Fusarium Oxysporum f. sp. Cannabis Isolated from Cannabis Sativa L.: In Vitro and In Planta Biocontrol by a Plant Growth Promoting-Bacteria Consortium
Abstract
:1. Introduction
2. Results
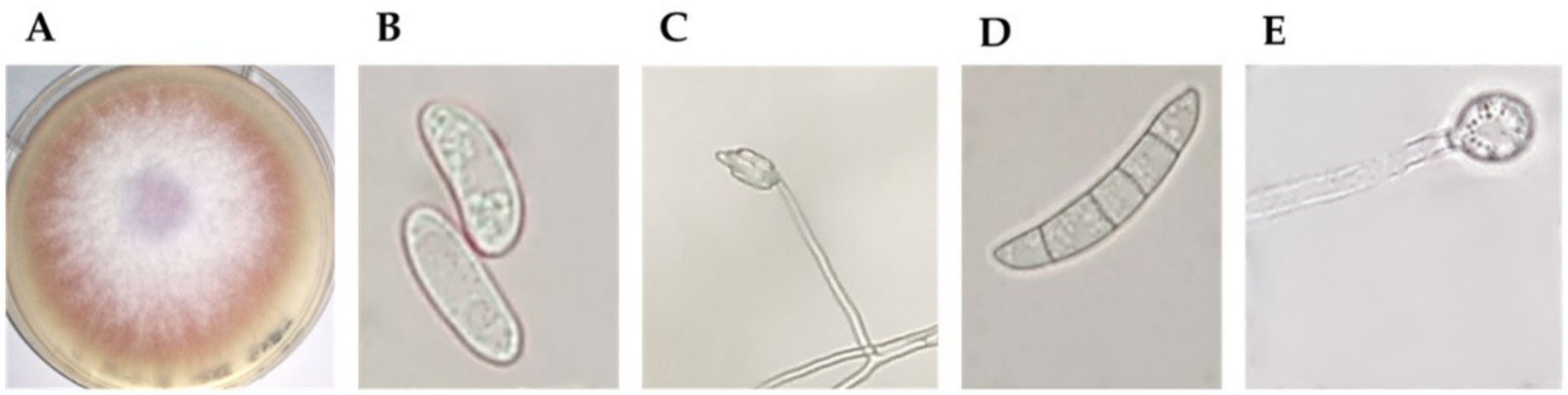
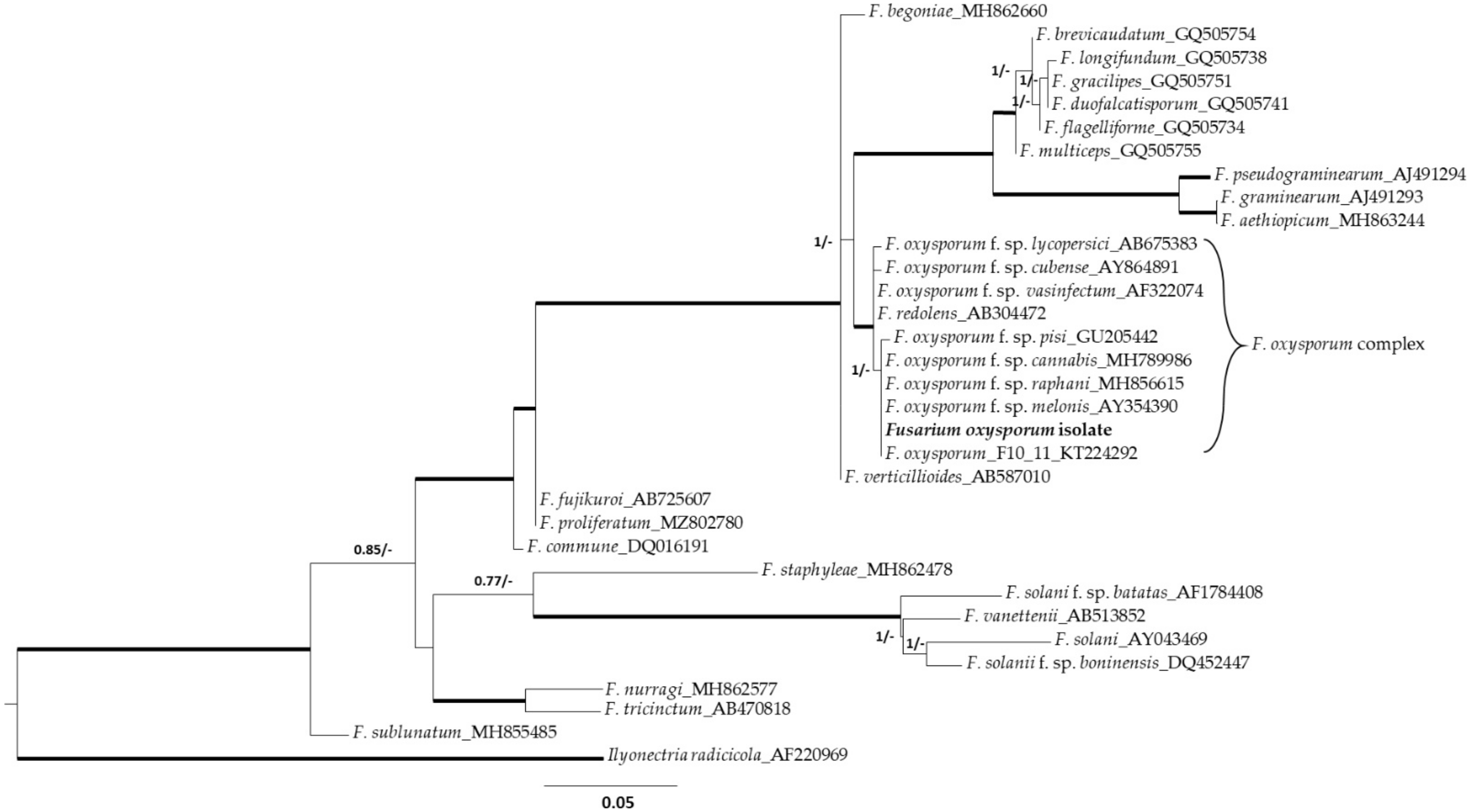
2.1. Fungal Isolate Morphological and Molecular Identification
2.2. In Vitro Antagonistic Activity
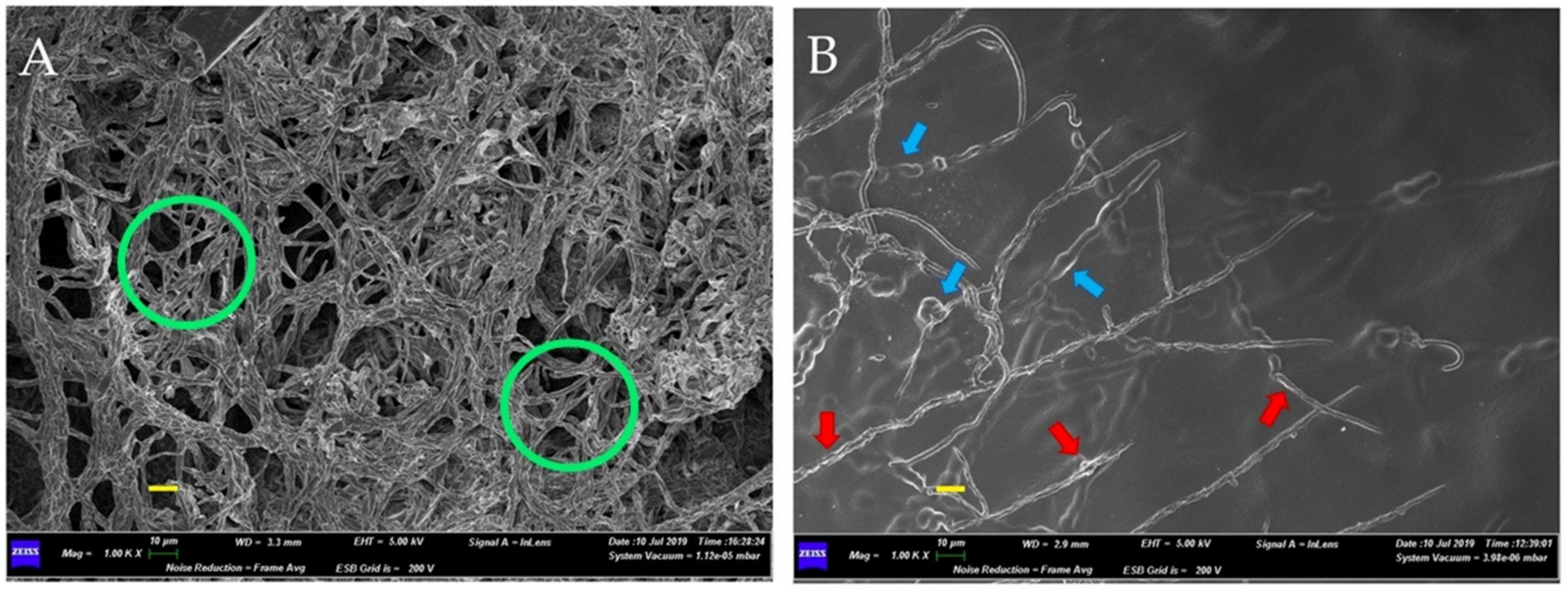
2.3. Bacterial Effects on Fungal Mycelium
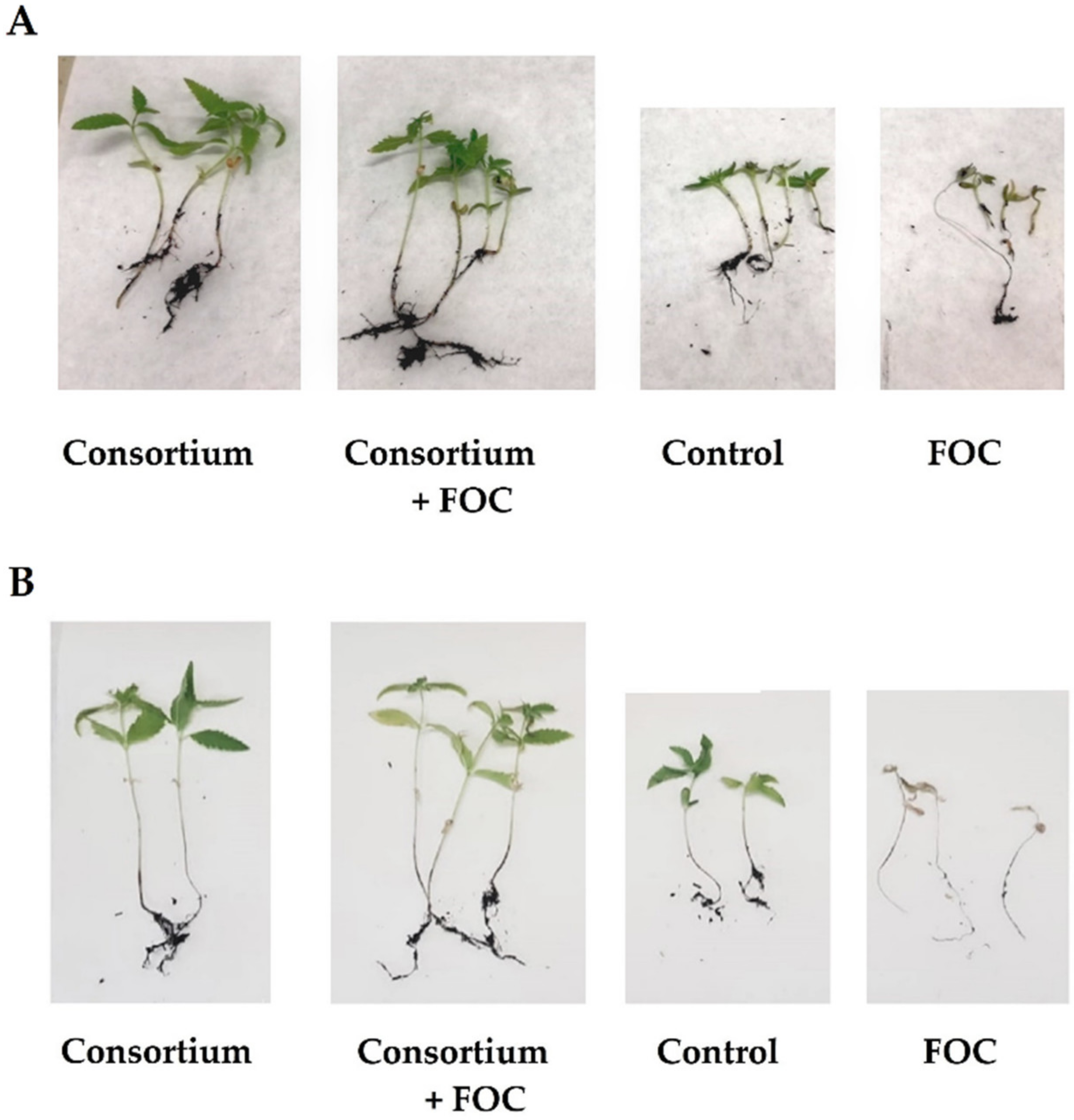
2.4. In Planta Biocontrol
3. Discussion
4. Materials and Methods
4.1. Fungal Strain Isolation and Growth Conditions
4.2. Fungal Strain Molecular Identification
4.3. Phylogenetic Analysis
4.4. Fungal Strain Formae Specialis Identification
4.5. Bacterial Strains and Growth Conditions
4.6. In Vitro Biocontrol Activity
4.7. Bacterial Consortium–Pathogen Interaction
4.8. In Planta Biocontrol Activity
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amaducci, S.; Gusovius, H.-J. Hemp—Cultivation, Extraction and Processing. In Industrial Applications of Natural Fibres; John Wiley & Sons, Ltd.: Chichester, UK, 2010; pp. 109–134. [Google Scholar]
- Aluko, R.E. Hemp Seed (Cannabis sativa L.) Proteins. In Sustainable Protein Sources; Elsevier: Amsterdam, The Netherlands, 2017; pp. 121–132. [Google Scholar]
- Fike, J. Industrial Hemp: Renewed Opportunities for an Ancient Crop. CRC Crit. Rev. Plant Sci. 2016, 35, 406–424. [Google Scholar] [CrossRef]
- Matteucci, F.; Saggio, A.; Terreri, M.; Fantozzi, D.; Servo, E.; Spera, D.M.; Del Gallo, M. Influence of Cannabis sativa cultivation on the soil microbial community in the Fucino plateau. In Proceedings of the Suoli di Qualità per una Vita di Qualità, Rome, Italy, 1–3 December 2015; Lo Papa, G., Benedetti, A., Eds.; SISS—Società Italiana della Scienza del Suolo: Florence, Italy, 2016; pp. 54–60. [Google Scholar]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Paulitz, T.C. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef] [Green Version]
