Concerto on Chromatin: Interplays of Different Epigenetic Mechanisms in Plant Development and Environmental Adaptation
Abstract
1. Introduction
2. Epigenetic Mechanisms Regulating Plant Gene Expression
3. Interplays of Different Epigenetic Marks Regulating Plant Gene Expression
4. Molecular Basis for Epigenetic Interplays in Plant Development and Environmental Adaptation
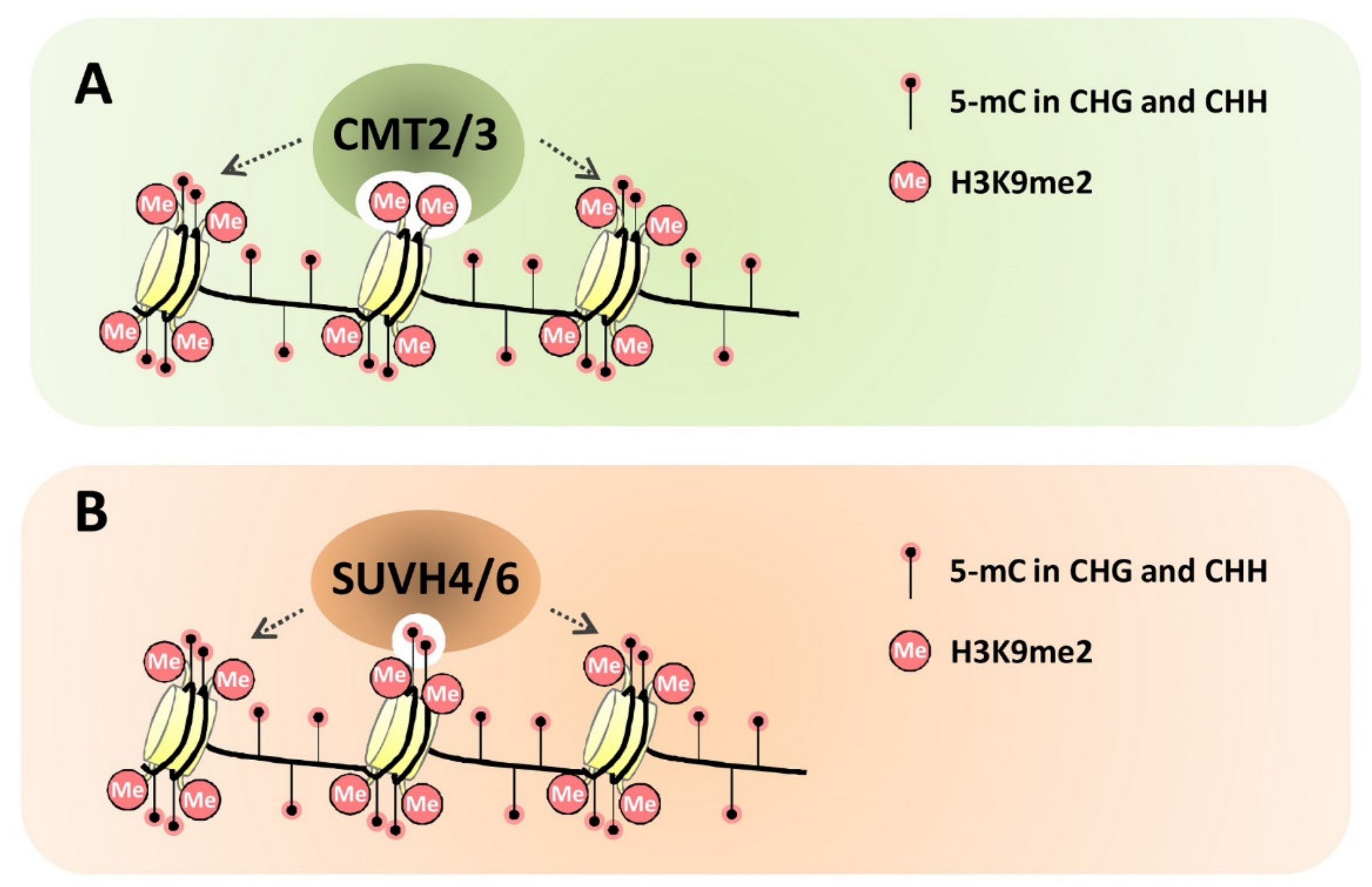
4.1. Structural Basis for the Direct Links between Non-CG DNA Methylation and Histone H3K9me2
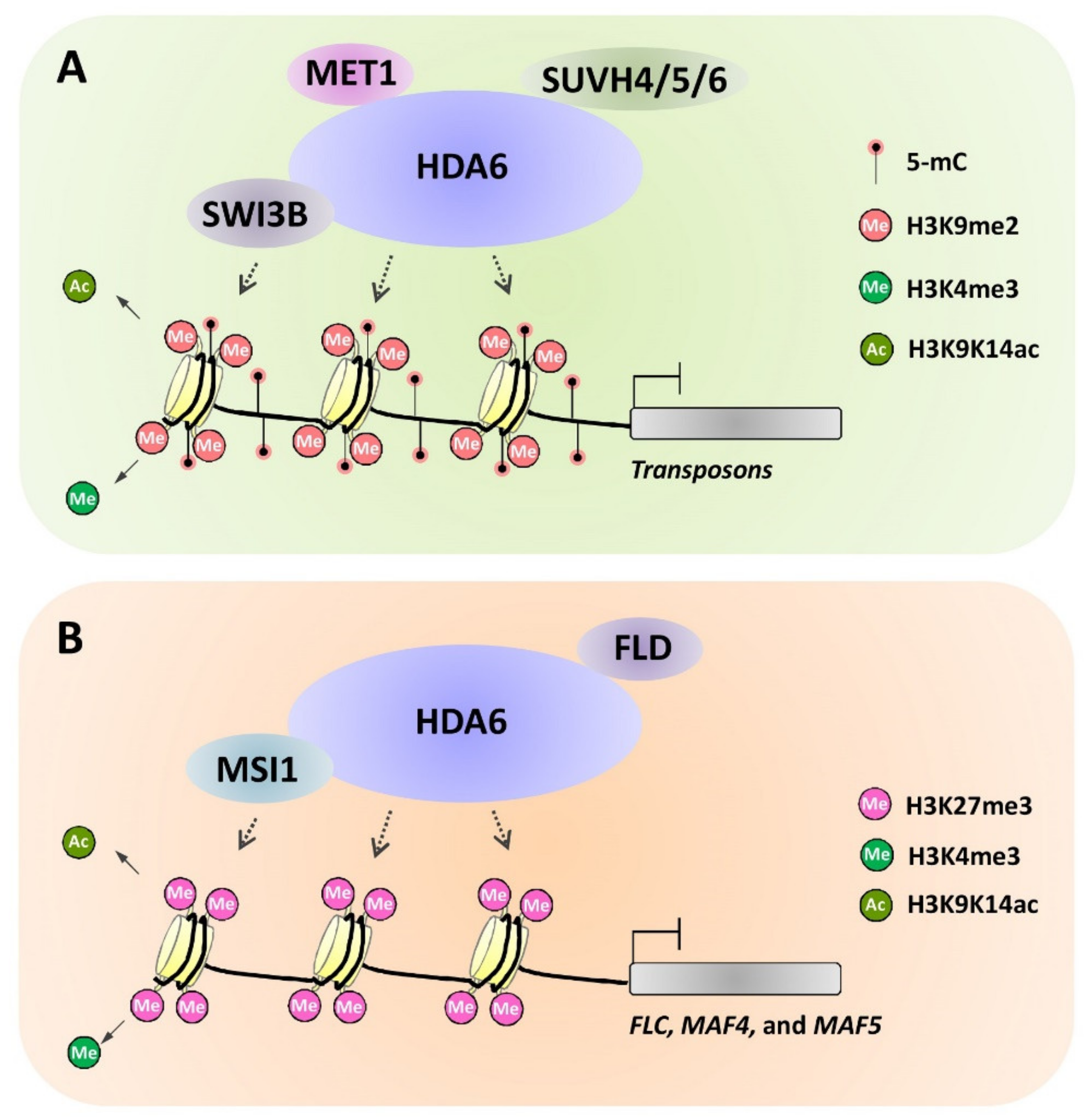
4.2. Physical Basis for the Epigenetic Interplayes Mediated by Chromatin Modifier Complex
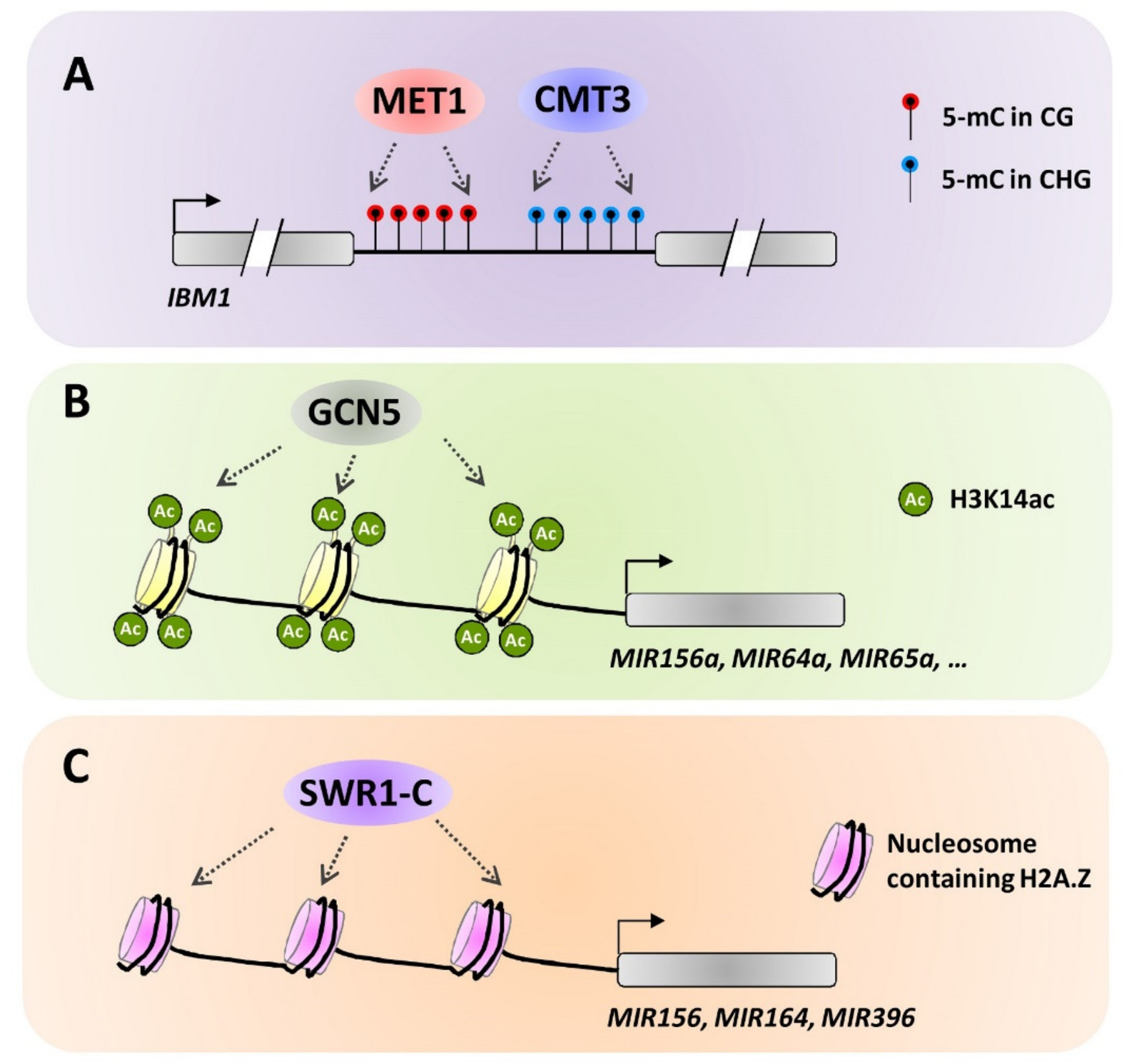
4.3. Trancriptional Basis for the Cross-Regulation of Different Epigenetic Mechanisms
4.4. Metabolic Basis for the Crosstalk of DNA and Histone Methylation
5. Concluding Remarks and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, J.H. Multifaceted chromatin structure and transcription changes in plant stress response. Int. J. Mol. Sci. 2021, 22, 2013. [Google Scholar] [CrossRef] [PubMed]
- Asensi-Fabado, M.A.; Amtmann, A.; Perrella, G. Plant responses to abiotic stress: The chromatin context of transcriptional regulation. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Song, X.; Wei, L.; Liu, C.; Cao, X. Epigenetic regulation and epigenomic landscape in rice. Natl. Sci. Rev. 2016, 3, 309–327. [Google Scholar] [CrossRef]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Kim, M.Y.; Vickers, M.; Park, J.S.; Hyun, Y.; Okamoto, T.; Zilberman, D.; Fischer, R.L.; Feng, X.; Choi, Y.; et al. DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc. Natl. Acad. Sci. USA 2016, 113, 15138–15143. [Google Scholar] [CrossRef]
- Zhi, P.; Chang, C. Exploiting epigenetic variations for crop disease resistance improvement. Front. Plant Sci. 2021, 12, 692328. [Google Scholar] [CrossRef]
- Kong, L.; Liu, Y.; Wang, X.; Chang, C. Insight into the role of epigenetic processes in abiotic and biotic stress response in wheat and barley. Int. J. Mol. Sci. 2020, 21, 1480. [Google Scholar] [CrossRef]
- Wang, J.; Meng, X.; Yuan, C.; Harrison, A.P.; Chen, M. The roles of cross-talk epigenetic patterns in Arabidopsis thaliana. Brief. Funct. Genom. 2016, 15, 278–287. [Google Scholar] [CrossRef]
