Nanopore-Based Comparative Transcriptome Analysis Reveals the Potential Mechanism of High-Temperature Tolerance in Cotton (Gossypium hirsutum L.)
Abstract
1. Introduction
2. Results
2.1. Overview of ONT Sequencing
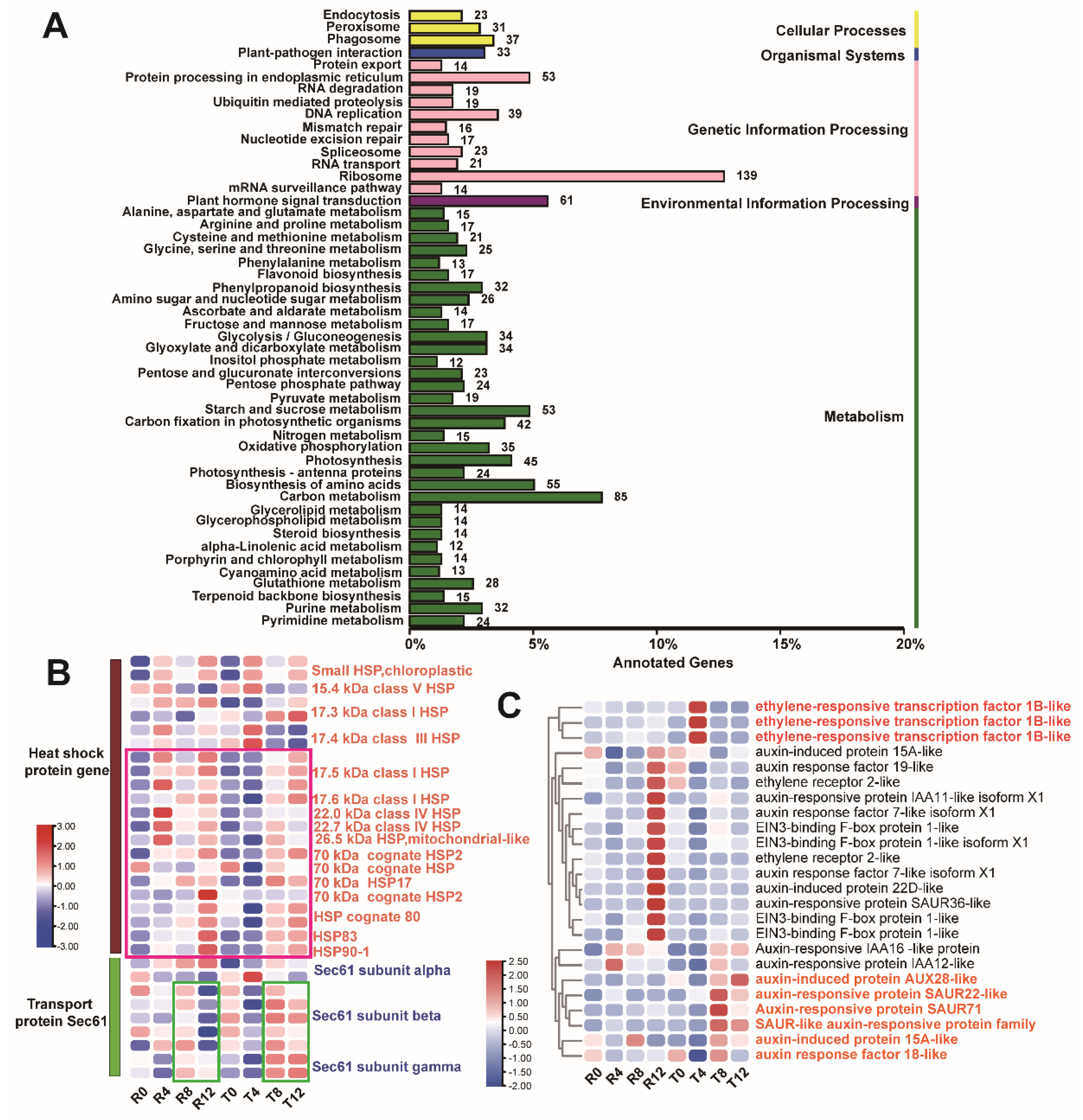
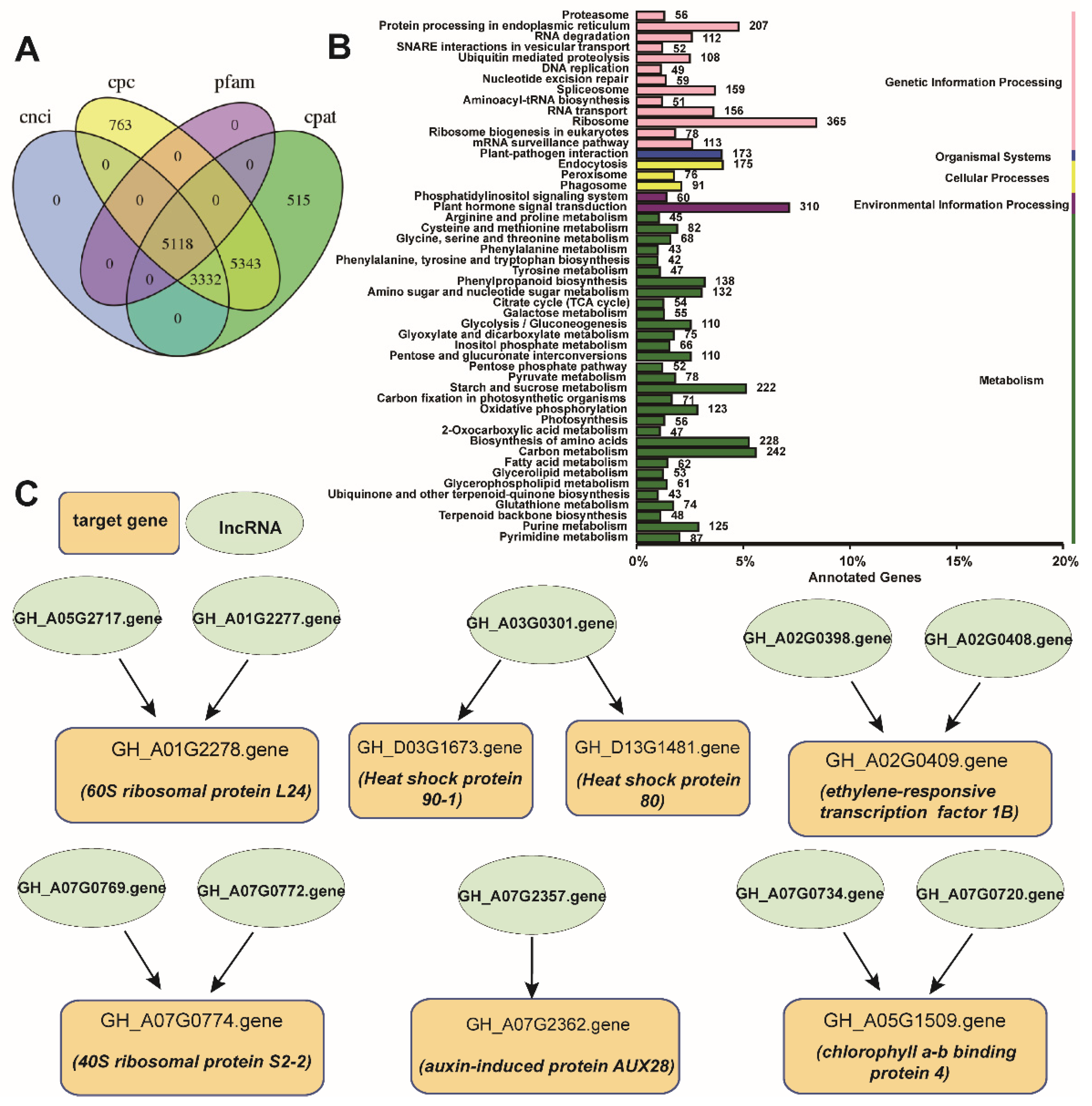
2.2. Enrichment Analysis of All the DEGs
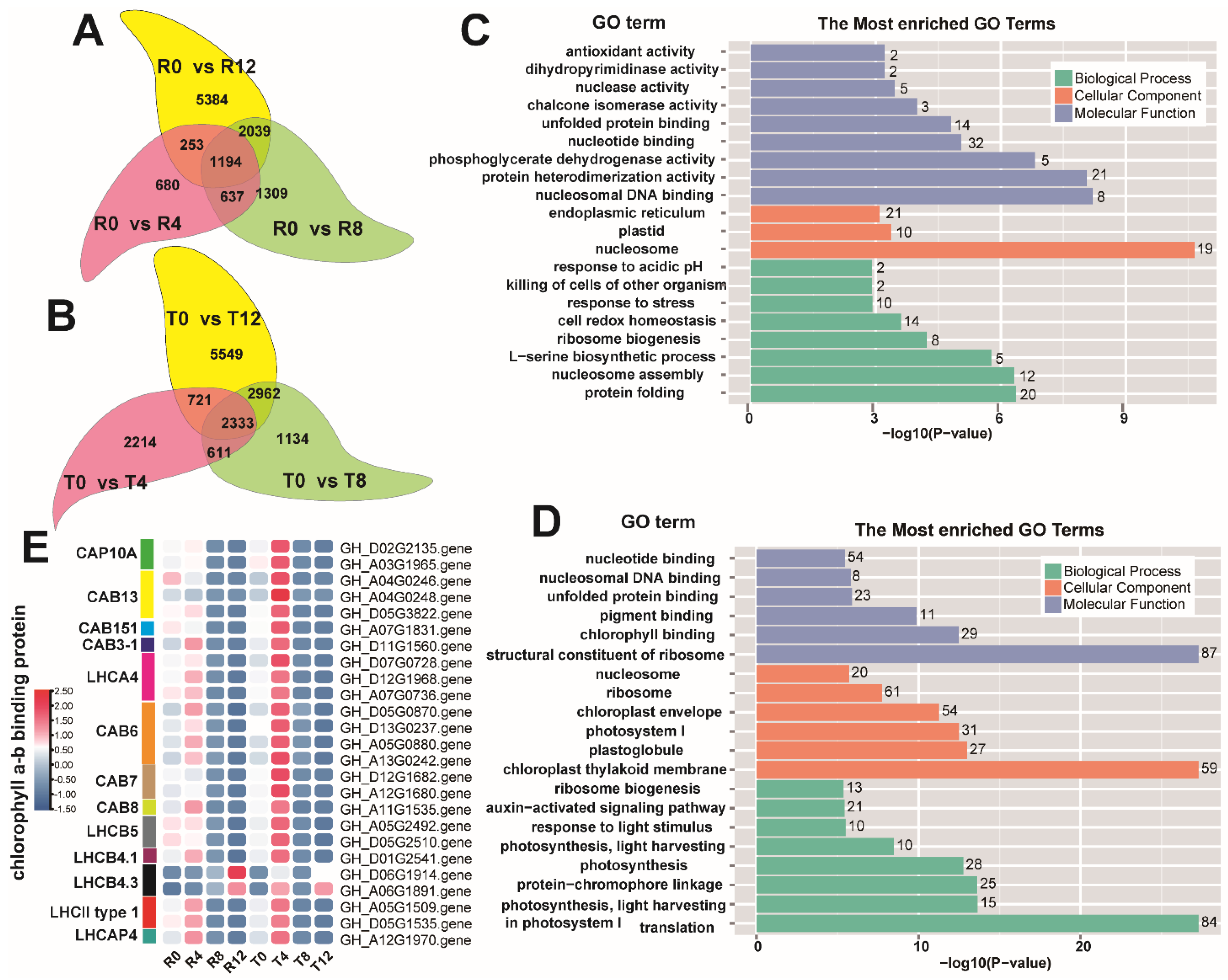
2.3. DEG Analysis between the Sensitive and Tolerant Types
2.4. Differential Analysis of DEG Enrichment in the Sensitive and Tolerant Types
2.5. LncRNA and Target Gene Prediction
2.6. WGCNA of the Screened DEGs
2.7. Validation of the Transcriptome Data via RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Library Preparation and ONT Sequencing
4.3. Functional Annotation of Genes
4.4. Quantification of Gene Expression Levels and Differential Expression Analysis
4.5. Prediction of lncRNAs and Their Target Genes
4.6. Weighted Gene Correlation Network Analysis (WGCNA)
4.7. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Frölicher, T.L.; Fischer, E.; Gruber, N. Marine heatwaves under global warming. Nature 2018, 560, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Yang, X.; Huang, G. Development of an integrated multivariate trend-frequency analysis method: Spatial-temporal characteristics of climate extremes under global warming for Central Asia. Environ. Res. 2021, 195, 110859. [Google Scholar] [CrossRef] [PubMed]
- De Lima, C.F.F.; Kleine-Vehn, J.; De Smet, I.; Feraru, E. Getting to the root of belowground high temperature responses in plants. J. Exp. Bot. 2021, 202. [Google Scholar] [CrossRef]
- Echer, F.; Oosterhuis, D.M.; Loka, D.A.; Rosolem, C.A. High Night Temperatures During the Floral Bud Stage Increase the Abscission of Reproductive Structures in Cotton. J. Agron. Crop. Sci. 2014, 200, 191–198. [Google Scholar] [CrossRef]
