Genes Associated with the Flax Plant Type (Oil or Fiber) Identified Based on Genome and Transcriptome Sequencing Data
Abstract
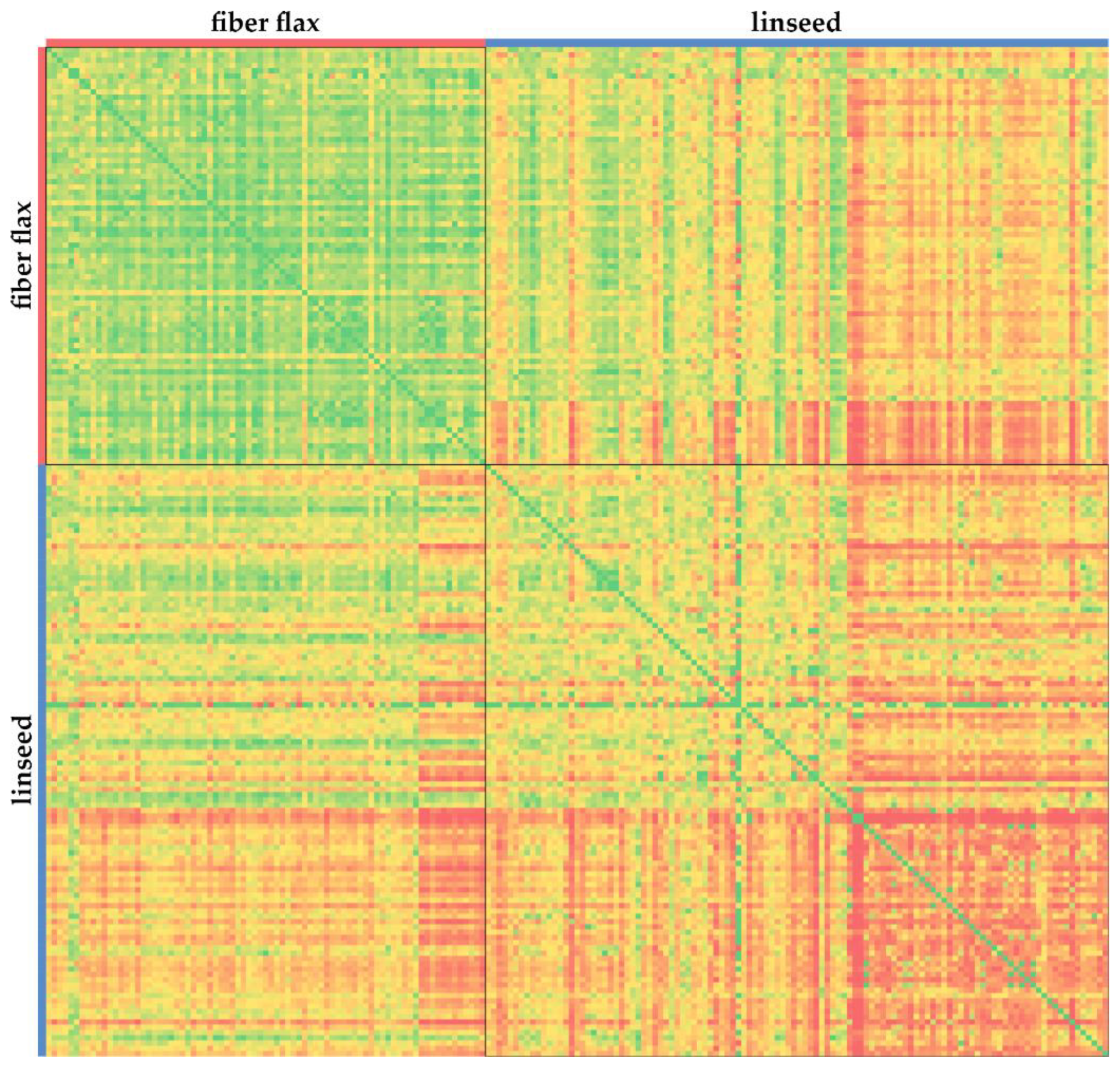
1. Introduction
2. Results
2.1. Gene Polymorphisms
2.2. Analysis of Genes Associated with Flax Type
2.2.1. Genes with Flax Type–Associated Polymorphisms
2.2.2. Genes Involved in Lignin Synthesis
2.2.3. Cellulose Synthases
2.2.4. Chitinase-Like Proteins
2.2.5. Tubulins
2.2.6. β-Galactosidases
2.2.7. Rhamnogalacturonan Lyases
2.2.8. Genes Involved in Lignan Synthesis
2.2.9. Genes Involved in Fatty Acid Synthesis
2.2.10. ABC Transporter and Heavy Metal-Associated Genes
3. Discussion
4. Materials and Methods
4.1. Variant Calling and Associative Analysis of Genome Sequencing Data
4.2. Analysis of Transcriptome Sequencing Data
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Muir, A.D.; Westcott, N.D. Flax: The Genus Linum; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- Weiss, E.; Zohary, D.; Hopf, M. Domestication of Plants in the Old World—The Origin and Spread of Domesticated Plants in South-West Asia, Europe, and the Mediterranean Basin; Oxford Scholarship: Oxford, UK, 2012. [Google Scholar]
- Diederichsen, A.; Richards, K. Cultivated Flax and the Genus linum L.: Taxonomy and Germplasm Conservation; CRC Press: Boca Raton, FL, USA, 2003; pp. 22–54. [Google Scholar]
- Diederichsen, A.; Ulrich, A. Variability in stem fibre content and its association with other characteristics in 1177 flax (Linum usitatissimum L.) genebank accessions. Ind. Crop. Prod. 2009, 30, 33–39. [Google Scholar] [CrossRef]
- Goyal, A.; Sharma, V.; Upadhyay, N.; Gill, S.; Sihag, M. Flax and flaxseed oil: An ancient medicine & modern functional food. J. Food Sci. Technol. 2014, 51, 1633–1653. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Ahmad, N.; Anjum, F.M.; Khan, M.K.; Mushtaq, Z.; Nadeem, M.; Hussain, S. Potential protective properties of flax lignan secoisolariciresinol diglucoside. Nutr. J. 2015, 14, 71. [Google Scholar] [CrossRef]
- Parikh, M.; Netticadan, T.; Pierce, G.N. Flaxseed: Its bioactive components and their cardiovascular benefits. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H146–H159. [Google Scholar] [CrossRef] [PubMed]
- Kezimana, P.; Dmitriev, A.A.; Kudryavtseva, A.V.; Romanova, E.V.; Melnikova, N.V. Secoisolariciresinol Diglucoside of Flaxseed and Its Metabolites: Biosynthesis and Potential for Nutraceuticals. Front. Genet. 2018, 9, 641. [Google Scholar] [CrossRef] [PubMed]
- Mali, A.V.; Padhye, S.B.; Anant, S.; Hegde, M.V.; Kadam, S.S. Anticancer and antimetastatic potential of enterolactone: Clinical, preclinical and mechanistic perspectives. Eur. J. Pharmacol. 2019, 852, 107–124. [Google Scholar] [CrossRef]
- Cullis, C.A. Genetics and Genomics of Linum; Springer International Publishing: Cham, Switzerland, 2019. [Google Scholar]
- Campos, J.R.; Severino, P.; Ferreira, C.S.; Zielinska, A.; Santini, A.; Souto, S.B.; Souto, E.B. Linseed Essential Oil—Source of Lipids as Active Ingredients for Pharmaceuticals and Nutraceuticals. Curr. Med. Chem. 2019, 26, 4537–4558. [Google Scholar] [CrossRef]
- Fombuena, V.; Petrucci, R.; Dominici, F.; Jorda-Vilaplana, A.; Montanes, N.; Torre, L. Maleinized Linseed Oil as Epoxy Resin Hardener for Composites with High Bio Content Obtained from Linen Byproducts. Polymers 2019, 11, 301. [Google Scholar] [CrossRef]
- Corino, C.; Rossi, R.; Cannata, S.; Ratti, S. Effect of dietary linseed on the nutritional value and quality of pork and pork products: Systematic review and meta-analysis. Meat Sci. 2014, 98, 679–688. [Google Scholar] [CrossRef]
- Singh, K.K.; Mridula, D.; Rehal, J.; Barnwal, P. Flaxseed: A Potential Source of Food, Feed and Fiber. Crit. Rev. Food Sci. Nutr. 2011, 51, 210–222. [Google Scholar] [CrossRef]
- Costa, S.M.; Ferreira, D.P.; Ferreira, A.; Vaz, F.; Fangueiro, R. Multifunctional Flax Fibres Based on the Combined Effect of Silver and Zinc Oxide (Ag/ZnO) Nanostructures. Nanomaterials 2018, 8, 1069. [Google Scholar] [CrossRef] [PubMed]
- Baley, C.; Gomina, M.; Breard, J.; Bourmaud, A.; Davies, P. Variability of mechanical properties of flax fibres for composite reinforcement. A review. Ind. Crop. Prod. 2019, 145, 111984. [Google Scholar] [CrossRef]
- Zhu, J.; Zhu, H.; Njuguna, J.; Abhyankar, H. Recent Development of Flax Fibres and Their Reinforced Composites Based on Different Polymeric Matrices. Materials 2013, 6, 5171–5198. [Google Scholar] [CrossRef]
