Genetic Diversity and Population Structure of Myanmar Rice (Oryza sativa L.) Varieties Using DArTseq-Based SNP and SilicoDArT Markers
Abstract
:1. Introduction
2. Results
2.1. Marker Quality Analysis
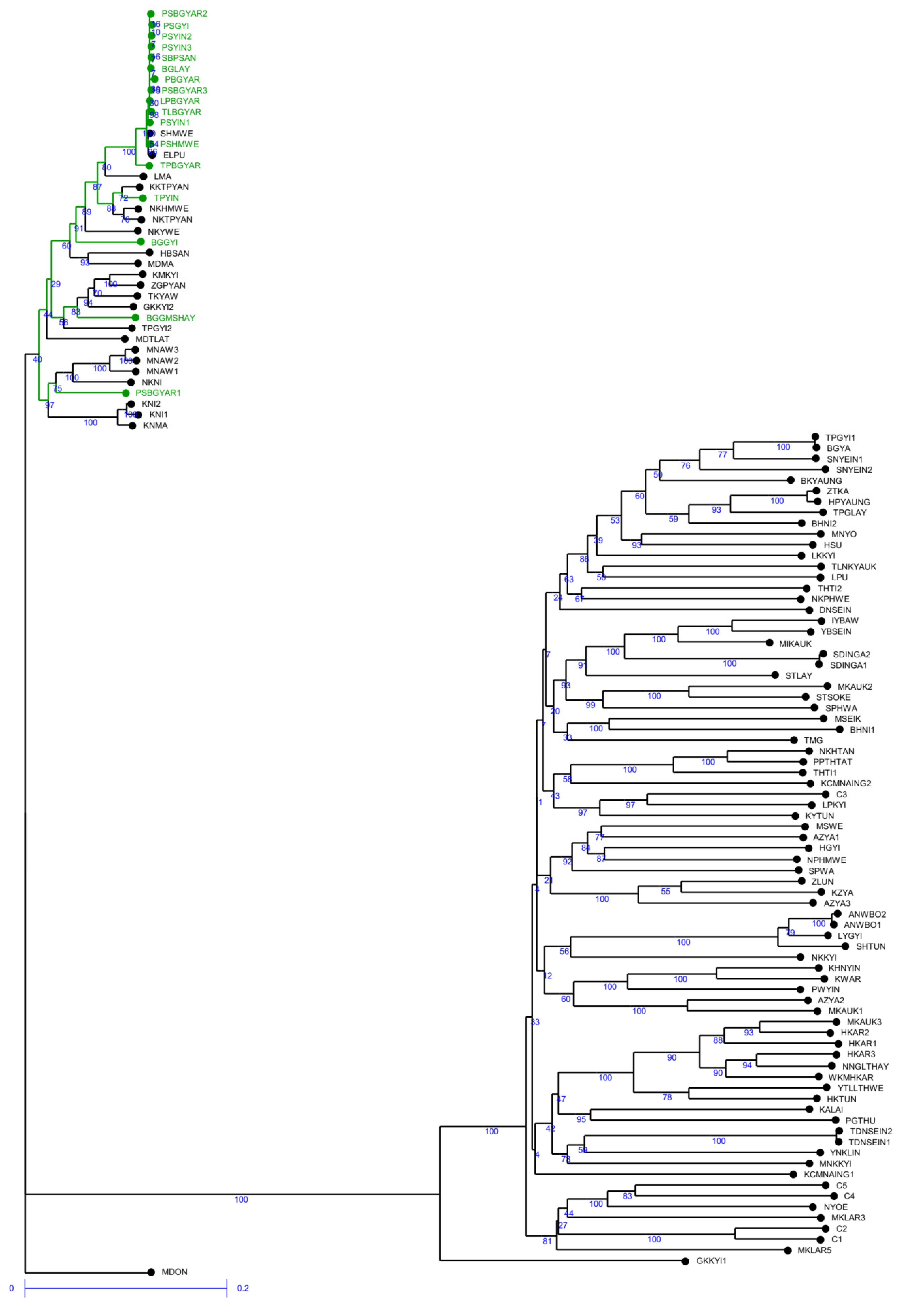
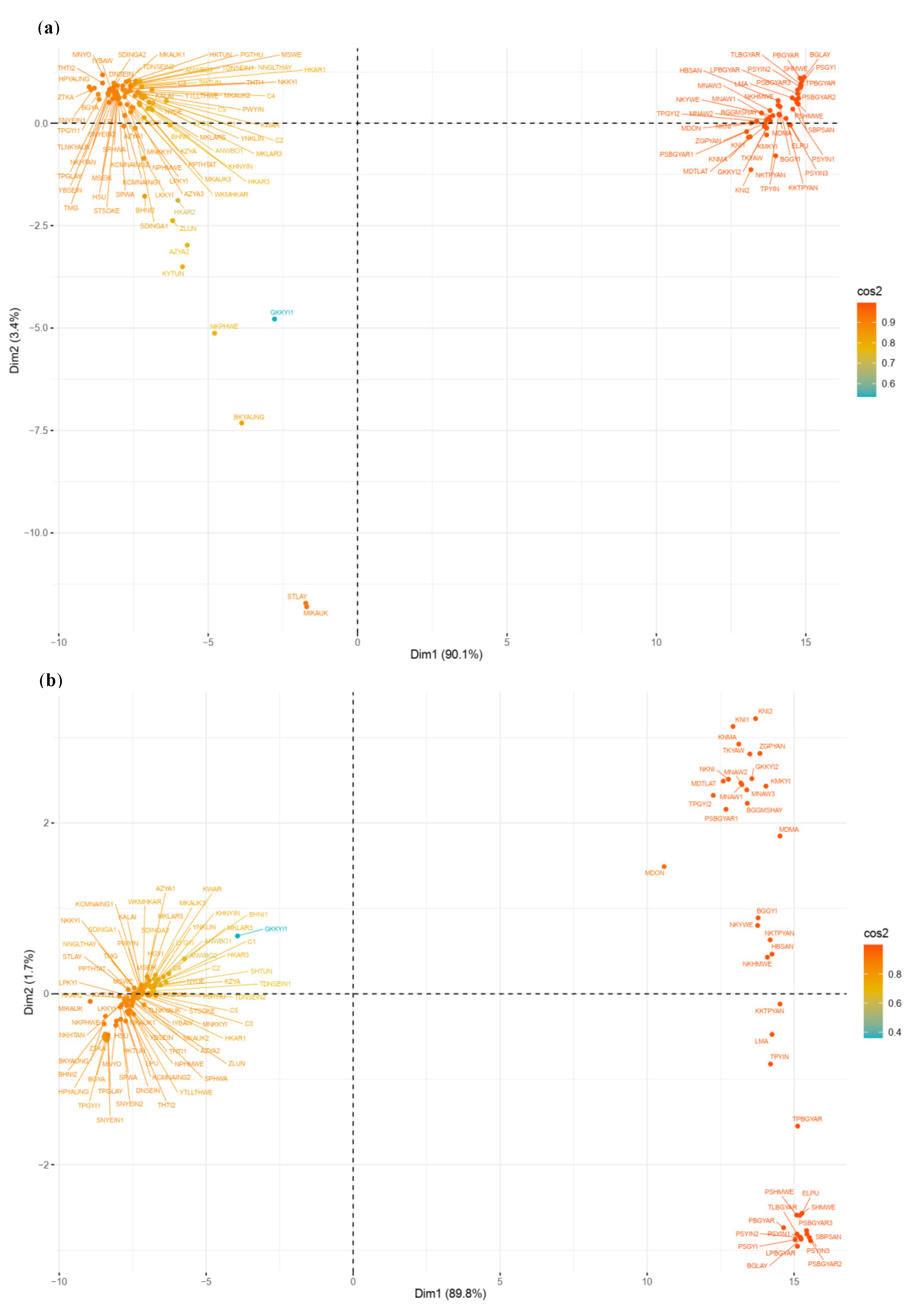
2.2. Genetic Relationships among Genotypes
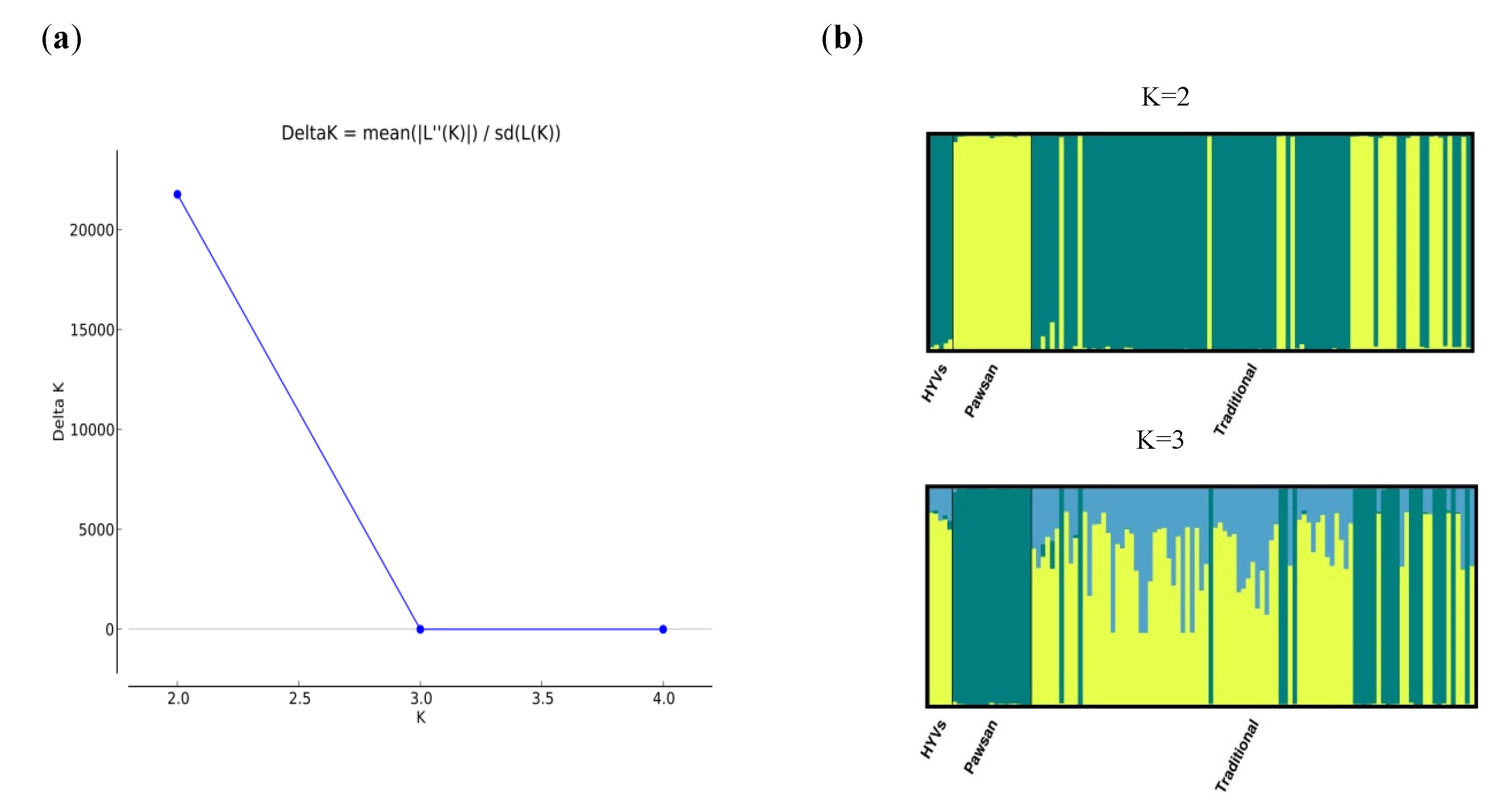
2.3. Population Structure