- Lee, J.S.; Smith, E.; Shilatifard, A. The language of histone crosstalk. Cell 2010, 142, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Li, A.; Yu, B.; Li, S. Interplay between miRNAs and lncRNAs: Mode of action and biological roles in plant development and stress adaptation. Comput. Struct. Biotechnol. J. 2021, 19, 2567–2574. [Google Scholar] [CrossRef]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications-writers that read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Elhamamsy, A.R. DNA methylation dynamics in plants and mammals: Overview of regulation and dysregulation. Cell Biochem. Funct. 2016, 34, 289–298. [Google Scholar] [CrossRef]
- Walker, J.; Zhang, J.; Liu, Y.; Vickers, M.; Dolan, L.; Nakajima, K.; Feng, X. Extensive N4 cytosine methylation is essential for Marchantia sperm function. bioRxiv 2021. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA Methylation. PLoS Genet. 2020, 16, e1009034. [Google Scholar] [CrossRef] [PubMed]
- Yaari, R.; Katz, A.; Domb, K.; Harris, K.D.; Zemach, A.; Ohad, N. RdDM-independent de novo and heterochromatin DNA methylation by plant CMT and DNMT3 orthologs. Nat. Commun. 2019, 10, 1613. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, H.; Dong, Y.; Hao, Y.; Zhang, X. DNA METHYLTRANSFERASE1-mediated shoot regeneration is regulated by cytokinin-induced cell cycle in Arabidopsis. New Phytol. 2018, 217, 219–232. [Google Scholar] [CrossRef]
- Zhu, J.K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef]
- Gehring, M.; Reik, W.; Henikoff, S. DNA demethylation by DNA repair. Trends Genet. 2009, 25, 82–90. [Google Scholar] [CrossRef]
- Tang, K.; Lang, Z.; Zhang, H.; Zhu, J.K. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat. Plants 2016, 2, 16169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Zilberman, D.; Gehring, M.; Tran, R.K.; Ballinger, T.; Henikoff, S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007, 39, 61–69. [Google Scholar] [CrossRef]
- Tessarz, P.; Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Janack, B.; Sosoi, P.; Krupinska, K.; Humbeck, K. Knockdown of WHIRLY1 affects drought stress-induced leaf senescence and histone modifications of the senescence-associated gene HvS40. Plants 2016, 5, 37. [Google Scholar] [CrossRef]