- Goudenhooft, C.; Bourmaud, A.; Baley, C. Flax (Linum usitatissimum L.) Fibers for Composite Reinforcement: Exploring the Link Between Plant Growth, Cell Walls Development, and Fiber Properties. Front. Plant Sci. 2019, 10, 411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, D.; Huang, T.; Hu, Q.; Lammer, H. Three-Dimensional Printing of Continuous Flax Fiber-Reinforced Thermoplastic Composites by Five-Axis Machine. Materials 2020, 13, 1678. [Google Scholar] [CrossRef]
- Mokhothu, T.H.; John, M.J. Review on hygroscopic aging of cellulose fibres and their biocomposites. Carbohydr. Polym. 2015, 131, 337–354. [Google Scholar] [CrossRef]
- Dhakal, H.N.; Sain, M. Enhancement of Mechanical Properties of Flax-Epoxy Composite with Carbon Fibre Hybridisation for Lightweight Applications. Materials 2019, 13, 109. [Google Scholar] [CrossRef]
- Wu, C.M.; Lai, W.Y.; Wang, C.Y. Effects of Surface Modification on the Mechanical Properties of Flax/beta-Polypropylene Composites. Materials 2016, 9, 314. [Google Scholar] [CrossRef]
- Kymäläinen, H.-R.; Sjöberg, A.-M. Flax and hemp fibres as raw materials for thermal insulations. Build. Environ. 2008, 43, 1261–1269. [Google Scholar] [CrossRef]
- Wang, Z.; Hobson, N.; Galindo, L.; Zhu, S.; Shi, D.; McDill, J.; Yang, L.; Hawkins, S.; Neutelings, G.; Datla, R.; et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. Cell Mol. Biol. 2012, 72, 461–473. [Google Scholar] [CrossRef]
- You, F.M.; Xiao, J.; Li, P.; Yao, Z.; Jia, G.; He, L.; Zhu, T.; Luo, M.C.; Wang, X.; Deyholos, M.K.; et al. Chromosome-scale pseudomolecules refined by optical, physical and genetic maps in flax. Plant J. Cell Mol. Biol. 2018, 95, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Sa, R.; Yi, L.; Siqin, B.; An, M.; Bao, H.; Song, X.; Wang, S.; Li, Z.; Zhang, Z.; Hazaisi, H.; et al. Chromosome-Level Genome Assembly and Annotation of the Fiber Flax (Linum usitatissimum) Genome. Front. Genet. 2021, 12, 735690. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qi, Y.; Wang, L.; Wang, L.; Yan, X.; Dang, Z.; Li, W.; Zhao, W.; Pei, X.; Li, X.; et al. Genomic Comparison and Population Diversity Analysis Provide Insights into the Domestication and Improvement of Flax. iScience 2020, 23, 100967. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, A.A.; Pushkova, E.N.; Novakovskiy, R.O.; Beniaminov, A.D.; Rozhmina, T.A.; Zhuchenko, A.A.; Bolsheva, N.L.; Muravenko, O.V.; Povkhova, L.V.; Dvorianinova, E.M.; et al. Genome Sequencing of Fiber Flax Cultivar Atlant Using Oxford Nanopore and Illumina Platforms. Front. Genet. 2020, 11, 590282. [Google Scholar] [CrossRef] [PubMed]
- Soto-Cerda, B.J.; Duguid, S.; Booker, H.; Rowland, G.; Diederichsen, A.; Cloutier, S. Genomic regions underlying agronomic traits in linseed (Linum usitatissimum L.) as revealed by association mapping. J. Integr. Plant Biol. 2014, 56, 75–87. [Google Scholar] [CrossRef]
- Xie, D.; Dai, Z.; Yang, Z.; Sun, J.; Zhao, D.; Yang, X.; Zhang, L.; Tang, Q.; Su, J. Genome-Wide Association Study Identifying Candidate Genes Influencing Important Agronomic Traits of Flax (Linum usitatissimum L.) Using SLAF-seq. Front. Plant Sci. 2017, 8, 2232. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Dai, Z.; Yang, Z.; Tang, Q.; Sun, J.; Yang, X.; Song, X.; Lu, Y.; Zhao, D.; Zhang, L.; et al. Genomic variations and association study of agronomic traits in flax. BMC Genom. 2018, 19, 512. [Google Scholar] [CrossRef] [PubMed]
- Soto-Cerda, B.J.; Cloutier, S.; Quian, R.; Gajardo, H.A.; Olivos, M.; You, F.M. Genome-Wide Association Analysis of Mucilage and Hull Content in Flax (Linum usitatissimum L.) Seeds. Int. J. Mol. Sci. 2018, 19, 2870. [Google Scholar] [CrossRef]
- Zhang, J.; Long, Y.; Wang, L.; Dang, Z.; Zhang, T.; Song, X.; Dang, Z.; Pei, X. Consensus genetic linkage map construction and QTL mapping for plant height-related traits in linseed flax (Linum usitatissimum L.). BMC Plant Biol. 2018, 18, 160. [Google Scholar] [CrossRef]
- You, F.M.; Xiao, J.; Li, P.; Yao, Z.; Jia, G.; He, L.; Kumar, S.; Soto-Cerda, B.; Duguid, S.D.; Booker, H.M.; et al. Genome-Wide Association Study and Selection Signatures Detect Genomic Regions Associated with Seed Yield and Oil Quality in Flax. Int. J. Mol. Sci. 2018, 19, 2303. [Google Scholar] [CrossRef]