- Song, G.; Wang, M.; Zeng, B.; Zhang, J.; Jiang, C.; Hu, Q.; Geng, G.; Tang, C. Anther response to high-temperature stress during development and pollen thermotolerance heterosis as revealed by pollen tube growth and in vitro pollen vigor analysis in upland cotton. Planta 2015, 241, 1271–1285. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ma, Y.; Liu, N.; Xu, J.; Hu, Q.; Li, Y.; Wu, Y.; Xie, S.; Zhu, L.; Min, L.; et al. microRNAs involved in auxin signalling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J. 2017, 91, 977–994. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, N.; Yang, X.; Tu, L.; Zhang, X. Small RNA-mediated responses to low- and high-temperature stresses in cotton. Sci. Rep. 2016, 6, 35558. [Google Scholar] [CrossRef]
- Chen, J.; Pan, A.; He, S.; Su, P.; Yuan, X.; Zhu, S.; Liu, Z. Different MicroRNA Families Involved in Regulating High Temperature Stress Response during Cotton (Gossypium hirsutum L.) Anther Development. Int. J. Mol. Sci. 2020, 21, 1280. [Google Scholar] [CrossRef]
- Cheng, G.; Zhang, L.; Wei, H.; Wang, H.; Lu, J.; Yu, S. Transcriptome Analysis Reveals a Gene Expression Pattern Associated with Fuzz Fiber Initiation Induced by High Temperature in Gossypium barbadense. Genes 2020, 11, 1066. [Google Scholar] [CrossRef]
- Shan, Q.; Ma, F.; Wei, J.; Li, H.; Ma, H.; Sun, P. Physiological Functions of Heat Shock Proteins. Curr. Protein Pept. Sci. 2020, 21, 751–760. [Google Scholar] [CrossRef]
- Haq, S.U.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.-X.; Zhang, H.-X.; Wei, A.-M.; Gong, Z.-H. Heat Shock Proteins: Dynamic Biomolecules to Counter Plant Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef]
- Heikema, A.; Horst-Kreft, D.; Boers, S.; Jansen, R.; Hiltemann, S.; De Koning, W.; Kraaij, R.; De Ridder, M.; Van Houten, C.; Bont, L.; et al. Comparison of Illumina versus Nanopore 16S rRNA Gene Sequencing of the Human Nasal Microbiota. Genes 2020, 11, 1105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Su, L.; Wang, Y.; Deng, S. Improved High-Throughput Sequencing of the Human Oral Microbiome: From Illumina to PacBio. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 1–13. [Google Scholar] [CrossRef]
- Cui, J.; Shen, N.; Lu, Z.; Xu, G.; Wang, Y.; Jin, B. Analysis and comprehensive comparison of PacBio and nanopore-based RNA sequencing of the Arabidopsis transcriptome. Plant Methods 2020, 16, 1–13. [Google Scholar] [CrossRef]