2.4. Genetic Differentiation of Populations
2.5. Allelic Pattern across Populations
3. Discussion
3.1. Marker Quality Analysis
3.2. Population Structure and Relationships
3.3. Genetic Differentiation of Populations
4. Materials and Methods
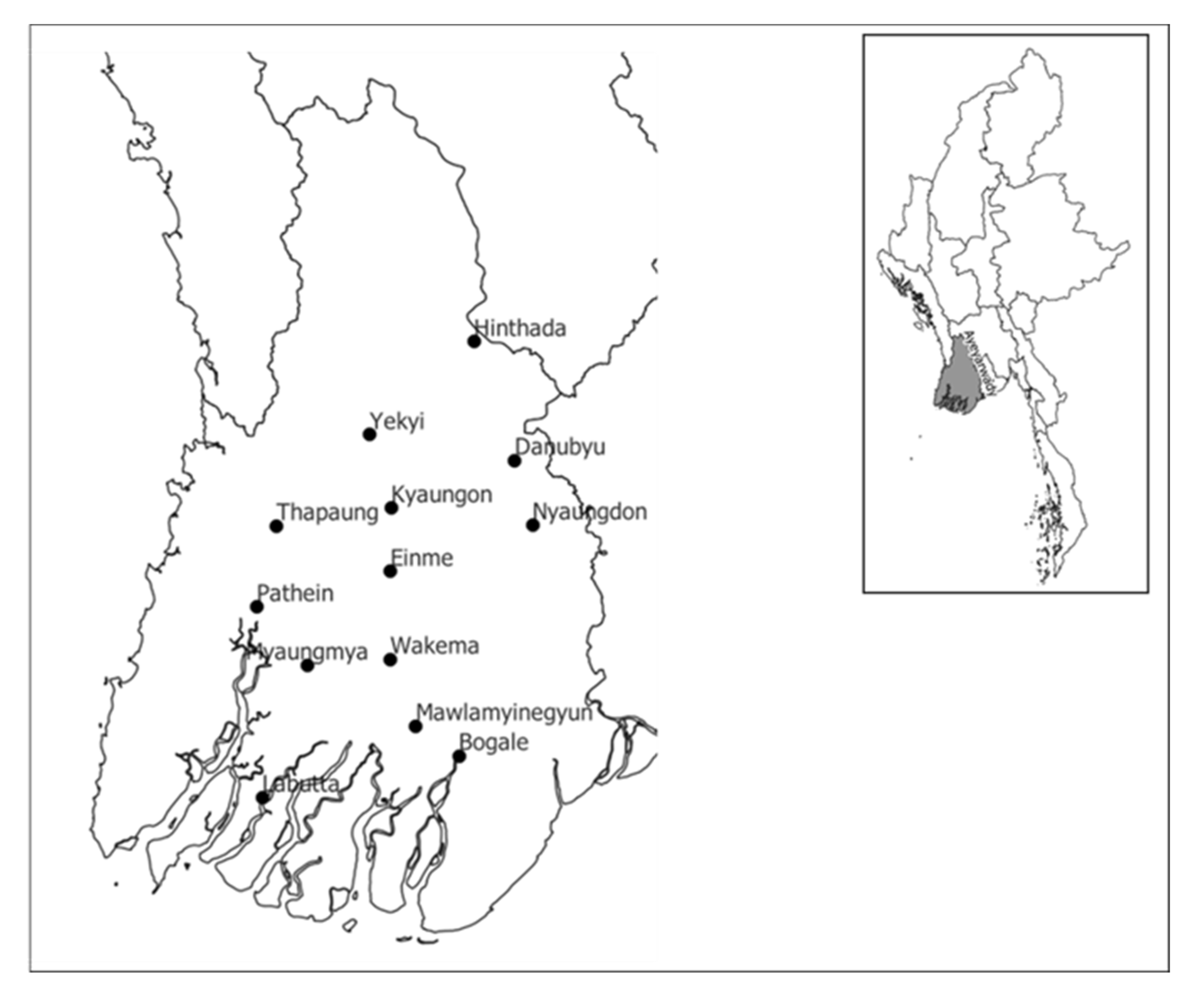
4.1. Plant Materials
4.2. DNA Extraction and Genotyping
4.3. Marker Quality Analysis
4.4. Genetic Relationships among Genotypes and Population Structure
4.5. Analysis of Molecular Variance (AMOVA) and Genetic Diversity Indices
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Xu, Q.; Yuan, X.; Wang, S.; Feng, Y.; Yu, H.; Wang, Y.; Yang, Y.; Wei, X.; Li, X. The genetic diversity and structure of indica rice in China as detected by single nucleotide polymorphism analysis. BMC Genet. 2016, 17, 53. [Google Scholar] [CrossRef] [Green Version]
- Ta, K.N.; Khong, N.G.; Ha, T.L.; Nguyen, D.T.; Mai, D.C.; Hoang, T.G.; Phung, T.P.N.; Bourrie, I.; Courtois, B.; Tran, T.T.H.; et al. A genome-wide association study using a Vietnamese landrace panel of rice (Oryza sativa) reveals new QTLs controlling panicle morphological traits. BMC Plant Biol. 2018, 18, 282. [Google Scholar] [CrossRef]
- FAOSTAT. FAO. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 25 June 2020).