- Ay, N.; Janack, B.; Fischer, A.; Reuter, G.; Humbeck, K. Alterations of histone modifications at the senescence-associated gene HvS40 in barley during senescence. Plant Mol. Biol. 2015, 89, 127–141. [Google Scholar] [CrossRef]
- Imhof, A.; Wolffe, A.P. Transcription: Gene control by targeted histone acetylation. Curr. Biol. 1998, 8, R422–R424. [Google Scholar] [CrossRef]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Jin, R.; Yu, X.; Shen, M.; Wagner, J.D.; Pai, A.; Song, C.; Zhuang, M.; Klasfeld, S.; He, C.; et al. Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 2017, 49, 1546–1552. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef] [PubMed]
- Casas-Mollano, J.A.; Jeong, B.R.; Xu, J.; Moriyama, H.; Cerutti, H. The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc. Natl. Acad. Sci. USA 2008, 105, 6486–6491. [Google Scholar] [CrossRef]
- Weake, V.M.; Workman, J.L. Histone ubiquitination: Triggering gene activity. Mol. Cell 2008, 29, 653–663. [Google Scholar] [CrossRef]
- Vaughan, R.M.; Kupai, A.; Rothbart, S.B. Chromatin regulation through ubiquitin and ubiquitin-like histone modifications. Trends Biochem. Sci. 2021, 46, 258–269. [Google Scholar] [CrossRef]
- Han, S.K.; Wu, M.F.; Cui, S.; Wagner, D. Roles and activities of chromatin remodeling ATPases in plants. Plant J. 2015, 83, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.T.; Liu, J.X.; Han, J.J. Chromatin remodeling factors regulate environmental stress responses in plants. J. Integr. Plant Biol. 2021, 63, 438–450. [Google Scholar] [CrossRef] [PubMed]
- Fenley, A.T.; Anandakrishnan, R.; Kidane, Y.H.; Onufriev, A.V. Modulation of nucleosomal DNA accessibility via charge-altering post-translational modifications in histone core. Epigenet. Chromatin 2018, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Li, S.; Landsman, D.; Panchenko, A.R. Histone tails as signaling antennas of chromatin. Curr. Opin. Struct. Biol. 2021, 67, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhi, P.; Fan, Q.; Zhang, M.; Chang, C. Wheat CHD3 protein TaCHR729 regulates the cuticular wax biosynthesis required for stimulating germination of Blumeria graminis f. sp. tritici. J. Exp. Bot. 2019, 70, 701–713. [Google Scholar] [CrossRef]
- Lu, Y.; Tan, F.; Zhao, Y.; Zhou, S.; Chen, X.; Hu, Y.; Zhou, D.X. A Chromodomain-helicase-DNA-binding factor functions in chromatin modification and gene regulation. Plant Physiol. 2020, 183, 1035–1046. [Google Scholar] [CrossRef]
