Bluster or Lustre: Can AI Improve Crops and Plant Health?
Abstract
:1. Introduction
2. AI in Plants
2.1. Phenotyping
2.2. Genotype-to-Phenotype
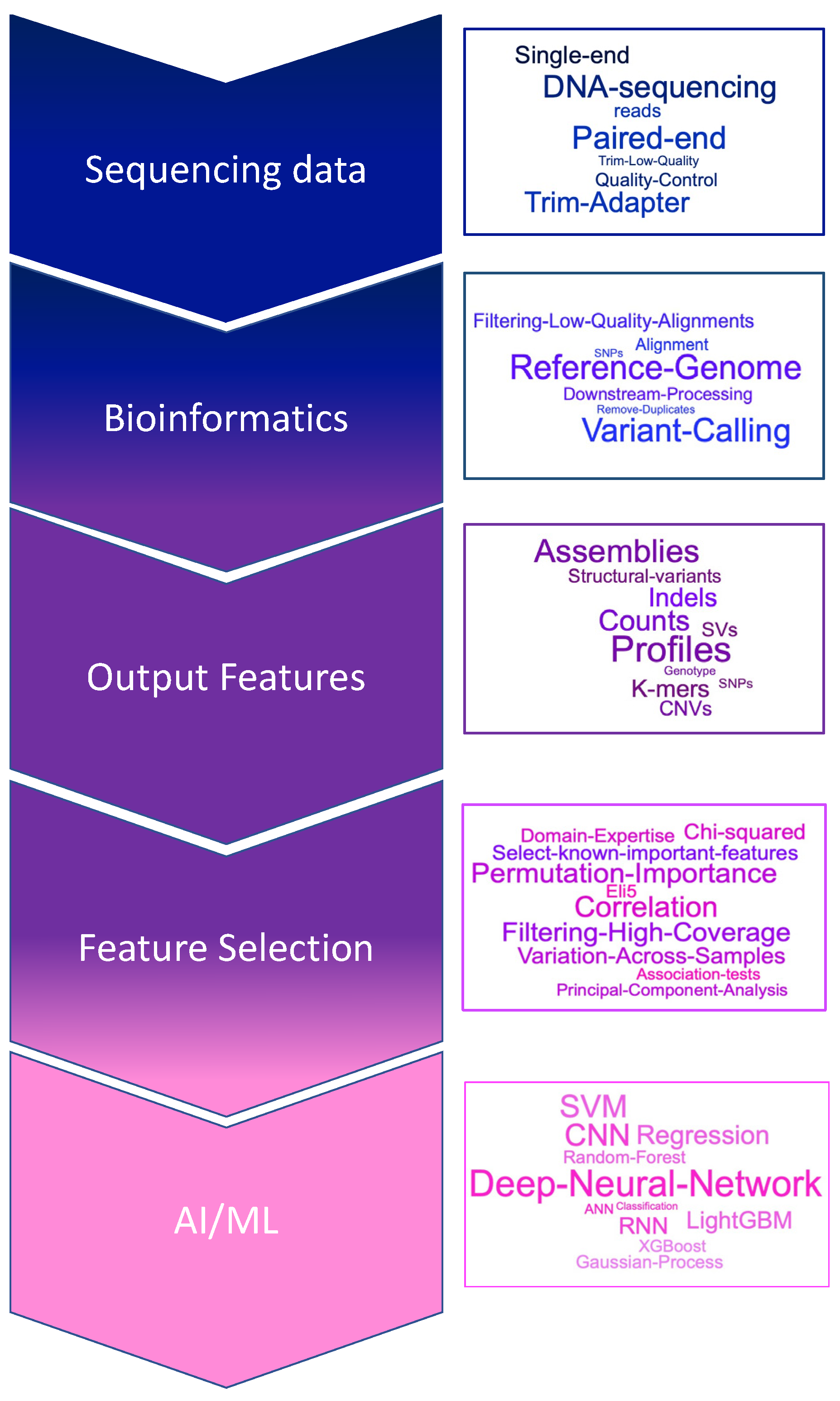
2.3. Omic Data
2.3.1. Challenges from Omic Data
2.3.2. Omic Data Integration
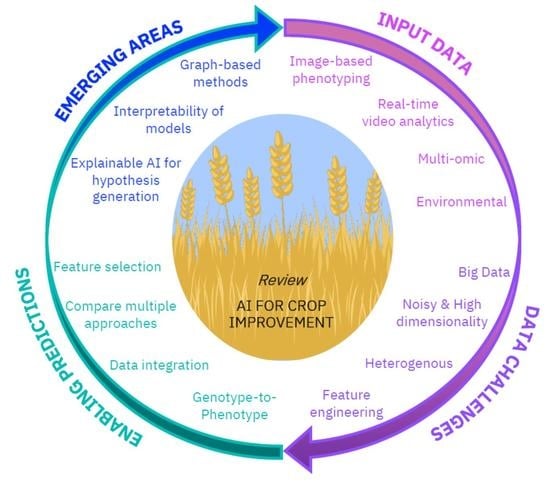
3. Emerging Areas of Interest in the Field
Explainability and Interpretability
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grafton, R.Q.; Daugbjerg, C.; Qureshi, M.E. Towards food security by 2050. Food Secur. 2015, 7, 179–183. [Google Scholar] [CrossRef]
- Corredor-Moreno, P.; Saunders, D.G.O. Expecting the unexpected: Factors influencing the emergence of fungal and oomycete plant pathogens. New Phytol. 2019, 225, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ye, W.; Wang, M.; Yan, X. Climate change and drought: A risk assessment of crop-yield impacts. Clim. Res. 2009, 39, 31–46. [Google Scholar] [CrossRef]
- Mall, R.; Gupta, A.; Sonkar, G. Effect of Climate Change on Agricultural Crops. In Current Developments in Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2017; pp. 23–46. [Google Scholar] [CrossRef]
- Reese, H. Understanding the Differences between AI, Machine Learning, and Deep Learning. February 2017. Available online: https://deeplearning.lipingyang.org/wp-content/uploads/2016/11/Understanding-the-differences-between-AI-machine-learning-and-deep-learning-TechRepublic.pdf (accessed on 8 December 2021).
- Wang, H.; Cimen, E.; Singh, N.; Buckler, E. Deep learning for plant genomics and crop improvement. Curr. Opin. Plant Biol. 2020, 54, 34–41. [Google Scholar] [CrossRef]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef] [Green Version]
- Tsouros, D.C.; Bibi, S.; Sarigiannidis, P.G. A review on UAV-based applications for precision agriculture. Information 2019, 10, 349. [Google Scholar] [CrossRef] [Green Version]
- Tsaftaris, S.A.; Minervini, M.; Scharr, H. Machine Learning for Plant Phenotyping Needs Image Processing. Trends Plant Sci. 2016, 21, 989–991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdulridha, J.; Ampatzidis, Y.; Roberts, P.; Kakarla, S.C. Detecting powdery mildew disease in squash at different stages using UAV-based hyperspectral imaging and artificial intelligence. Biosyst. Eng. 2020, 197, 135–148. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Uga, H.; Kagiwada, S.; Iyatomi, H. Basic Study of Automated Diagnosis of Viral Plant Diseases Using Convolutional Neural Networks. In Advances in Visual Computing; Bebis, G., Boyle, R., Parvin, B., Koracin, D., Pavlidis, I., Feris, R., McGraw, T., Elendt, M., Kopper, R., Ragan, E., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 638–645. [Google Scholar] [CrossRef]
- Fuentes, A.; Yoon, S.; Kim, S.C.; Park, D.S. A Robust Deep-Learning-Based Detector for Real-Time Tomato Plant Diseases and Pests Recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef] [Green Version]