- Wu, J.; Zhao, Q.; Zhang, L.; Li, S.; Ma, Y.; Pan, L.; Lin, H.; Wu, G.; Yuan, H.; Yu, Y.; et al. QTL Mapping of Fiber-Related Traits Based on a High-Density Genetic Map in Flax (Linum usitatissimum L.). Front. Plant Sci. 2018, 9, 885. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Jiang, H.; Yan, W.; Yang, L.; Ye, J.; Wang, Y.; Yan, Q.; Chen, J.; Gao, Y.; Duan, L.; et al. Resequencing 200 Flax Cultivated Accessions Identifies Candidate Genes Related to Seed Size and Weight and Reveals Signatures of Artificial Selection. Front. Plant Sci. 2019, 10, 1682. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Dai, Z.; Yang, Z.; Tang, Q.; Deng, C.; Xu, Y.; Wang, J.; Chen, J.; Zhao, D.; Zhang, S.; et al. Combined genome-wide association analysis and transcriptome sequencing to identify candidate genes for flax seed fatty acid metabolism. Plant Sci. Int. J. Exp. Plant Biol. 2019, 286, 98–107. [Google Scholar] [CrossRef]
- Galinousky, D.; Mokshina, N.; Padvitski, T.; Ageeva, M.; Bogdan, V.; Kilchevsky, A.; Gorshkova, T. The Toolbox for Fiber Flax Breeding: A Pipeline From Gene Expression to Fiber Quality. Front. Genet. 2020, 11, 589881. [Google Scholar] [CrossRef] [PubMed]
- Long, S.H.; Deng, X.; Wang, Y.F.; Li, X.; Qiao, R.Q.; Qiu, C.S.; Guo, Y.; Hao, D.M.; Jia, W.Q.; Chen, X.B. Analysis of 2297 expressed sequence tags (ESTs) from a cDNA library of flax (Linum ustitatissimum L.) bark tissue. Mol. Biol. Rep. 2012, 39, 6289–6296. [Google Scholar] [CrossRef]
- Guo, Y.; Qiu, C.; Long, S.; Chen, P.; Hao, D.; Preisner, M.; Wang, H.; Wang, Y. Digital gene expression profiling of flax (Linum usitatissimum L.) stem peel identifies genes enriched in fiber-bearing phloem tissue. Gene 2017, 626, 32–40. [Google Scholar] [CrossRef]
- Gorshkov, O.; Mokshina, N.; Gorshkov, V.; Chemikosova, S.; Gogolev, Y.; Gorshkova, T. Transcriptome portrait of cellulose-enriched flax fibres at advanced stage of specialization. Plant Mol. Biol. 2017, 93, 431–449. [Google Scholar] [CrossRef]
- Mokshina, N.; Gorshkov, O.; Ibragimova, N.; Chernova, T.; Gorshkova, T. Cellulosic fibres of flax recruit both primary and secondary cell wall cellulose synthases during deposition of thick tertiary cell walls and in the course of graviresponse. Funct. Plant Biol. FPB 2017, 44, 820–831. [Google Scholar] [CrossRef]
- Gorshkova, T.; Chernova, T.; Mokshina, N.; Gorshkov, V.; Kozlova, L.; Gorshkov, O. Transcriptome Analysis of Intrusively Growing Flax Fibers Isolated by Laser Microdissection. Sci. Rep. 2018, 8, 14570. [Google Scholar] [CrossRef]
- Gorshkov, O.; Chernova, T.; Mokshina, N.; Gogoleva, N.; Suslov, D.; Tkachenko, A.; Gorshkova, T. Intrusive Growth of Phloem Fibers in Flax Stem: Integrated Analysis of miRNA and mRNA Expression Profiles. Plants 2019, 8, 47. [Google Scholar] [CrossRef]
- Mokshina, N.; Gorshkov, O.; Galinousky, D.; Gorshkova, T. Genes with bast fiber-specific expression in flax plants—Molecular keys for targeted fiber crop improvement. Ind. Crop. Prod. 2020, 152, 112549. [Google Scholar] [CrossRef]
- Galindo-Gonzalez, L.; Deyholos, M.K. RNA-seq Transcriptome Response of Flax (Linum usitatissimum L.) to the Pathogenic Fungus Fusarium oxysporum f. sp. lini. Front. Plant Sci. 2016, 7, 1766. [Google Scholar] [CrossRef]
- Yu, Y.; Huang, W.; Chen, H.; Wu, G.; Yuan, H.; Song, X.; Kang, Q.; Zhao, D.; Jiang, W.; Liu, Y.; et al. Identification of differentially expressed genes in flax (Linum usitatissimum L.) under saline-alkaline stress by digital gene expression. Gene 2014, 549, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, A.A.; Krasnov, G.S.; Rozhmina, T.A.; Kishlyan, N.V.; Zyablitsin, A.V.; Sadritdinova, A.F.; Snezhkina, A.V.; Fedorova, M.S.; Yurkevich, O.Y.; Muravenko, O.V.; et al. Glutathione S-transferases and UDP-glycosyltransferases Are Involved in Response to Aluminum Stress in Flax. Front. Plant Sci. 2016, 7, 1920. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, A.A.; Kudryavtseva, A.V.; Krasnov, G.S.; Koroban, N.V.; Speranskaya, A.S.; Krinitsina, A.A.; Belenikin, M.S.; Snezhkina, A.V.; Sadritdinova, A.F.; Kishlyan, N.V.; et al. Gene expression profiling of flax (Linum usitatissimum L.) under edaphic stress. BMC Plant Biol. 2016, 16, 237. [Google Scholar] [CrossRef] [PubMed]
- Dash, P.K.; Cao, Y.; Jailani, A.K.; Gupta, P.; Venglat, P.; Xiang, D.; Rai, R.; Sharma, R.; Thirunavukkarasu, N.; Abdin, M.Z.; et al. Genome-wide analysis of drought induced gene expression changes in flax (Linum usitatissimum). GM Crop. Food 2014, 5, 106–119. [Google Scholar] [CrossRef]