- Hou, C.; Lian, H.; Cai, Y.; Wang, Y.; Liang, D.; He, B. Comparative Analyses of Full-Length Transcriptomes Reveal Gnetum luofuense Stem Developmental Dynamics. Front. Genet. 2021, 12, 615284. [Google Scholar] [CrossRef]
- Hou, C.; Tian, Y.; Wang, Y.; Lian, H.; Liang, D.; Shi, S.; Deng, N.; He, B. Revealing the developmental dynamics in male strobilus transcriptome of Gnetum luofuense using nanopore sequencing technology. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Kirov, I.; Omarov, M.; Merkulov, P.; Dudnikov, M.; Gvaramiya, S.; Kolganova, E.; Komakhin, R.; Karlov, G.; Soloviev, A. Genomic and Transcriptomic Survey Provides New Insight into the Organization and Transposition Activity of Highly Expressed LTR Retrotransposons of Sunflower (Helianthus annuus L.). Int. J. Mol. Sci. 2020, 21, 9331. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, Z.; Pei, H.; Guo, Z.; Wen, D.; Liu, R.; Song, B. Transcriptome profiling analysis of tea plant (Camellia sinensis) using Oxford Nanopore long-read RNA-Seq technology. Gene 2021, 769, 145247. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Lye, Z.N.; Groen, S.C.; Dai, X.; Rughani, P.; Zaaijer, S.; Harrington, E.D.; Juul, S.; Purugganan, M.D. Nanopore sequencing-based genome assembly and evolutionary genomics of circum-basmati rice. Genome Biol. 2020, 21, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Min, L.; Wang, J.; Li, Y.; Wu, Y.; Hu, Q.; Ding, Y.; Wang, M.; Liang, Y.; Gong, Z.; et al. A combination of genome-wide and transcriptome-wide association studies reveals genetic elements leading to male sterility during high temperature stress in cotton. New Phytol. 2021, 231, 165–181. [Google Scholar] [CrossRef] [PubMed]
- Nissen, P.; Hansen, J.; Ban, N.; Moore, P.B.; Steitz, T.A. The Structural Basis of Ribosome Activity in Peptide Bond Synthesis. Science 2000, 289, 920–930. [Google Scholar] [CrossRef]
- Kressler, D.; Hurt, E.; Baßler, J. A Puzzle of Life: Crafting Ribosomal Subunits. Trends Biochem. Sci. 2017, 42, 640–654. [Google Scholar] [CrossRef]
- Sáez-Vásquez, J.; Delseny, M. Ribosome Biogenesis in Plants: From Functional 45S Ribosomal DNA Organization to Ribosome Assembly Factors. Plant Cell 2019, 31, 1945–1967. [Google Scholar] [CrossRef]