- Jeon, H.Y.; Tian, L.F.; Zhu, H. Robust Crop and Weed Segmentation under Uncontrolled Outdoor Illumination. Sensors 2011, 11, 6270–6283. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liao, W.; Nuyttens, D.; Lootens, P.; Vangeyte, J.; Pižurica, A.; He, Y.; Pieters, J.G. Fusion of pixel and object-based features for weed mapping using unmanned aerial vehicle imagery. Int. J. Appl. Earth Obs. Geoinf. 2018, 67, 43–53. [Google Scholar] [CrossRef]
- An, J.; Li, W.; Li, M.; Cui, S.; Yue, H. Identification and Classification of Maize Drought Stress Using Deep Convolutional Neural Network. Symmetry 2019, 11, 256. [Google Scholar] [CrossRef] [Green Version]
- Jiang, B.; Wang, P.; Zhuang, S.; Li, M.; Gong, Z. Drought Stress Detection in the Middle Growth Stage of Maize Based on Gabor Filter and Deep Learning. In Proceedings of the 2019 Chinese Control Conference (CCC), Guangzhou, China, 27–30 July 2019; pp. 7751–7756. [Google Scholar] [CrossRef]
- Zhuang, S.; Wang, P.; Jiang, B.; Li, M. Learned features of leaf phenotype to monitor maize water status in the fields. Comput. Electron. Agric. 2020, 172, 105347. [Google Scholar] [CrossRef]
- Chandel, N.S.; Chakraborty, S.K.; Rajwade, Y.A.; Dubey, K.; Tiwari, M.K.; Jat, D. Identifying crop water stress using deep learning models. Neural Comput. Appl. 2021, 33, 5353–5367. [Google Scholar] [CrossRef]
- Li, H.; Yin, Z.; Manley, P.; Burken, J.G.; Shakoor; Fahlgren, N.; Mockler, T. Early Drought Plant Stress Detection with Bi-Directional Long-Term Memory Networks. Photogramm. Eng. Remote Sens. 2018, 84, 459–468. [Google Scholar] [CrossRef]
- You, J.; Li, X.; Low, M.; Lobell, D.; Ermon, S. Deep Gaussian Process for Crop Yield Prediction Based on Remote Sensing Data. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; Volume 31, pp. 4559–4565. [Google Scholar]
- García-Martínez, H.; Flores-Magdaleno, H.; Ascencio-Hernández, R.; Khalil-Gardezi, A.; Tijerina-Chávez, L.; Mancilla-Villa, O.; Vázquez-Peña, M. Corn Grain Yield Estimation from Vegetation Indices, Canopy Cover, Plant Density, and a Neural Network Using Multispectral and RGB Images Acquired with Unmanned Aerial Vehicles. Agriculture 2020, 10, 277. [Google Scholar] [CrossRef]
- Yang, Q.; Shi, L.; Han, J.; Zha, Y.; Zhu, P. Deep convolutional neural networks for rice grain yield estimation at the ripening stage using UAV-based remotely sensed images. Field Crop. Res. 2019, 235, 142–153. [Google Scholar] [CrossRef]
- Khaki, S.; Wang, L.; Archontoulis, S.V. A CNN-RNN Framework for Crop Yield Prediction. Front. Plant Sci. 2020, 10, 1750. [Google Scholar] [CrossRef]
- Ubbens, J.R.; Stavness, I. Deep Plant Phenomics: A Deep Learning Platform for Complex Plant Phenotyping Tasks. Front. Plant Sci. 2017, 8, 1190. [Google Scholar] [CrossRef] [Green Version]
- Pound, M.P.; Atkinson, J.A.; Wells, D.M.; Pridmore, T.P.; French, A.P. Deep Learning for Multi-task Plant Phenotyping. In Proceedings of the 2017 IEEE International Conference on Computer Vision Workshops (ICCVW), Venice, Italy, 22–29 October 2017; pp. 2055–2063. [Google Scholar]