- Dash, P.K.; Rai, R.; Mahato, A.K.; Gaikwad, K.; Singh, N.K. Transcriptome Landscape at Different Developmental Stages of a Drought Tolerant Cultivar of Flax (Linum usitatissimum). Front. Chem. 2017, 5, 82. [Google Scholar] [CrossRef]
- Dmitriev, A.A.; Krasnov, G.S.; Rozhmina, T.A.; Novakovskiy, R.O.; Snezhkina, A.V.; Fedorova, M.S.; Yurkevich, O.Y.; Muravenko, O.V.; Bolsheva, N.L.; Kudryavtseva, A.V.; et al. Differential gene expression in response to Fusarium oxysporum infection in resistant and susceptible genotypes of flax (Linum usitatissimum L.). BMC Plant Biol. 2017, 17, 253. [Google Scholar] [CrossRef]
- Preisner, M.; Wojtasik, W.; Kostyn, K.; Boba, A.; Czuj, T.; Szopa, J.; Kulma, A. The cinnamyl alcohol dehydrogenase family in flax: Differentiation during plant growth and under stress conditions. J. Plant Physiol. 2018, 221, 132–143. [Google Scholar] [CrossRef]
- Dmitriev, A.A.; Krasnov, G.S.; Rozhmina, T.A.; Zyablitsin, A.V.; Snezhkina, A.V.; Fedorova, M.S.; Pushkova, E.N.; Kezimana, P.; Novakovskiy, R.O.; Povkhova, L.V.; et al. Flax (Linum usitatissimum L.) response to non-optimal soil acidity and zinc deficiency. BMC Plant Biol. 2019, 19, 54. [Google Scholar] [CrossRef]
- Wu, J.; Zhao, Q.; Wu, G.; Yuan, H.; Ma, Y.; Lin, H.; Pan, L.; Li, S.; Sun, D. Comprehensive Analysis of Differentially Expressed Unigenes under NaCl Stress in Flax (Linum usitatissimum L.) Using RNA-Seq. Int. J. Mol. Sci. 2019, 20, 369. [Google Scholar] [CrossRef]
- Le Roy, J.; Blervacq, A.S.; Creach, A.; Huss, B.; Hawkins, S.; Neutelings, G. Spatial regulation of monolignol biosynthesis and laccase genes control developmental and stress-related lignin in flax. BMC Plant Biol. 2017, 17, 124. [Google Scholar] [CrossRef] [PubMed]
- Mokshina, N.; Makshakova, O.; Nazipova, A.; Gorshkov, O.; Gorshkova, T. Flax rhamnogalacturonan lyases: Phylogeny, differential expression and modeling of protein structure. Physiol. Plant 2019, 167, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Hobson, N.; Deyholos, M.K. Genomic and expression analysis of the flax (Linum usitatissimum) family of glycosyl hydrolase 35 genes. BMC Genom. 2013, 14, 344. [Google Scholar] [CrossRef] [PubMed]
- Morello, L.; Pydiura, N.; Galinousky, D.; Blume, Y.; Breviario, D. Flax tubulin and CesA superfamilies represent attractive and challenging targets for a variety of genome- and base-editing applications. Funct. Integr. Genom. 2020, 20, 163–176. [Google Scholar] [CrossRef]
- Mokshina, N.; Gorshkova, T.; Deyholos, M.K. Chitinase-like (CTL) and cellulose synthase (CESA) gene expression in gelatinous-type cellulosic walls of flax (Linum usitatissimum L.) bast fibers. PLoS ONE 2014, 9, e97949. [Google Scholar] [CrossRef]
- Chantreau, M.; Chabbert, B.; Billiard, S.; Hawkins, S.; Neutelings, G. Functional analyses of cellulose synthase genes in flax (Linum usitatissimum) by virus-induced gene silencing. Plant Biotechnol. J. 2015, 13, 1312–1324. [Google Scholar] [CrossRef]
- Galinousky, D.; Padvitski, T.; Bayer, G.; Pirko, Y.; Pydiura, N.; Anisimova, N.; Nikitinskaya, T.; Khotyleva, L.; Yemets, A.; Kilchevsky, A.; et al. Expression analysis of cellulose synthase and main cytoskeletal protein genes in flax (Linum usitatissimum L.). Cell Biol. Int. 2019, 43, 1065–1071. [Google Scholar] [CrossRef]
- Yurkevich, O.Y.; Kirov, I.V.; Bolsheva, N.L.; Rachinskaya, O.A.; Grushetskaya, Z.E.; Zoschuk, S.A.; Samatadze, T.E.; Bogdanova, M.V.; Lemesh, V.A.; Amosova, A.V.; et al. Integration of Physical, Genetic, and Cytogenetic Mapping Data for Cellulose Synthase (CesA) Genes in Flax (Linum usitatissimum L.). Front. Plant Sci. 2017, 8, 1467. [Google Scholar] [CrossRef]
- Pydiura, N.; Pirko, Y.; Galinousky, D.; Postovoitova, A.; Yemets, A.; Kilchevsky, A.; Blume, Y. Genome-wide identification, phylogenetic classification, and exon-intron structure characterization of the tubulin and actin genes in flax (Linum usitatissimum). Cell Biol. Int. 2019, 43, 1010–1019. [Google Scholar] [CrossRef]
- Dalisay, D.S.; Kim, K.W.; Lee, C.; Yang, H.; Rubel, O.; Bowen, B.P.; Davin, L.B.; Lewis, N.G. Dirigent Protein-Mediated Lignan and Cyanogenic Glucoside Formation in Flax Seed: Integrated Omics and MALDI Mass Spectrometry Imaging. J. Nat. Prod. 2015, 78, 1231–1242. [Google Scholar] [CrossRef] [PubMed]