- Horiguchi, G.; Mollá-Morales, A.; Pérez-Pérez, J.M.; Kojima, K.; Robles, P.; Ponce, M.R.; Micol, J.L.; Tsukaya, H. Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 2011, 65, 724–736. [Google Scholar] [CrossRef]
- Zhang, H.; Luo, M.; Day, R.C.; Talbot, M.J.; Ivanova, A.; Ashton, A.R.; Chaudhury, A.M.; Macknight, R.C.; Hrmova, M.; Koltunow, A.M. Developmentally regulated HEART STOPPER, a mitochondrially targeted L18 ribosomal protein gene, is required for cell division, differentiation, and seed development in Arabidopsis. J. Exp. Bot. 2015, 66, 5867–5880. [Google Scholar] [CrossRef]
- Kang, C.H.; Lee, Y.M.; Park, J.H.; Nawkar, G.M.; Oh, H.T.; Kim, M.G.; Lee, S.I.; Kim, W.Y.; Yun, D.; Lee, S.Y. Ribosomal P3 protein AtP3B of Arabidopsis acts as both protein and RNA chaperone to increase tolerance of heat and cold stresses. Plant Cell Environ. 2016, 39, 1631–1642. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yuan, H.; Yang, Y.; Fish, T.; Lyi, S.M.; Thannhauser, T.W.; Zhang, L.; Li, L. Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J. Exp. Bot. 2016, 67, 2731–2744. [Google Scholar] [CrossRef]
- Juntawong, P.; Girke, T.; Bazin, J.; Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, E203–E212. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Kurihara, Y.; Iwasaki, S. The Plant Translatome Surveyed by Ribosome Profiling. Plant Cell Physiol. 2019, 60, 1917–1926. [Google Scholar] [CrossRef] [PubMed]
- Mikhaylina, A.O.; Nikonova, E.Y.; Kostareva, O.S.; Piendl, W.; Erlacher, M.; Tishchenko, S.V. Characterization of Regulatory Elements of L11 and L1 Operons in Thermophilic Bacteria and Archaea. Biochemistry 2021, 86, 397–408. [Google Scholar] [CrossRef]
- Swindell, W.R.; Huebner, M.; Weber, A.P. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genom. 2007, 8, 125. [Google Scholar] [CrossRef]
- Neal, S.J.; Karunanithi, S.; Best, A.; So, A.K.-C.; Tanguay, R.M.; Atwood, H.L.; Westwood, J.T. Thermoprotection of synaptic transmission in aDrosophilaheat shock factor mutant is accompanied by increased expression of Hsp83 and DnaJ-1. Physiol. Genom. 2006, 25, 493–501. [Google Scholar] [CrossRef]