- MoALI (Ministry of Agriculture, Livestock and Irrigation). Myanmar Agriculture Sector in Brief; Ministry of Agriculture, Livestock and Irrigation: Nay Pyi Taw, Myanmar, 2019; pp. 14–16.
- Garcia, A.G.; Ramanatha, R.V.; Sajise, P.E.; Than, T.U. (Eds.) Plant Genetic Resources Conservation and Use in Myanmar. In Proceedings of the National Seminar Workshop on Conservation and Utilization of Plant Genetic Resources in Myanmar for Sustainable Agriculture Development and Food Security, Serdang, Malaysia, 11 October 2002; Myanmar Agriculture Office Headquarters (IPGRI-APO): Yangon, Myanmar, 2003. [Google Scholar]
- Jatoi, S.A.; Fujimura, T.; Yamanaka, S.; Watanabe, J.; Watanabe, K.N. Potential loss of unique genetic diversity in tomato landraces by genetic colonization of modern cultivars at a non-center of origin. Plant Breed. 2008, 127, 189–196. [Google Scholar] [CrossRef]
- Oo, K.S.; An, V.N.; Takeuchi, W.; Lin, S.; Win, Z. Relating the KBDI with sea water intrusion to farm land. In Proceedings of the 33rd Asian Conference on Remote Sensing (ACRS), Bangkok, Thailand, 26–30 November 2012. [Google Scholar]
- Thant, A.A.; Teutscherova, N.; Vazquez, E.; Kalousova, M.; Phyo, A.; Singh, R.K.; Lojka, B. On-farm rice diversity and farmers’ preferences for varietal attributes in Ayeyarwady Delta, Myanmar. J. Crop. Improv. 2020, 34, 549–570. [Google Scholar] [CrossRef]
- Irie, K.; Aye, K.; Myint, Y.Y.; Kha, L.N.; Tun, Y.T.; Nagamine, T.; Fujimaki, H.; Kikuchi, F. Regional variations of grain characters observed in rice landraces of Myanmar. Jpn. J. Trop. Agric. 2004, 48, 101–110. [Google Scholar]
- Thein, M.S.; Aung, H.H.; Aung, Y.W.; San, N.S.; Tin, T.; Lwin, W.W.; Maung, K.K.; Aung, P.P.; Thet, K.M. Variation of some Pawsan rice (Oryza. sativa L.) cultivars of Myanmar using morphological, physiochemical and molecular characters. In Proceedings of the Eighth Agricultural Research Conference in Commemoration of the 90th Anniversary of Yezin Agricultural University, Nay Pyi Taw, Myanmar, 23–24 December 2014. [Google Scholar]
- Myint, P.; Napasintuw, O. Economic Analysis of Paw San Rice Adoption in Myanmar. Asian J. Agric. Res. 2016, 10, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Wunna; Watanabe, K.N.; Ohsawa, R.; Obara, M.; Yanagihara, S.; Aung, P.P.; Yoshimichi, F. Genetic variation of rice (Oryza sativa L.) germplasm in Myanmar based on genomic compositions of DNA markers. Breed. Sci. 2016, 66, 762–767. [Google Scholar] [CrossRef] [Green Version]
- Mogga, M.; Sibiya, J.; Shimelis, H.; Lamo, J.; Yao, N. Diversity analysis and genome-wide association studies of grain shape and eating quality traits in rice (Oryza sativa L.) using DArT markers. PLoS ONE 2018, 13, e0198012. [Google Scholar] [CrossRef] [Green Version]
- Winter, P.; Kahl, G. Molecular marker technologies for plant improvement. World J. Microbiol. Biotechnol. 1995, 11, 438–448. [Google Scholar] [CrossRef]
- Adhikari, S.; Saha, S.; Biswas, A.; Rana, T.S.; Bandyopadhyay, T.K.; Ghosh, P. Application of molecular markers in plant genome analysis: A review. Nucleus 2017, 60, 283–297. [Google Scholar] [CrossRef]
- Saw, O.M.; Doi, K.; Khin, A.; Irie, K.; Yoshimura, A. Genetic Diversity of Myanmar Rice Cultivars Detected by DNA markers. J. Fac. Agric. Kyushu Univ. 2006, 51, 181–187. [Google Scholar] [CrossRef]