- Falk, K.G.; Jubery, T.Z.; Mirnezami, S.V.; Parmley, K.A.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B.; Singh, A.K. Computer vision and machine learning enabled soybean root phenotyping pipeline. Plant Methods 2020, 16, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Namin, S.T.; Esmaeilzadeh, M.; Najafi, M.; Brown, T.B.; Borevitz, J.O. Deep phenotyping: Deep learning for temporal phenotype/genotype classification. Plant Methods 2018, 14, 66. [Google Scholar] [CrossRef] [Green Version]
- Al-Shakarji, N.M.; Kassim, Y.M.; Palaniappan, K. Unsupervised Learning Method for Plant and Leaf Segmentation. In Proceedings of the 2017 IEEE Applied Imagery Pattern Recognition Workshop (AIPR), Washington, DC, USA, 10–12 October 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Douarre, C.; Schielein, R.; Frindel, C.; Gerth, S.; Rousseau, D. Transfer Learning from Synthetic Data Applied to Soil–Root Segmentation in X-Ray Tomography Images. J. Imaging 2018, 4, 65. [Google Scholar] [CrossRef] [Green Version]
- Gardiner, L.-J.; Bansept-Basler, P.; El-Soda, M.; Hall, A.; O’Sullivan, D.M. A framework for gene mapping in wheat demonstrated using the Yr7 yellow rust resistance gene. PLoS ONE 2020, 15, e0231157. [Google Scholar] [CrossRef] [Green Version]
- Joynson, R.; Molero, G.; Coombes, B.; Gardiner, L.; Rivera-Amado, C.; Piñera-Chávez, F.J.; Evans, J.R.; Furbank, R.T.; Reynolds, M.P.; Hall, A. Uncovering candidate genes involved in photosynthetic capacity using unexplored genetic variation in Spring Wheat. Plant Biotechnol. J. 2021, 19, 1537–1552. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Usai, M.G.; Goddard, M.E.; Hayes, B.J. LASSO with cross-validation for genomic selection. Genet. Res. 2009, 91, 427–436. [Google Scholar] [CrossRef] [Green Version]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, G.D.L.; Naya, H.; Gianola, D.; Crossa, J.; Legarra, A.; Manfredi, E.; Weigel, K.; Cotes, J.M. Predicting Quantitative Traits with Regression Models for Dense Molecular Markers and Pedigree. Genetics 2009, 182, 375–385. [Google Scholar] [CrossRef] [Green Version]
- Crain, J.; Mondal, S.; Rutkoski, J.; Singh, R.P.; Poland, J. Combining High-Throughput Phenotyping and Genomic Information to Increase Prediction and Selection Accuracy in Wheat Breeding. Plant Genome 2018, 11, 170043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holliday, J.A.; Wang, T.; Aitken, S. Predicting Adaptive Phenotypes from Multilocus Genotypes in Sitka Spruce (Picea sitchensis) Using Random Forest. G3 Genes Genomes Genet. 2012, 2, 1085–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, N.; Gianola, D.; Rosa, G.J.M.; Weigel, K.A. Application of support vector regression to genome-assisted prediction of quantitative traits. Theor. Appl. Genet. 2011, 123, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- González-Camacho, J.M.; Campos, G.D.L.; Pérez, P.; Gianola, D.; Cairns, J.E.; Mahuku, G.; Babu, R.; Crossa, J. Genome-enabled prediction of genetic values using radial basis function neural networks. Theor. Appl. Genet. 2012, 125, 759–771. [Google Scholar] [CrossRef] [Green Version]