- Ghose, K.; Selvaraj, K.; McCallum, J.; Kirby, C.W.; Sweeney-Nixon, M.; Cloutier, S.J.; Deyholos, M.; Datla, R.; Fofana, B. Identification and functional characterization of a flax UDP-glycosyltransferase glucosylating secoisolariciresinol (SECO) into secoisolariciresinol monoglucoside (SMG) and diglucoside (SDG). BMC Plant Biol. 2014, 14, 82. [Google Scholar] [CrossRef] [PubMed]
- Hemmati, S.; von Heimendahl, C.B.; Klaes, M.; Alfermann, A.W.; Schmidt, T.J.; Fuss, E. Pinoresinol-lariciresinol reductases with opposite enantiospecificity determine the enantiomeric composition of lignans in the different organs of Linum usitatissimum L. Planta Med. 2010, 76, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Corbin, C.; Drouet, S.; Markulin, L.; Auguin, D.; Laine, E.; Davin, L.B.; Cort, J.R.; Lewis, N.G.; Hano, C. A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: From gene identification and evolution to differential regulation. Plant Mol. Biol. 2018, 97, 73–101. [Google Scholar] [CrossRef] [PubMed]
- Fofana, B.; Duguid, S.; Cloutier, S. Cloning of fatty acid biosynthetic genes β-ketoacyl CoA synthase, fatty acid elongase, stearoyl-ACP desaturase, and fatty acid desaturase and analysis of expression in the early developmental stages of flax (Linum usitatissimum L.) seeds. Plant Sci. 2004, 166, 1487–1496. [Google Scholar] [CrossRef]
- Vrinten, P.; Hu, Z.; Munchinsky, M.A.; Rowland, G.; Qiu, X. Two FAD3 desaturase genes control the level of linolenic acid in flax seed. Plant Physiol. 2005, 139, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Thambugala, D.; Duguid, S.; Loewen, E.; Rowland, G.; Booker, H.; You, F.M.; Cloutier, S. Genetic variation of six desaturase genes in flax and their impact on fatty acid composition. Theor. Appl. Genet. 2013, 126, 2627–2641. [Google Scholar] [CrossRef]
- Fofana, B.; Cloutier, S.; Duguid, S.; Ching, J.; Rampitsch, C. Gene expression of stearoyl-ACP desaturase and delta12 fatty acid desaturase 2 is modulated during seed development of flax (Linum usitatissimum). Lipids 2006, 41, 705–712. [Google Scholar] [CrossRef]
- Khan, N.; You, F.M.; Datla, R.; Ravichandran, S.; Jia, B.; Cloutier, S. Genome-wide identification of ATP binding cassette (ABC) transporter and heavy metal associated (HMA) gene families in flax (Linum usitatissimum L.). BMC Genom. 2020, 21, 722. [Google Scholar] [CrossRef]
- Dmitriev, A.A.; Novakovskiy, R.O.; Pushkova, E.N.; Rozhmina, T.A.; Zhuchenko, A.A.; Bolsheva, N.L.; Beniaminov, A.D.; Mitkevich, V.A.; Povkhova, L.V.; Dvorianinova, E.M.; et al. Transcriptomes of Different Tissues of Flax (Linum usitatissimum L.) Cultivars With Diverse Characteristics. Front. Genet. 2020, 11, 565146. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin engineering. Curr. Opin. Plant Biol. 2008, 11, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Rohde, A.; Morreel, K.; Ralph, J.; Goeminne, G.; Hostyn, V.; De Rycke, R.; Kushnir, S.; Van Doorsselaere, J.; Joseleau, J.P.; Vuylsteke, M.; et al. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 2004, 16, 2749–2771. [Google Scholar] [CrossRef]
- Chen, F.; Srinivasa Reddy, M.S.; Temple, S.; Jackson, L.; Shadle, G.; Dixon, R.A. Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant J. Cell Mol. Biol. 2006, 48, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Matthews, M.L.; Williams, C.M.; Shi, R.; Yang, C.; Tunlaya-Anukit, S.; Chen, H.C.; Li, Q.; Liu, J.; Lin, C.Y.; et al. Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 2018, 9, 1579. [Google Scholar] [CrossRef]
- Bate, N.J.; Orr, J.; Ni, W.; Meromi, A.; Nadler-Hassar, T.; Doerner, P.W.; Dixon, R.A.; Lamb, C.J.; Elkind, Y. Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc. Natl. Acad. Sci. USA 1994, 91, 7608–7612. [Google Scholar] [CrossRef]
- Sewalt, V.; Ni, W.; Blount, J.W.; Jung, H.G.; Masoud, S.A.; Howles, P.A.; Lamb, C.; Dixon, R.A. Reduced Lignin Content and Altered Lignin Composition in Transgenic Tobacco Down-Regulated in Expression of L-Phenylalanine Ammonia-Lyase or Cinnamate 4-Hydroxylase. Plant Physiol. 1997, 115, 41–50. [Google Scholar] [CrossRef]
- Cao, S.; Huang, C.; Luo, L.; Zheng, S.; Zhong, Y.; Sun, J.; Gui, J.; Li, L. Cell-Specific Suppression of 4-Coumarate-CoA Ligase Gene Reveals Differential Effect of Lignin on Cell Physiological Function in Populus. Front. Plant Sci. 2020, 11, 589729. [Google Scholar] [CrossRef]