- Zou, B.; Sun, Q.; Zhang, W.; Ding, Y.; Yang, D.L.; Shi, Z.; Hua, J. The Arabidopsis chromatin-remodeling factor CHR5 regulates plant immune responses and nucleosome occupancy. Plant Cell Physiol. 2017, 58, 2202–2216. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Hong, Y.; Ren, Z.; Tang, K.; Zhang, H.; Zhu, J.K.; Zhao, C. A role for PICKLE in the regulation of cold and salt stress tolerance in Arabidopsis. Front. Plant Sci. 2019, 10, 900. [Google Scholar] [CrossRef] [PubMed]
- Archacki, R.; Yatuseich, R.; Buszewicz, D.; Krzyczmonik, K.; Patryn, J.; Iwanicka-Nowicka, R.; Biecek, P.; Wilczynski, B.; Koblowska, M.; Jerzmanowski, A. Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucleic Acids Res. 2017, 45, 3116–3129. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Meng, X.; Dobrovolskaya, O.B.; Orlov, Y.L.; Chen, M. Non-coding RNAs and their roles in stress response in plants. Genom. Proteom. Bioinform. 2017, 15, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Chen, R.; Qu, L.; Cao, X. Noncoding RNA: From dark matter to bright star. Sci. China Life Sci. 2020, 63, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant noncoding RNAs: Hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, D. The role of long noncoding rnas in plant stress tolerance. Methods Mol. Biol. 2017, 1631, 41–68. [Google Scholar]
- Sun, X.; Zhang, H.; Sui, N. Regulation mechanism of long non-coding RNA in plant response to stress. Biochem. Biophys. Res. Commun. 2018, 503, 402–407. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Chen, L.; Zhu, Q.H.; Kaufmann, K. Long non-coding RNAs in plants: Emerging modulators of gene activity in development and stress responses. Planta 2020, 252, 92. [Google Scholar] [CrossRef]
- Wu, L.; Liu, S.; Qi, H.; Cai, H.; Xu, M. Research progress on plant long non-coding RNA. Plants 2020, 9, 408. [Google Scholar] [CrossRef] [PubMed]
- Brant, E.J.; Budak, H. Plant small non-coding RNAs and their roles in biotic stresses. Front. Plant Sci. 2018, 9, 1038. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.J.; Harrison, J.; Paul, C.L.; Frommer, M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994, 22, 2990–2997. [Google Scholar]