- González-Camacho, J.M.; Crossa, J.; Pérez-Rodríguez, P.; Ornella, L.; Gianola, D. Genome-enabled prediction using probabilistic neural network classifiers. BMC Genom. 2016, 17, 208. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Qiu, Z.; Song, J.; Li, J.; Cheng, Q.; Zhai, J.; Ma, C. A deep convolutional neural network approach for predicting phenotypes from genotypes. Planta 2018, 248, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, I.C.; Nascimento, M.; Silva, G.N.; Nascimento, A.C.C.; Cruz, C.D.; Silva, F.F.E.; De Almeida, D.P.; Pestana, K.N.; Azevedo, C.F.; Zambolim, L.; et al. Genomic prediction of leaf rust resistance to Arabica coffee using machine learning algorithms. Sci. Agric. 2021, 78, 20200021. [Google Scholar] [CrossRef]
- Zingaretti, L.M.; Gezan, S.A.; Ferrão, L.F.V.; Osorio, L.F.; Monfort, A.; Muñoz, P.R.; Whitaker, V.M.; Pérez-Enciso, M. Exploring Deep Learning for Complex Trait Genomic Prediction in Polyploid Outcrossing Species. Front. Plant Sci. 2020, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandhu, K.S.; Lozada, D.N.; Zhang, Z.; Pumphrey, M.O.; Carter, A.H. Deep Learning for Predicting Complex Traits in Spring Wheat Breeding Program. Front. Plant Sci. 2021, 11, 613325. [Google Scholar] [CrossRef]
- Azodi, C.B.; Bolger, E.; McCarren, A.; Roantree, M.; Campos, G.D.L.; Shiu, S.-H. Benchmarking Parametric and Machine Learning Models for Genomic Prediction of Complex Traits. G3 Genes Genomes Genet. 2019, 9, 3691–3702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spindel, J.; Begum, H.; Akdemir, D.; Virk, P.; Collard, B.; Redoña, E.; Atlin, G.; Jannink, J.-L.; McCouch, S.R. Genomic Selection and Association Mapping in Rice (Oryza sativa): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines. PLoS Genet. 2015, 11, e1004982. [Google Scholar] [CrossRef] [Green Version]
- You, Q.; Yang, X.; Peng, Z.; Xu, L.; Wang, J. Development and Applications of a High Throughput Genotyping Tool for Polyploid Crops: Single Nucleotide Polymorphism (SNP) Array. Front. Plant Sci. 2018, 9, 104. [Google Scholar] [CrossRef] [Green Version]
- Korani, W.; Clevenger, J.; Chu, Y.; Ozias-Akins, P. Machine Learning as an Effective Method for Identifying True Single Nucleotide Polymorphisms in Polyploid Plants. Plant Genome 2019, 12, 180023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mochida, K.; Koda, S.; Inoue, K.; Nishii, R. Statistical and Machine Learning Approaches to Predict Gene Regulatory Networks from Transcriptome Datasets. Front. Plant Sci. 2018, 9, 1770. [Google Scholar] [CrossRef]
- Huynh-Thu, V.A.; Irrthum, A.; Wehenkel, L.; Geurts, P. Inferring Regulatory Networks from Expression Data Using Tree-Based Methods. PLoS ONE 2010, 5, e12776. [Google Scholar] [CrossRef]
- Agarwal, V.; Shendure, J. Predicting mRNA Abundance Directly from Genomic Sequence Using Deep Convolutional Neural Networks. Cell Rep. 2020, 31, 107663. [Google Scholar] [CrossRef]
- Beer, M.A.; Tavazoie, S. Predicting Gene Expression from Sequence. Cell 2004, 117, 185–198. [Google Scholar] [CrossRef] [Green Version]