- Aung, H.H. Genetic Diversity Assessment of Myanmar Rice (Oryza sativa L.) Varieties Using Morphological Characters and SSR Markers. Master’s Thesis, University of the Philippines, Los Baños (UPLB), Los Baños, Philippines, 2007. [Google Scholar]
- Oo, M.M.; Myint, T.; Wunna Thet, K.M.; Yamanaka, S.; Watanabe, K.N. A study on Myanmar rice varieties from isozyme classification Group-V. Myanmar Academy of Agricultural, Forestry, Livestock and Fishery Sciences. In Proceedings of the Third Agricultural Research Conference, Nay Pyi Taw, Myanmar, 28−29 November 2008; pp. 45–73. [Google Scholar]
- Yamanaka, S.; Jatoi, S.A.; Yi, S.S.; Kothari, S.L.; Htut, T.; Watanabe, K.N. Genetic diversity of Myanmar rice and their implementation on management methods. Afr. J. Biotechnol. 2011, 10, 1290–1298. [Google Scholar]
- Thein, M.S.; Lee, G.A.; Cho, K.T.; Sung, J.S.; Jeong, J.W.; Park, J.H.; Baek, H.J. Assessment of genetic diversity in Meedon rice (Oryza sativa L.) germplasm. J. Korean Soc. Int. Agric. 2012, 24, 232–240. [Google Scholar]
- Rafalski, A. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 2002, 5, 94–100. [Google Scholar] [CrossRef]
- Thant, A.A.; Htun, T.M.; Kalousova, M.; Singh, R.K.; Lojka, B. Microsatellite markers and their application on genetic diversity studies of rice landraces (Oryza sativa L.) in Myanmar—A Review. IJERD 2018, 9, 8–13. [Google Scholar] [CrossRef]
- Kumar, S.; Banks, T.W.; Cloutier, S. SNP Discovery through Next-Generation Sequencing and Its Applications. Int. J. Plant Genom. 2012, 2012, 831460. [Google Scholar] [CrossRef]
- Jaccoud, D.; Peng, K.; Feinstein, D.; Kilian, A. Diversity Arrays: A solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001, 29, e25. [Google Scholar] [CrossRef] [Green Version]
- Kilian, A.; Huttner, E.; Wenzl, P.E.; Jaccoud, D.; Carling, J.; Caig, V.; Evers, M.; Heller-Uszynska, K.A.; Cayla, C.; Patarapuwadol, S.; et al. The Fast and the Cheap: SNP and DArT-based whole genome profiling for crop improvement. In Proceedings of the International Congress in the Wake of the Double Helix: From the Green Revolution to the Gene Revolution, Bologna, Italy, 27–31 May 2003; pp. 443–461. [Google Scholar]
- Adeboye, K.A.; Oyedeji, O.E.; Alqudah, A.M.; Börner, A.; Oduwaye, O.; Adebambo, O.; Daniel, I.O. Genetic structure and diversity of upland rice germplasm using diversity array technology (DArT)-based single nucleotide polymorphism (SNP) markers. Plant Genet. Resour. 2020, 18, 343–350. [Google Scholar] [CrossRef]
- Alam, M.; Neal, J.; O’Connor, K.; Kilian, A.; Topp, B. Ultra-high-throughput DArTseq-based SilicoDArT and SNP markers for genomic studies in Macadamia. PLoS ONE 2018, 13, e0203465. [Google Scholar] [CrossRef] [Green Version]
- Mace, E.S.; Xia, L.; Jordan, D.R.; Halloran, K.; Parh, D.K.; Huttner, E.; Wenzl, P.; Kilian, A. DArT markers: Diversity analyses and mapping in Sorghum bicolor. BMC Genom. 2008, 9, 26. [Google Scholar] [CrossRef] [Green Version]
- Xia, L.; Peng, K.; Yang, S.; Wenzl, P.; De Vicente, M.C.; Fregene, M.; Kilian, A. DArT for high-throughput genotyping of Cassava (Manihot esculenta) and its wild relatives. Theor. Appl. Genet. 2005, 110, 1092–1098. [Google Scholar] [CrossRef] [PubMed]
- Akbari, M.; Wenzl, P.; Caig, V.; Carling, J.; Xia, L.; Yang, S.; Uszynski, G.; Mohler, V.; Lehmensiek, A.; Kuchel, H.; et al. Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor. Appl. Genet. 2006, 113, 1409–1420. [Google Scholar] [CrossRef] [PubMed]