- Feng, L.; Lou, J. DNA methylation analysis. Methods Mol. Biol. 2019, 1894, 181–227. [Google Scholar] [PubMed]
- Guevara, M.Á.; de María, N.; Sáez-Laguna, E.; Vélez, M.D.; Cervera, M.T.; Cabezas, J.A. Analysis of DNA cytosine methylation patterns using methylation-sensitive amplification polymorphism (MSAP). Methods Mol. Biol. 2017, 1456, 99–112. [Google Scholar]
- Hsu, H.K.; Weng, Y.I.; Hsu, P.Y.; Huang, T.H.; Huang, Y.W. Detection of DNA methylation by MeDIP and MBDCap assays: An overview of techniques. Methods Mol. Biol. 2020, 2102, 225–234. [Google Scholar]
- Li, Q.; Hermanson, P.J.; Springer, N.M. Detection of DNA methylation by whole-genome bisulfite sequencing. Methods Mol. Biol. 2018, 1676, 185–196. [Google Scholar]
- Roudier, F.; Ahmed, I.; Bérard, C.; Sarazin, A.; Mary-Huard, T.; Cortijo, S.; Bouyer, D.; Caillieux, E.; Duvernois-Berthet, E.; Al-Shikhley, L.; et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011, 30, 1928–1938. [Google Scholar] [CrossRef]
- Ha, M.; Ng, D.W.; Li, W.H.; Chen, Z.J. Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res. 2011, 21, 590–598. [Google Scholar] [CrossRef]
- To, T.K.; Kim, J.M.; Matsui, A.; Kurihara, Y.; Morosawa, T.; Ishida, J.; Tanaka, M.; Endo, T.; Kakutani, T.; Toyoda, T.; et al. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 2011, 7, e1002055. [Google Scholar] [CrossRef]
- Trejo-Arellano, M.S.; Mahrez, W.; Nakamura, M.; Moreno-Romero, J.; Nanni, P.; Köhler, C.; Hennig, L. H3K23me1 is an evolutionarily conserved histone modification associated with CG DNA methylation in Arabidopsis. Plant J. 2017, 90, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, V.V.; Kapoor, A.; Zhang, K.; Zhu, J.; Zhou, T.; Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 2007, 447, 735–738. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zhang, W.; Marand, A.P.; Zhu, B.; Buell, C.R.; Jiang, J. Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato. Genome Biol. 2019, 20, 123. [Google Scholar] [CrossRef]
- Kumar, S.V. H2A.Z at the core of transcriptional regulation in plants. Mol. Plant 2018, 11, 1112–1114. [Google Scholar] [CrossRef]
- Su, Y.; Wang, S.; Zhang, F.; Zheng, H.; Liu, Y.; Huang, T.; Ding, Y. Phosphorylation of histone H2A at serine 95: A plant-specific mark involved in flowering time regulation and H2A.Z deposition. Plant Cell 2017, 29, 2197–2213. [Google Scholar] [CrossRef]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef]