- McPartland, J.M.; Clarke, R.C.; Watson, D.P. Hemp Diseases and Pests: Management and Biological Control—An Advanced Treatise; CABI: Wallingford, UK, 2000; ISBN 9780851994543. [Google Scholar]
- Noviello, C.; Snyder, W.C. Fusarium wilt of Hemp. Phytopathology 1962, 52, 1315–1317. [Google Scholar]
- Tan, T.; Zhu, J.; Shen, A.; Li, J.; Yu, Y.; Zhang, M. Isolation and identification of a Bacillus subtilis HZ-72 exhibiting biocontrol activity against flax seedling blight. Eur. J. Plant Pathol. 2019, 153, 825–836. [Google Scholar] [CrossRef]
- Khoshru, B.; Mitra, D.; Khoshmanzar, E.; Myo, E.M.; Uniyal, N.; Mahakur, B.; Das Mohapatra, P.K.; Panneerselvam, P.; Boutaj, H.; Alizadeh, M.; et al. Current scenario and future prospects of plant growth-promoting rhizobacteria: An economic valuable resource for the agriculture revival under stressful conditions. J. Plant Nutr. 2020, 43, 3062–3092. [Google Scholar] [CrossRef]
- Pellegrini, M.; Ercole, C.; Di Zio, C.; Matteucci, F.; Pace, L.; Del Gallo, M. In vitro and in planta antagonistic effects of plant growth-promoting rhizobacteria consortium against soilborne plant pathogens of Solanum tuberosum and Solanum lycopersicum. FEMS Microbiol. Lett. 2020, 367, fna099. [Google Scholar] [CrossRef]
- Pagnani, G.; Pellegrini, M.; Galieni, A.; D’Egidio, S.; Matteucci, F.; Ricci, A.; Stagnari, F.; Sergi, M.; Lo Sterzo, C.; Pisante, M.; et al. Plant growth-promoting rhizobacteria (PGPR) in Cannabis sativa ‘Finola’ cultivation: An alternative fertilization strategy to improve plant growth and quality characteristics. Ind. Crops Prod. 2018, 123, 75–83. [Google Scholar] [CrossRef]