- Hafez, D.; Karabacak, A.; Krueger, S.; Hwang, Y.-C.; Wang, L.-S.; Zinzen, R.P.; Ohler, U. McEnhancer: Predicting gene expression via semi-supervised assignment of enhancers to target genes. Genome Biol. 2017, 18, 199. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Lanchantin, J.; Robins, G.; Qi, Y. DeepChrome: Deep-learning for predicting gene expression from histone modifications. Bioinformatics 2016, 32, i639–i648. [Google Scholar] [CrossRef]
- Natarajan, A.; Yardımcı, G.G.; Sheffield, N.C.; Crawford, G.E.; Ohler, U. Predicting cell-type–specific gene expression from regions of open chromatin. Genome Res. 2012, 22, 1711–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Washburn, J.D.; Mejia-Guerra, M.K.; Ramstein, G.; Kremling, K.A.; Valluru, R.; Buckler, E.S.; Wang, H. Evolutionarily informed deep learning methods for predicting relative transcript abundance from DNA sequence. Proc. Natl. Acad. Sci. USA 2019, 116, 5542–5549. [Google Scholar] [CrossRef] [Green Version]
- Gardiner, L.-J.; Rusholme-Pilcher, R.; Colmer, J.; Rees, H.; Crescente, J.M.; Carrieri, A.P.; Duncan, S.; Pyzer-Knapp, E.O.; Krishna, R.; Hall, A. Interpreting machine learning models to investigate circadian regulation and facilitate exploration of clock function. Genomics 2021. preprint. [Google Scholar] [CrossRef]
- Ramírez-González, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; van Ex, F.; Pasha, A.; et al. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardiner, L.-J.; Joynson, R.; Omony, J.; Rusholme-Pilcher, R.; Olohan, L.; Lang, D.; Bai, C.; Hawkesford, M.; Salt, D.; Spannagl, M.; et al. Hidden variation in polyploid wheat drives local adaptation. Genome Res. 2018, 28, 1319–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardiner, L.-J. Understanding DNA Methylation Patterns in Wheat. In Plant Epigenetics and Epigenomics; Spillane, C., McKeown, P., Eds.; Springer: New York, NY, USA, 2020; Volume 2093, pp. 33–46. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Kuhn, M.; Vizcaíno, J.A.; Hitz, M.-P.; Audain, E. Accurate and fast feature selection workflow for high-dimensional omics data. PLoS ONE 2017, 12, e0189875. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Zhang, N.; Wang, Y.-G.; George, A.W.; Reverter, A.; Li, Y. Genomic Prediction of Breeding Values Using a Subset of SNPs Identified by Three Machine Learning Methods. Front. Genet. 2018, 9, 237. [Google Scholar] [CrossRef]
- Degenhardt, F.; Seifert, S.; Szymczak, S. Evaluation of variable selection methods for random forests and omics data sets. Brief. Bioinform. 2019, 20, 492–503. [Google Scholar] [CrossRef] [Green Version]
- Haas, R.; Zelezniak, A.; Iacovacci, J.; Kamrad, S.; Townsend, S.; Ralser, M. Designing and interpreting ‘multi-omic’ experiments that may change our understanding of biology. Curr. Opin. Syst. Biol. 2017, 6, 37–45. [Google Scholar] [CrossRef]