- Salem, K.; Sallam, A. Analysis of population structure and genetic diversity of Egyptian and exotic rice (Oryza sativa L.) genotypes. Comptes Rendus Biol. 2016, 339, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Dierig, D.; Ray, D.T. New crops breeding: Lesquerella. In Oil Crops; Vollmann, J., Rajcan, I., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 507–516. [Google Scholar]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity arrays technology: A generic genome profiling technology on open platforms. Methods Mol. Biol. 2012, 888, 67–89. [Google Scholar] [CrossRef]
- Phung, N.T.P.; Mai, C.D.; Mournet, P.; Frouin, J.; Droc, G.; Ta, N.K.; Jouannic, S.; Lê, L.T.; Do, V.N.; Gantet, P.; et al. Characterization of a panel of Vietnamese rice varieties using DArT and SNP markers for association mapping purposes. BMC Plant Biol. 2014, 14, 371. [Google Scholar] [CrossRef] [Green Version]
- Eltaher, S.; Sallam, A.; Belamkar, V.; Emara, H.A.; Nower, A.A.; Salem, K.F.; Poland, J.; Baenziger, P.S. Genetic diversity and population structure of F3:6 Nebraska winter wheat genotypes using genotyping-by-sequencing. Front. Genet. 2018, 9, 76. [Google Scholar] [CrossRef]
- Myint, T.T.; former deputy director general of Department of Agricultural Research, Nay Pyi Taw, Myanmar. Personal communication, 2021.
- Thant, A.A. Morphological and Genetic Diversity, Agronomic Traits of Indigenous Landraces of Rice (Oryza sativa L.) in Myanmar. Ph.D. Thesis, Czech University of Life Sciences Prague, Prague, Czech Republic, 2021. in preparation. [Google Scholar]
- Khush, G.S.; Brar, D.; Virk, P.S.; Tang, S.X.; Malik, S.S.; Busto, G.A.; Lee, Y.T.; McNally, R.; Trinh, L.N.; Jiang, Y.; et al. Classifying rice germplasm by isozyme polymorphism and origin of cultivated rice. In IRRI Discussion Paper Series No.46; International Rice Research Institute: Los Baños, Philippines, 2003; p. 279. [Google Scholar]
- Wunna. Genetic Diversity of Rice (Oryza sativa L.) Germplasm from Different Agro-Ecological Zones in Myanmar. Ph.D. Thesis, University of Tsukuba, Tsukuba, Japan, 2015. [Google Scholar]
- Wright, S. Evolution and the Genetics of Populations; Volume 4, Variability within and among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1987. [Google Scholar]
- Arora, A.; Kundu, S.; Dilbaghi, N.; Sharma, I.; Tiwari, R. Population structure and genetic diversity among Indian wheat varieties using microsatellite (SSR) markers. Aust. J. Crop Sci. 2014, 8, 1281–1289. [Google Scholar]
- Wright, S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Nuijten, E.; Richards, P. Gene flow in farmers’ field. In Realizing Africa’s Rice Promise; Wopereis, M.C.S., Johnson, D.E., Ahmadi, N., Tollens, E., Jalloh, A., Eds.; CABI Publishing: Wallingford, UK, 2013; pp. 95–107. [Google Scholar]
- Luo, Z.; Brock, J.; Dyer, J.M.; Kutchan, T.; Schachtman, D.; Augustin, M.; Ge, Y.; Fahlgren, N.; Abdel-Haleem, H. Genetic Diversity and Population Structure of a Camelina sativa Spring Panel. Front. Plant Sci. 2019, 10, 184. [Google Scholar] [CrossRef] [Green Version]