- Groenhagen, U.; Baumgartner, R.; Bailly, A.; Gardiner, A.; Eberl, L.; Schulz, S.; Weisskopf, L. Production of Bioactive Volatiles by Different Burkholderia ambifaria Strains. J. Chem. Ecol. 2013, 39, 892–906. [Google Scholar] [CrossRef] [PubMed]
- Logeshwarn, P.; Thangaraju, M.; Rajasundari, K. Archives of Phytopathology and Plant Protection Antagonistic potential of Gluconacetobacter diazotrophicus against Fusarium oxysporum in sweet potato (Ipomea batatus). Arch. Phytopathol. Plant Prot. 2011, 44, 216–223. [Google Scholar] [CrossRef]
- Do Amaral, F.P.; Bueno, J.C.F.; Hermes, V.S.; Arisi, A.C.M. Gene expression analysis of maize seedlings (DKB240 variety) inoculated with plant growth promoting bacterium Herbaspirillum seropedicae. Symbiosis 2014, 62, 41–50. [Google Scholar] [CrossRef]
- Bashan, Y.; De-Bashan, L.E. How the Plant Growth-Promoting Bacterium Azospirillum Promotes Plant Growth—A Critical Assessment. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2010; pp. 77–136. [Google Scholar]
- Leslie, J.; Summerell, B. The Fusarium Laboratory Manual; Blackwell Publishing: Ames, IA, USA, 2006. [Google Scholar]
- McPartland, J.M.; Hillig, K.W. Cannabis clinic Fusarium Wilt. J. Ind. Hemp 2004, 9, 67–77. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 871, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonetti, A.E.; Roberts, I.N.; Marcela, S.; Gutierrez-Boem, F.H.; Gomez, F.M.; Ruiz, J.A. A novel Burkholderia ambifaria strain able to degrade the mycotoxin fusaric acid and to inhibit Fusarium spp. growth. Microbiol. Res. 2018, 206, 50–59. [Google Scholar] [CrossRef]
- Lu, S.-E.; Novak, J.; Austin, F.W.; Gu, G.; Ellis, D.; Kirk, M.; Wilson-Stanford, S.; Tonelli, M.; Smith, L. Occidiofungin, a Unique Antifungal Glycopeptide Produced by a Strain of Burkholderia contaminans. Biochemistry 2009, 48, 8312–8321. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, S.; Blom, J.F.; Pernthaler, J.; Berg, G.; Baldwin, A.; Mahenthiralingam, E.; Eberl, L. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ. Microbiol. 2009, 11, 1422–1437. [Google Scholar] [CrossRef]