- Ozparpucu, M.; Gierlinger, N.; Burgert, I.; Van Acker, R.; Vanholme, R.; Boerjan, W.; Pilate, G.; Dejardin, A.; Ruggeberg, M. The effect of altered lignin composition on mechanical properties of CINNAMYL ALCOHOL DEHYDROGENASE (CAD) deficient poplars. Planta 2018, 247, 887–897. [Google Scholar] [CrossRef]
- Zhang, S.; Jia, T.; Zhang, Z.; Zou, X.; Fan, S.; Lei, K.; Jiang, X.; Niu, D.; Yuan, Y.; Shang, H. Insight into the relationship between S-lignin and fiber quality based on multiple research methods. Plant Physiol. Biochem. PPB 2020, 147, 251–261. [Google Scholar] [CrossRef]
- Chantreau, M.; Grec, S.; Gutierrez, L.; Dalmais, M.; Pineau, C.; Demailly, H.; Paysant-Leroux, C.; Tavernier, R.; Trouve, J.P.; Chatterjee, M.; et al. PT-Flax (phenotyping and TILLinG of flax): Development of a flax (Linum usitatissimum L.) mutant population and TILLinG platform for forward and reverse genetics. BMC Plant Biol. 2013, 13, 159. [Google Scholar] [CrossRef]
- Liu, W.; Jiang, Y.; Wang, C.; Zhao, L.; Jin, Y.; Xing, Q.; Li, M.; Lv, T.; Qi, H. Lignin synthesized by CmCAD2 and CmCAD3 in oriental melon (Cucumis melo L.) seedlings contributes to drought tolerance. Plant Mol. Biol. 2020, 103, 689–704. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Jiang, Y.; Jin, Y.; Wang, C.; Yang, J.; Qi, H. Drought-induced ABA, H2O2 and JA positively regulate CmCAD genes and lignin synthesis in melon stems. BMC Plant Biol. 2021, 21, 83. [Google Scholar] [CrossRef] [PubMed]
- Novakovskiy, R.O.; Povkhova, L.V.; Krasnov, G.S.; Rozhmina, T.A.; Zhuchenko, A.A.; Kudryavtseva, L.P.; Pushkova, E.N.; Kezimana, P.; Kudryavtseva, A.V.; Dmitriev, A.A.; et al. The cinnamyl alcohol dehydrogenase gene family is involved in the response to Fusarium oxysporum in resistant and susceptible flax genotypes. Vavilov J. Genet. Breed. 2019, 23, 896–901. [Google Scholar] [CrossRef]
- Bagniewska-Zadworna, A.; Barakat, A.; Lakomy, P.; Smolinski, D.J.; Zadworny, M. Lignin and lignans in plant defence: Insight from expression profiling of cinnamyl alcohol dehydrogenase genes during development and following fungal infection in Populus. Plant Sci. Int. J. Exp. Plant Biol. 2014, 229, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Zhang, J.; Tschaplinski, T.J.; Tuskan, G.A.; Chen, J.G.; Muchero, W. Regulation of Lignin Biosynthesis and Its Role in Growth-Defense Tradeoffs. Front. Plant Sci. 2018, 9, 1427. [Google Scholar] [CrossRef] [PubMed]
- Robert, S.; Mouille, G.; Höfte, H. The mechanism and regulation of cellulose synthesis in primary walls: Lessons from cellulose-deficient Arabidopsis mutants. Cellulose 2004, 11, 351–364. [Google Scholar] [CrossRef]
- Grover, A. Plant Chitinases: Genetic Diversity and Physiological Roles. Crit. Rev. Plant Sci. 2012, 31, 57–73. [Google Scholar] [CrossRef]
- Kombrink, E.; Schroder, M.; Hahlbrock, K. Several “pathogenesis-related” proteins in potato are 1,3-beta-glucanases and chitinases. Proc. Natl. Acad. Sci. USA 1988, 85, 782–786. [Google Scholar] [CrossRef]
- Hermans, C.; Porco, S.; Verbruggen, N.; Bush, D.R. Chitinase-like protein CTL1 plays a role in altering root system architecture in response to multiple environmental conditions. Plant Physiol. 2010, 152, 904–917. [Google Scholar] [CrossRef]
- Levy, I.; Shani, Z.; Shoseyov, O. Modification of polysaccharides and plant cell wall by endo-1,4-beta-glucanase and cellulose-binding domains. Biomol. Eng. 2002, 19, 17–30. [Google Scholar] [CrossRef]
- Chandrasekar, B.; van der Hoorn, R.A. Beta galactosidases in Arabidopsis and tomato—A mini review. Biochem. Soc. Trans. 2016, 44, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Roach, M.J.; Mokshina, N.Y.; Badhan, A.; Snegireva, A.V.; Hobson, N.; Deyholos, M.K.; Gorshkova, T.A. Development of cellulosic secondary walls in flax fibers requires beta-galactosidase. Plant Physiol. 2011, 156, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Silva, I.R.; Jers, C.; Meyer, A.S.; Mikkelsen, J.D. Rhamnogalacturonan I modifying enzymes: An update. Nat. Biotechnol. 2016, 33, 41–54. [Google Scholar] [CrossRef]
- Hano, C.; Martin, I.; Fliniaux, O.; Legrand, B.; Gutierrez, L.; Arroo, R.R.; Mesnard, F.; Lamblin, F.; Laine, E. Pinoresinol-lariciresinol reductase gene expression and secoisolariciresinol diglucoside accumulation in developing flax (Linum usitatissimum) seeds. Planta 2006, 224, 1291–1301. [Google Scholar] [CrossRef]