- Johnson, L.M.; Bostick, M.; Zhang, X.; Kraft, E.; Henderson, I.; Callis, J.; Jacobsen, S.E. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 2007, 17, 379–384. [Google Scholar] [CrossRef]
- Du, J.; Zhong, X.; Bernatavichute, Y.V.; Stroud, H.; Feng, S.; Caro, E.; Vashisht, A.A.; Terragni, J.; Chin, H.G.; Tu, A.; et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 2012, 151, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Stroud, H.; Do, T.; Du, J.; Zhong, X.; Feng, S.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2014, 21, 64–72. [Google Scholar] [CrossRef]
- Du, J.; Johnson, L.M.; Groth, M.; Feng, S.; Hale, C.J.; Li, S.; Vashisht, A.A.; Wohlschlegel, J.A.; Patel, D.J.; Jacobsen, S.E. Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol. Cell 2014, 55, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Harris, C.J.; Zhong, Z.; Chen, W.; Liu, R.; Jia, B.; Wang, Z.; Li, S.; Jacobsen, S.E.; Du, J. Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation. Proc. Natl. Acad. Sci. USA 2018, 115, E8793–E8802. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Luo, M.; Wu, K. Epigenetic interplay of histone modifications and DNA methylation mediated by HDA6. Plant Signal. Behav. 2012, 7, 633–635. [Google Scholar] [CrossRef]
- Liu, X.; Yu, C.W.; Duan, J.; Luo, M.; Wang, K.; Tian, G.; Cui, Y.; Wu, K. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 2012, 158, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Tian, J.; Luo, R.; Kang, Y.; Lu, Y.; Hu, Y.; Liu, N.; Zhang, J.; Cheng, H.; Niu, S.; et al. MiR399d and epigenetic modification comodulate anthocyanin accumulation in Malus leaves suffering from phosphorus deficiency. Plant Cell Environ. 2020, 43, 1148–1159. [Google Scholar] [CrossRef]
- Wang, B.; Yang, X.; Wang, Y.; Xie, Y.; Zhou, X. Tomato yellow leaf curl virus V2 interacts with host histone deacetylase 6 to suppress methylation-mediated transcriptional gene silencing in plants. J. Virol. 2018, 92, e00036-18. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Wöhrmann, H.J.; Raissig, M.T.; Arand, J.; Gheyselinck, J.; Gagliardini, V.; Heichinger, C.; Walter, J.; Grossniklaus, U. The Polycomb group protein MEDEA and the DNA methyltransferase MET1 interact to repress autonomous endosperm development in Arabidopsis. Plant J. 2013, 73, 776–787. [Google Scholar] [CrossRef]