- Tawfik, K.A.; Jeffs, P.; Bray, B.; Dubay, G.; Falkinham, J.O.; Mesbah, M.; Youssef, D.; Khalifa, S.; Schmidt, E.W. Burkholdines 1097 and 1229, Potent Antifungal Peptides from Burkholderia ambifaria 2.2N. Org. Lett. 2010, 12, 664–666. [Google Scholar] [CrossRef] [Green Version]
- Mehnaz, S.; Lazarovits, G. Inoculation Effects of Pseudomonas putida, Gluconacetobacter azotocaptans, and Azospirillum lipoferum on Corn Plant Growth Under Greenhouse Conditions. Microb. Ecol. 2006, 51, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Weber, O.B.; Muniz, C.R.; Vitor, A.O. Interaction of endophytic diazotrophic bacteria and Fusarium oxysporum f. sp. cubense on plantlets of banana ‘Maça’. Plant Soil 2007, 298, 47–56. [Google Scholar] [CrossRef]
- Brusamarello-Santos, L.C.C.; Pacheco, F.; Aljanabi, S.M.M.; Monteiro, R.A.; Cruz, L.M.; Baura, V.A.; Pedrosa, F.O.; Souza, E.M.; Wassem, R. Differential gene expression of rice roots inoculated with the diazotroph Herbaspirillum seropedicae. Plant Soil 2012, 356, 113–125. [Google Scholar] [CrossRef]
- Rosconi, F.; Davyt, D.; Martínez, V.; Martínez, M.; Abin-Carriquiry, J.A.; Zane, H.; Butler, A.; de Souza, E.M.; Fabiano, E. Identification and structural characterization of serobactins, a suite of lipopeptide siderophores produced by the grass endophyte Herbaspirillum seropedicae. Environ. Microbiol. 2013, 15, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Rosconi, F.; Trovero, M.F.; de Souza, E.M.; Fabiano, E. Serobactins-mediated iron acquisition systems optimize competitive fitness of H erbaspirillum seropedicae inside rice plants. Environ. Microbiol. 2016, 18, 2523–2533. [Google Scholar] [CrossRef]