- Lee, S.; Lee, D.W.; Lee, Y.; Mayer, U.; Stierhof, Y.-D.; Lee, S.; Jürgens, G.; Hwang, I. Heat Shock Protein Cognate 70-4 and an E3 Ubiquitin Ligase, CHIP, Mediate Plastid-Destined Precursor Degradation through the Ubiquitin-26S Proteasome System in Arabidopsis. Plant Cell 2010, 21, 3984–4001. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Bernard, C.; Van De Cotte, B.; Van Montagu, M.; Verbruggen, N. At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J. 2001, 27, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Tretter, T.; Pereira, F.P.; Ulucan, O.; Helms, V.; Allan, S.; Kalies, K.-U.; Römisch, K. ERAD and protein import defects in a sec61 mutant lacking ER-lumenal loop 7. BMC Cell Biol. 2013, 14, 56. [Google Scholar] [CrossRef] [PubMed]
- Pilon, M.; Schekman, R.; Römisch, K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997, 16, 4540–4548. [Google Scholar] [CrossRef]
- Min, L.; Li, Y.; Hu, Q.; Zhu, L.; Gao, W.; Wu, Y.; Ding, Y.; Liu, S.; Yang, X.; Zhang, X. Sugar and Auxin Signaling Pathways Respond to High-Temperature Stress during Anther Development as Revealed by Transcript Profiling Analysis in Cotton. Plant Physiol. 2014, 164, 1293–1308. [Google Scholar] [CrossRef]
- Müller, M.; Munné-Bosch, S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Yoong, F.-Y.; O’Brien, L.K.; Truco, M.J.; Huo, H.; Sideman, R.; Hayes, R.; Michelmore, R.W.; Bradford, K.J. Genetic Variation for Thermotolerance in Lettuce Seed Germination Is Associated with Temperature-Sensitive Regulation of ETHYLENE RESPONSE FACTOR1 (ERF1). Plant Physiol. 2016, 170, 472–488. [Google Scholar] [CrossRef]
- Majeed, S.; Rana, I.A.; Mubarik, M.S.; Atif, R.M.; Yang, S.-H.; Chung, G.; Jia, Y.; Du, X.; Hinze, L.; Azhar, M.T. Heat Stress in Cotton: A Review on Predicted and Unpredicted Growth-Yield Anomalies and Mitigating Breeding Strategies. Agronomy 2021, 11, 1825. [Google Scholar] [CrossRef]
- Gupta, N.K.; Agarwal, S.; Agarwal, V.P.; Nathawat, N.S.; Gupta, S.; Singh, G. Effect of short-term heat stress on growth, physiology and antioxidative defence system in wheat seedlings. Acta Physiol. Plant. 2013, 35, 1837–1842. [Google Scholar] [CrossRef]