- Tsykun, T.; Rellstab, C.; Dutech, C.; Sipos, G.; Prospero, S. Comparative assessment of SSR and SNP markers for inferring the population genetic structure of the common fungus Armillaria cepistipes. Heredity 2017, 119, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Guichoux, E.; Lagache, L.; Wagner, S.; Chaumeil, P.; Léger, P.; Lepais, O.; Lepoittevin, C.; Malausa, T.; Revardel, E.; Salin, F.; et al. Current trends in microsatellite genotyping. Mol. Ecol. Resour. 2011, 11, 591–611. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small amounts of fresh leaf issue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Faleiro, F.G.; Araújo, I.S.; Bahia, R.C.; Santos, R.F.; Yamada, M.M.; Anhert, D. Optimization of DNA extraction and amplification of Theobroma cacao L. aiming obtainment of RAPD markers. Agrotrópica 2002, 14, 31–34. [Google Scholar]
- Barilli, E.; Cobos, M.J.; Carrillo, E.; Kilian, A.; Carling, J.; Rubiales, D. A High-Density Integrated DArTseq SNP-Based Genetic Map of Pisum fulvum and Identification of QTLs Controlling Rust Resistance. Front. Plant Sci. 2018, 9, 167. [Google Scholar] [CrossRef] [Green Version]
- Perrier, X.; Flori, A.; Bonnot, F. Data analysis methods. In Genetic Diversity of Cultivated Tropical Plants; Hamon, P., Seguin, M., Perrier, X., Glaszmann, J.C., Eds.; Enfield, Science Publishers: Montpellier, France, 2003; pp. 43–76. [Google Scholar]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1.0.7. 2017. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 1 April 2020).
- Pritchard, J.K.; Wen, W.; Falush, D. Documentation for Structure Software: Version 2.3; University of Chicago: Chicago, IL, USA, 2010. [Google Scholar]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]

| Population | Inferred Clusters | Mean Fst 1 | Exp. Het. 2 | No. of Genotypes |
|---|---|---|---|---|
| pop1 3 | 0.449 | 0.832 | 0.128 | 40 |
| pop2 4 | 0.551 | 0.687 | 0.169 | 77 |
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among populations | 1 | 209,102.877 | 209,102.877 | 1973.534 | 74 |
| Among individuals | 115 | 148,509.820 | 1291.390 | 585.915 | 22 |
| Within individuals | 117 | 13,988.500 | 119.560 | 119.560 | 4 |
| Total | 233 | 371,601.197 | 2679.009 | 100 | |
| Fst 1 | 0.737 (P = 0.001) | ||||
| Nm 2 | 0.089 | ||||
| Pop | Na 1 | Ne 2 | I 3 | Ho 4 | He 5 | uHe 6 | F 7 | PPL (%) 8 |
|---|---|---|---|---|---|---|---|---|
| pop1 | 1.411 | 1.146 | 0.137 | 0.025 | 0.087 | 0.088 | 0.515 | 41.07 |
| pop2 | 1.797 | 1.358 | 0.327 | 0.035 | 0.214 | 0.215 | 0.776 | 79.73 |
| Mean | 1.604 | 1.252 | 0.232 | 0.030 | 0.150 | 0.152 | 0.688 | 60.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thant, A.A.; Zaw, H.; Kalousova, M.; Singh, R.K.; Lojka, B. Genetic Diversity and Population Structure of Myanmar Rice (Oryza sativa L.) Varieties Using DArTseq-Based SNP and SilicoDArT Markers. Plants 2021, 10, 2564. https://doi.org/10.3390/plants10122564
Thant AA, Zaw H, Kalousova M, Singh RK, Lojka B. Genetic Diversity and Population Structure of Myanmar Rice (Oryza sativa L.) Varieties Using DArTseq-Based SNP and SilicoDArT Markers. Plants. 2021; 10(12):2564. https://doi.org/10.3390/plants10122564
Chicago/Turabian StyleThant, Aye Aye, Hein Zaw, Marie Kalousova, Rakesh Kumar Singh, and Bohdan Lojka. 2021. "Genetic Diversity and Population Structure of Myanmar Rice (Oryza sativa L.) Varieties Using DArTseq-Based SNP and SilicoDArT Markers" Plants 10, no. 12: 2564. https://doi.org/10.3390/plants10122564