- Vert, J.-P. Reconstruction of Biological Networks by Supervised Machine Learning Approaches. In Elements of Computational Systems Biology; Lodhi, H.M., Muggleton, S.H., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010; pp. 163–188. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Wei, S.; Huang, Z.; Tang, H.; Zhang, J.; Ding, Z.; Huang, K. MORONET: Multi-omics Integration via Graph Convolutional Networks for Biomedical Data Classification. bioRxiv 2020. [Google Scholar] [CrossRef]
- Xiaoxue, L.; Xuesong, B.; Longhe, W.; Bingyuan, R.; Shuhan, L.; Lin, L. Review and Trend Analysis of Knowledge Graphs for Crop Pest and Diseases. IEEE Access 2019, 7, 62251–62264. [Google Scholar] [CrossRef]
- Timón-Reina, S.; Rincón, M.; Martínez-Tomás, R. An overview of graph databases and their applications in the biomedical domain. Database 2021, 2021, baab026. [Google Scholar] [CrossRef]
- Dai, X.; Li, J.; Liu, T.; Zhao, P.X. HRGRN: A Graph Search-Empowered Integrative Database of Arabidopsis Signaling Transduction, Metabolism and Gene Regulation Networks. Plant Cell Physiol. 2015, 57, e12. [Google Scholar] [CrossRef] [Green Version]
- Venkatesan, A.; Ngompe, G.T.; El Hassouni, N.; Chentli, I.; Guignon, V.; Jonquet, C.; Ruiz, M.; Larmande, P. Agronomic Linked Data (AgroLD): A knowledge-based system to enable integrative biology in agronomy. PLoS ONE 2018, 13, e0198270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassani-Pak, K.; Singh, A.; Brandizi, M.; Hearnshaw, J.; Parsons, J.D.; Amberkar, S.; Phillips, A.L.; Doonan, J.H.; Rawlings, C. KnetMiner: A comprehensive approach for supporting evidence-based gene discovery and complex trait analysis across species. Plant Biotechnol. J. 2021, 19, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Harfouche, A.J.; Jacobson, D.A.; Kainer, D.; Romero, J.C.; Harfouche, A.H.; Mugnozza, G.S.; Moshelion, M.; Tuskan, G.A.; Keurentjes, J.J.; Altman, A. Accelerating Climate Resilient Plant Breeding by Applying Next-Generation Artificial Intelligence. Trends Biotechnol. 2019, 37, 1217–1235. [Google Scholar] [CrossRef]
- Ghosal, S.; Blystone, D.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. An explainable deep machine vision framework for plant stress phenotyping. Proc. Natl. Acad. Sci. USA 2018, 115, 4613–4618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, S.J.; Furbank, R.T. Explainable machine learning models of major crop traits from satellite-monitored continent-wide field trial data. Nat. Plants 2021, 7, 1354–1363. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gardiner, L.-J.; Krishna, R. Bluster or Lustre: Can AI Improve Crops and Plant Health? Plants 2021, 10, 2707. https://doi.org/10.3390/plants10122707
Gardiner L-J, Krishna R. Bluster or Lustre: Can AI Improve Crops and Plant Health? Plants. 2021; 10(12):2707. https://doi.org/10.3390/plants10122707
Chicago/Turabian StyleGardiner, Laura-Jayne, and Ritesh Krishna. 2021. "Bluster or Lustre: Can AI Improve Crops and Plant Health?" Plants 10, no. 12: 2707. https://doi.org/10.3390/plants10122707
APA StyleGardiner, L.-J., & Krishna, R. (2021). Bluster or Lustre: Can AI Improve Crops and Plant Health? Plants, 10(12), 2707. https://doi.org/10.3390/plants10122707