- Lynham, S.; Freile, F.G.; Puri, N.M.; O’Reilly, N.; Mitchell, G.H.; Wells, T.N.C.; Willcox, M.; Beatson, R. Identification of chlorophyll a-b binding protein AB96 as a novel TGFβ1 neutralizing agent. Sci. Rep. 2021, 11, 1–7. [Google Scholar] [CrossRef]
- Spence, A.K.; Boddu, J.; Wang, D.; James, B.; Swaminathan, K.; Moose, S.P.; Long, S.P. Transcriptional responses indicate maintenance of photosynthetic proteins as key to the exceptional chilling tolerance of C4 photosynthesis in Miscanthus × giganteus. J. Exp. Bot. 2014, 65, 3737–3747. [Google Scholar] [CrossRef] [PubMed]
- Zygadlo, A.; Jensen, P.E.; Leister, D.; Scheller, H.V. Photosystem I lacking the PSI-G subunit has a higher affinity for plastocyanin and is sensitive to photodamage. Biochim. Biophys. Acta Bioenerg. 2005, 1708, 154–163. [Google Scholar] [CrossRef][Green Version]
- Yi, X.; Hargett, S.R.; Frankel, L.K.; Bricker, T.M. The PsbQ Protein Is Required in Arabidopsis for Photosystem II Assembly/Stability and Photoautotrophy under Low Light Conditions. J. Biol. Chem. 2006, 281, 26260–26267. [Google Scholar] [CrossRef]
- Jensen, P.E.; Haldrup, A.; Zhang, S.; Scheller, H.V. The PSI-O Subunit of Plant Photosystem I Is Involved in Balancing the Excitation Pressure between the Two Photosystems. J. Biol. Chem. 2004, 279, 24212–24217. [Google Scholar] [CrossRef]
- Do, T.; Qu, Z.; Searle, I. Purification and Functional Analysis of Plant Long Noncoding RNAs (lncRNA). Methods Mol. Biol. 2019, 1933, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cheng, C.; Feng, X.; Lai, R.; Gao, M.; Chen, W.; Wu, R. Integrated analysis of lncRNA and mRNA transcriptomes reveals the potential regulatory role of lncRNA in kiwifruit ripening and softening. Sci. Rep. 2021, 11, 1671. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Chen, P.; Liu, P.; Bu, C.; Zhang, D. High-Temperature-Responsive poplar lncRNAs modulate target gene expression via RNA interference and act as RNA scaffolds to enhance heat tolerance. Int. J. Mol. Sci. 2020, 21, 6808. [Google Scholar] [CrossRef]