- Heydari, A.; Pessarakli, M. A Review on Biological Control of Fungal Plant Pathogens Using Microbial Antagonists. J. Biol. Sci. 2010, 10, 273–290. [Google Scholar] [CrossRef] [Green Version]
- Frey-Klett, P.; Burlinson, P.; Deveau, A.; Barret, M.; Tarkka, M.; Sarniguet, A. Bacterial-Fungal Interactions: Hyphens between Agricultural, Clinical, Environmental, and Food Microbiologists. Microbiol. Mol. Biol. Rev. 2011, 75, 583–609. [Google Scholar] [CrossRef] [Green Version]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of Action of Microbial Biological Control Agents Against Plant Diseases: Relevance Beyond Efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef] [Green Version]
- Santoyo, G.; Guzmán-Guzmán, P.; Parra-Cota, F.I.; de los Santos-Villalobos, S.; Orozco-Mosqueda, M.D.C.; Glick, B.R. Plant Growth Stimulation by Microbial Consortia. Agronomy 2021, 11, 219. [Google Scholar] [CrossRef]
- Pellegrini, M.; Pagnani, G.; Rossi, M.; D’Egidio, S.; Del Gallo, M.; Forni, C. Daucus carota L. Seed Inoculation with a Consortium of Bacteria Improves Plant Growth, Soil Fertility Status and Microbial Community. Appl. Sci. 2021, 11, 3274. [Google Scholar] [CrossRef]
- Mateo-Sagasta, J.; Marjani, S.; Turral, H.; Burke, J. Water Pollution from Agriculture: A Global Review Executive Summary; The Food and Agriculture Organization of the United Nations: Rome, Italy, 2017. [Google Scholar]
- Weaver, M.A.; Lyn, M.E.; Boyette, C.D.; Hoagland, R.E. Bioherbicides for Weed Control. In Non-Chemical Weed Management; Updhyaya, M.K., Blackshaw, R., Eds.; CABI, International: Cambridge, MA, USA, 2007; pp. 93–110. [Google Scholar]
- O’Donnell, K.; Ward, T.J.; Robert, V.A.R.G.; Crous, P.W.; Geiser, D.M.; Kang, S. DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica 2015, 43, 583–595. [Google Scholar] [CrossRef] [Green Version]