- Corbin, C.; Drouet, S.; Mateljak, I.; Markulin, L.; Decourtil, C.; Renouard, S.; Lopez, T.; Doussot, J.; Lamblin, F.; Auguin, D.; et al. Functional characterization of the pinoresinol-lariciresinol reductase-2 gene reveals its roles in yatein biosynthesis and flax defense response. Planta 2017, 246, 405–420. [Google Scholar] [CrossRef]
- Renouard, S.; Tribalatc, M.A.; Lamblin, F.; Mongelard, G.; Fliniaux, O.; Corbin, C.; Marosevic, D.; Pilard, S.; Demailly, H.; Gutierrez, L.; et al. RNAi-mediated pinoresinol lariciresinol reductase gene silencing in flax (Linum usitatissimum L.) seed coat: Consequences on lignans and neolignans accumulation. J. Plant Physiol. 2014, 171, 1372–1377. [Google Scholar] [CrossRef]
- Hamade, K.; Fliniaux, O.; Fontaine, J.X.; Molinie, R.; Otogo Nnang, E.; Bassard, S.; Guenin, S.; Gutierrez, L.; Laine, E.; Hano, C.; et al. NMR and LC-MS-Based Metabolomics to Study Osmotic Stress in Lignan-Deficient Flax. Molecules 2021, 26, 767. [Google Scholar] [CrossRef]
- Rajwade, A.V.; Kadoo, N.Y.; Borikar, S.P.; Harsulkar, A.M.; Ghorpade, P.B.; Gupta, V.S. Differential transcriptional activity of SAD, FAD2 and FAD3 desaturase genes in developing seeds of linseed contributes to varietal variation in alpha-linolenic acid content. Phytochemistry 2014, 98, 41–53. [Google Scholar] [CrossRef]
- Zhang, Y.; Maximova, S.N.; Guiltinan, M.J. Characterization of a stearoyl-acyl carrier protein desaturase gene family from chocolate tree, Theobroma cacao L. Front. Plant Sci. 2015, 6, 239. [Google Scholar] [CrossRef][Green Version]
- Dmitriev, A.A.; Kezimana, P.; Rozhmina, T.A.; Zhuchenko, A.A.; Povkhova, L.V.; Pushkova, E.N.; Novakovskiy, R.O.; Pavelek, M.; Vladimirov, G.N.; Nikolaev, E.N.; et al. Genetic diversity of SAD and FAD genes responsible for the fatty acid composition in flax cultivars and lines. BMC Plant Biol. 2020, 20, 301. [Google Scholar] [CrossRef]
- Kezimana, P.; Rozhmina, T.A.; Krasnov, G.S.; Povkhova, L.V.; Novakovskiy, R.O.; Pushkova, E.N.; Zhuchenko, A.A.; Bjelková, M.; Pavelek, M.; Dmitriev, A.A.; et al. Evaluation of polymorphism of SAD and FAD genes in flax (Linum usitatissimum L.) cultivars and lines using deep sequencing. In Proceedings of the Theory and Practice of Adaptive Plant Breeding (Zhuchenkov’s Readings VI), Krasnodar, Russia, 25 September 2020; pp. 49–52. [Google Scholar]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC Transporters. Arab. Book 2011, 9, e0153. [Google Scholar] [CrossRef]
- D’yakov, A.B. Flax Physiology and Ecology; LAP LAMBERT Academic Publishing: Sunnyvale, CA, USA, 2006. [Google Scholar]
- Kumar, S.; You, F.M.; Duguid, S.; Booker, H.; Rowland, G.; Cloutier, S. QTL for fatty acid composition and yield in linseed (Linum usitatissimum L.). Theor. Appl. Genet. 2015, 128, 965–984. [Google Scholar] [CrossRef] [PubMed]
- Cloutier, S.; Ragupathy, R.; Niu, Z.; Duguid, S. SSR-based linkage map of flax (Linum usitatissimum L.) and mapping of QTLs underlying fatty acid composition traits. Mol. Breed. 2011, 28, 437–451. [Google Scholar] [CrossRef]
- Soto-Cerda, B.J.; Duguid, S.; Booker, H.; Rowland, G.; Diederichsen, A.; Cloutier, S. Association mapping of seed quality traits using the Canadian flax (Linum usitatissimum L.) core collection. Theor. Appl. Genet. 2014, 127, 881–896. [Google Scholar] [CrossRef]
- Asgarinia, P.; Cloutier, S.; Duguid, S.; Rashid, K.; Mirlohi, A.; Banik, M.; Saeidi, G. Mapping Quantitative Trait Loci for Powdery Mildew Resistance in Flax (Linum usitatissimum L.). Crop Sci. 2013, 53, 2462–2472. [Google Scholar] [CrossRef]
- He, L.; Xiao, J.; Rashid, K.Y.; Yao, Z.; Li, P.; Jia, G.; Wang, X.; Cloutier, S.; You, F.M. Genome-Wide Association Studies for Pasmo Resistance in Flax (Linum usitatissimum L.). Front. Plant Sci. 2018, 9, 1982. [Google Scholar] [CrossRef]
- Lan, S.; Zheng, C.; Hauck, K.; McCausland, M.; Duguid, S.D.; Booker, H.M.; Cloutier, S.; You, F.M. Genomic Prediction Accuracy of Seven Breeding Selection Traits Improved by QTL Identification in Flax. Int. J. Mol. Sci. 2020, 21, 1577. [Google Scholar] [CrossRef]
- Sertse, D.; You, F.M.; Ravichandran, S.; Soto-Cerda, B.J.; Duguid, S.; Cloutier, S. Loci harboring genes with important role in drought and related abiotic stress responses in flax revealed by multiple GWAS models. Theor. Appl. Genet. 2021, 134, 191–212. [Google Scholar] [CrossRef] [PubMed]