- He, X.; Guo, S.; Wang, Y.; Wang, L.; Shu, S.; Sun, J. Systematic identification and analysis of heat-stress-responsive lncRNAs, circRNAs and miRNAs with associated co-expression and ceRNA networks in cucumber ( Cucumis sativus L.). Physiol. Plant. 2020, 168, 736–754. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.E.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.-P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Li, J.; Ma, W.; Zeng, P.; Wang, J.; Geng, B.; Yang, J.; Cui, Q. LncTar: A tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 2015, 16, 806–812. [Google Scholar] [CrossRef]




| Annotation Database. | COG | GO | KEGG | KOG | Pfam | Swissprot | eggNOG | Nr | All |
|---|---|---|---|---|---|---|---|---|---|
| Annotated Number | 25,469 | 55,690 | 26,860 | 40,413 | 58,269 | 53,832 | 69,684 | 78,569 | 78,601 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Y.; Gong, Z.; Wang, J.; Zheng, J.; Ma, Y.; Min, L.; Chen, Q.; Li, Z.; Qu, Y.; Chen, Q.; et al. Nanopore-Based Comparative Transcriptome Analysis Reveals the Potential Mechanism of High-Temperature Tolerance in Cotton (Gossypium hirsutum L.). Plants 2021, 10, 2517. https://doi.org/10.3390/plants10112517
Liang Y, Gong Z, Wang J, Zheng J, Ma Y, Min L, Chen Q, Li Z, Qu Y, Chen Q, et al. Nanopore-Based Comparative Transcriptome Analysis Reveals the Potential Mechanism of High-Temperature Tolerance in Cotton (Gossypium hirsutum L.). Plants. 2021; 10(11):2517. https://doi.org/10.3390/plants10112517
Chicago/Turabian StyleLiang, Yajun, Zhaolong Gong, Junduo Wang, Juyun Zheng, Yizan Ma, Ling Min, Qin Chen, Zhiqiang Li, Yanying Qu, Quanjia Chen, and et al. 2021. "Nanopore-Based Comparative Transcriptome Analysis Reveals the Potential Mechanism of High-Temperature Tolerance in Cotton (Gossypium hirsutum L.)" Plants 10, no. 11: 2517. https://doi.org/10.3390/plants10112517
APA StyleLiang, Y., Gong, Z., Wang, J., Zheng, J., Ma, Y., Min, L., Chen, Q., Li, Z., Qu, Y., Chen, Q., & Li, X. (2021). Nanopore-Based Comparative Transcriptome Analysis Reveals the Potential Mechanism of High-Temperature Tolerance in Cotton (Gossypium hirsutum L.). Plants, 10(11), 2517. https://doi.org/10.3390/plants10112517