- Khan, P.; Bora, L.C.; Borah, P.K. Management of lettuce rot caused by Fusarium oxysporum f. sp. lactucae in hydroponically grown crop using microbial. Indian Phytopathol. 2017, 70, 232–237. [Google Scholar] [CrossRef] [Green Version]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A.; Ludwig, T.; Meier, H. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 2005, 21, 456–463. [Google Scholar] [CrossRef] [Green Version]
- Botta, A.L.; Santacecilia, A.; Ercole, C.; Cacchio, P.; Del Gallo, M. In vitro and in vivo inoculation of four endophytic bacteria on Lycopersicon esculentum. New Biotechnol. 2013, 30, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Pace, L.; Pellegrini, M.; Palmieri, S.; Rocchi, R.; Lippa, L.; Del Gallo, M. Plant growth-promoting rhizobacteria for in vitro and ex vitro performance enhancement of Apennines’ Genepì (Artemisia umbelliformis subsp. eriantha), an endangered phytotherapeutic plant. Vitr. Cell. Dev. Biol. Plant 2020, 56, 134–142. [Google Scholar] [CrossRef]

| Strains | Inhibition (%) |
|---|---|
| Azospirillum brasilense | <20% |
| Burkholderia ambifaria | 65.0 a |
| Gluconacetobacter diazotrophicus | 64.1 a |
| Herbaspirillum seropedicae | 66.9 a |
| Consortium | 70.6 a |
| LSD | 6.7 |
| Germination | Cg | Damages | Cg | Roots | Cg | Shoots | Cg | Leaves | Cg | Chl tot | Cg | Chl a/b Ratio | Cg | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Consortium | Pre | 100 a | A | - | - | 3.3 c | A | 8.7 a | A | 4.3 c | A | 2.23 a | A | 4.99 d | AB |
| Post | 100 a | - | 6.0 a | 6.5 c | 9.5 a | 0.49 d | 5.43 c | ||||||||
| Control | Pre | 100 a | A | - | - | 2.1 e | C | 4.0 e | C | 3.5 cd | B | 1.11 c | C | 3.26 f | B |
| Post | 100 a | - | 3.6 c | 4.2 e | 5.5 b | 0.20 f | 6.70 a | ||||||||
