R, Youssef HM, Pillen K, Börner A. Detection and Verification of QTL for Salinity Tolerance at Germination and Seedling Stages Using Wild Barley Introgression Lines. Plants. 2021; 10(11):2246. https://doi.org/10.3390/plants10112246
Chicago/Turabian StyleSayed, Mohammed Abdelaziz, Rasha Tarawneh, Helmy Mohamed Youssef, Klaus Pillen, and Andreas Börner. 2021. "Detection and Verification of QTL for Salinity Tolerance at Germination and Seedling Stages Using Wild Barley Introgression Lines" Plants 10, no. 11: 2246. https://doi.org/10.3390/plants10112246