- Soto-Cerda, B.J.; Aravena, G.; Cloutier, S. Genetic dissection of flowering time in flax (Linum usitatissimum L.) through single- and multi-locus genome-wide association studies. Mol. Genet. Genom. MGG 2021, 296, 877–891. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997v2. [Google Scholar]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907v2. [Google Scholar]
- You, F.M.; Cloutier, S. Mapping Quantitative Trait Loci onto Chromosome-Scale Pseudomolecules in Flax. Methods Protoc. 2020, 3, 28. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]


| Gene | Number of FTA Polymorphisms | Gene | Number of FTA Polymorphisms | Gene | Number of FTA Polymorphisms | Gene | Number of FTA Polymorphisms |
|---|---|---|---|---|---|---|---|
| Lignin synthesis | BGAL30 | 21 | ABCB45 | 19 | ABCG68 | 2 | |
| 4CL1 | 32 | BGAL31 | 1 | ABCB46 | 7 | ABCG69 | 4 |
| 4CL4 | 2 | BGAL32 | 4 | ABCB47 | 30 | ABCG71 | 30 |
| 4CL5 | 3 | BGAL33 | 6 | ABCB48 | 5 | ABCG72 | 2 |
| C4H3 | 2 | BGAL35 | 1 | ABCB7 | 6 | ABCG73 | 15 |
| C4H4 | 9 | BGAL37 | 7 | ABCC10 | 19 | ABCG75 | 1 |
| CAD1A | 1 | BGAL40 | 25 | ABCC16 | 1 | ABCG79 | 15 |
| CAD1B | 2 | BGAL41 | 1 | ABCC18 | 2 | ABCG8 | 19 |
| CAD4A | 1 | BGAL6 | 12 | ABCC4 | 5 | ABCG80 | 39 |
| CAD4B | 4 | BGAL7 | 4 | ABCC6 | 1 | ABCG83 | 10 |
| CAD7 | 2 | BGAL9 | 4 | ABCC5 | 1 | ABCH1 | 3 |
| CCR11 | 1 | ABC and HMA | ABCF8 | 2 | ABCH10 | 15 | |
| CCR4 | 3 | ABCA1 | 32 | ABCG1 | 2 | ABCH11 | 4 |
| CCoAOMT5 | 2 | ABCA2 | 16 | ABCG11 | 3 | ABCH12 | 1 |
| COMT2 | 5 | ABCA3 | 1 | ABCG12 | 2 | ABCH8 | 1 |
| COMT3 | 2 | ABCA4 | 1 | ABCG13 | 3 | HMA12 | 18 |
| F5H1 | 2 | ABCA5 | 11 | ABCG14 | 8 | HMA2 | 2 |
| F5H7 | 1 | ABCA6 | 2 | ABCG16 | 15 | HMA3 | 2 |
| PAL1 | 2 | ABCA7 | 27 | ABCG22 | 6 | HMA4 | 9 |
| PAL3 | 3 | ABCA8 | 12 | ABCG24 | 1 | HMA6 | 13 |
| CTL | ABCB1 | 3 | ABCG25 | 7 | Lignan synthesis | ||
| CTL1 | 16 | ABCB12 | 1 | ABCG33 | 4 | PLR1 | 50 |
| CTL10 | 6 | ABCB13 | 10 | ABCG35 | 15 | TUB | |
| CTL13 | 8 | ABCB16 | 2 | ABCG36 | 4 | Alfa_TUB2 | 1 |
| CTL18 | 11 | ABCB2 | 1 | ABCG37 | 1 | Beta_TUB13 | 1 |
| CTL2 | 7 | ABCB22 | 2 | ABCG4 | 3 | Beta_TUB3 | 8 |
| CTL22 | 1 | ABCB23 | 1 | ABCG40 | 1 | Beta_TUB6 | 6 |
| CTL23 | 7 | ABCB25 | 1 | ABCG47 | 8 | Beta_TUB7 | 2 |
| CTL24 | 3 | ABCB26 | 1 | ABCG52 | 2 | CESA | |
| CTL26 | 2 | ABCB29 | 23 | ABCG56 | 13 | CESA1-B | 1 |
| CTL35 | 2 | ABCB3 | 2 | ABCG57 | 2 | CESA3-A | 1 |
| CTL4 | 1 | ABCB32 | 2 | ABCG58 | 12 | CESA4 | 2 |
| BGAL | ABCB33 | 3 | ABCG59 | 1 | CESA8-A | 1 | |
| BGAL1 | 2 | ABCB37 | 1 | ABCG6 | 4 | RGL | |
| BGAL10 | 1 | ABCB39 | 1 | ABCG60 | 2 | RGL1_B | 1 |
| BGAL2 | 11 | ABCB40 | 22 | ABCG61 | 1 | RGL4_B | 1 |
| BGAL23 | 1 | ABCB42 | 27 | ABCG62 | 2 | Fatty acid synthesis | |
| BGAL27 | 13 | ABCB43 | 2 | ABCG64 | 4 | FAD2A | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Povkhova, L.V.; Melnikova, N.V.; Rozhmina, T.A.; Novakovskiy, R.O.; Pushkova, E.N.; Dvorianinova, E.M.; Zhuchenko, A.A.; Kamionskaya, A.M.; Krasnov, G.S.; Dmitriev, A.A. Genes Associated with the Flax Plant Type (Oil or Fiber) Identified Based on Genome and Transcriptome Sequencing Data. Plants 2021, 10, 2616. https://doi.org/10.3390/plants10122616
Povkhova LV, Melnikova NV, Rozhmina TA, Novakovskiy RO, Pushkova EN, Dvorianinova EM, Zhuchenko AA, Kamionskaya AM, Krasnov GS, Dmitriev AA. Genes Associated with the Flax Plant Type (Oil or Fiber) Identified Based on Genome and Transcriptome Sequencing Data. Plants. 2021; 10(12):2616. https://doi.org/10.3390/plants10122616
Chicago/Turabian StylePovkhova, Liubov V., Nataliya V. Melnikova, Tatiana A. Rozhmina, Roman O. Novakovskiy, Elena N. Pushkova, Ekaterina M. Dvorianinova, Alexander A. Zhuchenko, Anastasia M. Kamionskaya, George S. Krasnov, and Alexey A. Dmitriev. 2021. "Genes Associated with the Flax Plant Type (Oil or Fiber) Identified Based on Genome and Transcriptome Sequencing Data" Plants 10, no. 12: 2616. https://doi.org/10.3390/plants10122616
APA StylePovkhova, L. V., Melnikova, N. V., Rozhmina, T. A., Novakovskiy, R. O., Pushkova, E. N., Dvorianinova, E. M., Zhuchenko, A. A., Kamionskaya, A. M., Krasnov, G. S., & Dmitriev, A. A. (2021). Genes Associated with the Flax Plant Type (Oil or Fiber) Identified Based on Genome and Transcriptome Sequencing Data. Plants, 10(12), 2616. https://doi.org/10.3390/plants10122616