| Consortium + FOC | Pre | 89 b | B | 2 c | B | 2.6 d | B | 7.6 b | B | 4.0 c | B | 1.78 b | B | 4.61 de | A |
| Post | 85 c | 2 b | 5.2 b | 5.5 d | 6.0 b | 0.21 f | 6.12 b | ||||||||
| FOC | Pre | 55 d | C | 5 a | A | 1.4 f | D | 2.2 g | D | 1.5 e | C | 0.43 e | D | 1.02 g | C |
| Post | 42 e | 5 a | 2.8 d | 3.0 f | 2.8 d | 0.02 g | 4.36 e | ||||||||
| LSD Condition | 1.4 | 0.3 | 0.2 | 0.4 | 0.6 | 0.04 | 0.31 | ||||||||
| LSD Trial | 0.9 * | 0.2 ns | 0.2 * | 0.3 * | 0.4 * | 0.03 * | 0.22 * | ||||||||
| LSD Condition x Trial | 1.9 | 0.4 | 0.3 | 0.5 | 0.9 | 0.06 | 0.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pellegrini, M.; Ercole, C.; Gianchino, C.; Bernardi, M.; Pace, L.; Del Gallo, M. Fusarium Oxysporum f. sp. Cannabis Isolated from Cannabis Sativa L.: In Vitro and In Planta Biocontrol by a Plant Growth Promoting-Bacteria Consortium. Plants 2021, 10, 2436. https://doi.org/10.3390/plants10112436
Pellegrini M, Ercole C, Gianchino C, Bernardi M, Pace L, Del Gallo M. Fusarium Oxysporum f. sp. Cannabis Isolated from Cannabis Sativa L.: In Vitro and In Planta Biocontrol by a Plant Growth Promoting-Bacteria Consortium. Plants. 2021; 10(11):2436. https://doi.org/10.3390/plants10112436
Chicago/Turabian StylePellegrini, Marika, Claudia Ercole, Carmelo Gianchino, Matteo Bernardi, Loretta Pace, and Maddalena Del Gallo. 2021. "Fusarium Oxysporum f. sp. Cannabis Isolated from Cannabis Sativa L.: In Vitro and In Planta Biocontrol by a Plant Growth Promoting-Bacteria Consortium" Plants 10, no. 11: 2436. https://doi.org/10.3390/plants10112436
APA StylePellegrini, M., Ercole, C., Gianchino, C., Bernardi, M., Pace, L., & Del Gallo, M. (2021). Fusarium Oxysporum f. sp. Cannabis Isolated from Cannabis Sativa L.: In Vitro and In Planta Biocontrol by a Plant Growth Promoting-Bacteria Consortium. Plants, 10(11), 2436. https://doi.org/10.3390/plants10112436