- Han, Y.F.; Dou, K.; Ma, Z.Y.; Zhang, S.W.; Huang, H.W.; Li, L.; Cai, T.; Chen, S.; Zhu, J.K.; He, X.J. SUVR2 is involved in transcriptional gene silencing by associating with SNF2-related chromatin-remodeling proteins in Arabidopsis. Cell Res. 2014, 24, 1445–1465. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, J.M.; To, T.K.; Seki, M. An epigenetic integrator: New insights into genome regulation, environmental stress responses and developmental controls by histone deacetylase 6. Plant Cell Physiol. 2012, 53, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Wang, Y.Y.; Liu, X.; Yang, S.; Lu, Q.; Cui, Y.; Wu, K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012, 63, 3297–3306. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhi, P.; Wang, X.; Fan, Q.; Chang, C. Wheat WD40-repeat protein TaHOS15 functions in a histone deacetylase complex to fine-tune defense responses to Blumeria graminis f.sp. tritici. J. Exp. Bot. 2019, 70, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Zhi, P.; Kong, L.; Liu, J.; Zhang, X.; Wang, X.; Li, H.; Sun, M.; Li, Y.; Chang, C. Histone deacetylase TaHDT701 functions in TaHDA6-TaHOS15 complex to regulate wheat defense responses to Blumeriagraminis f.sp. tritici. Int. J. Mol. Sci. 2020, 21, 2640. [Google Scholar] [CrossRef]
- Yu, C.W.; Tai, R.; Wang, S.C.; Yang, P.; Luo, M.; Yang, S.; Cheng, K.; Wang, W.C.; Cheng, Y.S.; Wu, K. HISTONE DEACETYLASE6 acts in concert with Histone Methyltransferases SUVH4, SUVH5, and SUVH6 to regulate transposon silencing. Plant Cell 2017, 29, 1970–1983. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yuan, L.; Yen, M.R.; Zheng, F.; Ji, R.; Peng, T.; Gu, D.; Yang, S.; Cui, Y.; Chen, P.Y.; et al. SWI3B and HDA6 interact and are required for transposon silencing in Arabidopsis. Plant J. 2020, 102, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.W.; Liu, X.; Luo, M.; Chen, C.; Lin, X.; Tian, G.; Lu, Q.; Cui, Y.; Wu, K. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 2011, 156, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Qing, L.; Yuan, L.; Huang, Y.; Hung, F.Y.; Wu, K.; Yang, S. MSI1 and HDA6 function interdependently to control flowering time via chromatin modifications. Plant J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Buszewicz, D.; Archacki, R.; Palusiński, A.; Kotliński, M.; Fogtman, A.; Iwanicka-Nowicka, R.; Sosnowska, K.; Kuciński, J.; Pupel, P.; Olędzki, J.; et al. HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 2016, 39, 2108–2122. [Google Scholar] [CrossRef]
- Rigal, M.; Kevei, Z.; Pélissier, T.; Mathieu, O. DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J. 2012, 31, 2981–2993. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Tian, M.; Ci, D.; Zhang, D. Methylation of microRNA genes regulates gene expression in bisexual flower development in andromonoecious poplar. J. Exp. Bot. 2015, 66, 1891–1905. [Google Scholar] [CrossRef]
- Kim, W.; Benhamed, M.; Servet, C.; Latrasse, D.; Zhang, W.; Delarue, M.; Zhou, D.X. Histone acetyltransferase GCN5 interferes with the miRNA pathway in Arabidopsis. Cell Res. 2009, 19, 899–909. [Google Scholar] [CrossRef]
- Choi, K.; Kim, J.; Müller, S.Y.; Oh, M.; Underwood, C.; Henderson, I.; Lee, I. Regulation of microRNA-mediated developmental changes by the SWR1 chromatin remodeling complex. Plant Physiol. 2016, 171, 1128–1143. [Google Scholar] [PubMed]
- Hou, N.; Cao, Y.; Li, F.; Yuan, W.; Bian, H.; Wang, J.; Zhu, M.; Han, N. Epigenetic regulation of miR396 expression by SWR1-C and the effect of miR396 on leaf growth and developmental phase transition in Arabidopsis. J. Exp. Bot. 2019, 70, 5217–5229. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.R.; Zhang, F.F.; Ma, Z.Y.; Huang, H.W.; Jiang, L.; Cai, T.; Zhu, J.K.; Zhang, C.; He, X.J. Folate polyglutamylation is involved in chromatin silencing by maintaining global DNA methylation and histone H3K9 dimethylation in Arabidopsis. Plant Cell 2013, 25, 2545–2559. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Ma, L.; Pang, H.; Wang, P.; Liu, L.; Cheng, Y.; Cheng, J.; Guo, Y.; Li, Q. Methionine synthase1 is involved in chromatin silencing by maintaining DNA and histone methylation. Plant Physiol. 2019, 181, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Wang, L.; Wang, J.; Zhao, X.; Cheng, J.; Yu, W.; Jin, D.; Li, Q.; Gong, Z. Methionine adenosyltransferase4 mediates DNA and histone methylation. Plant Physiol. 2018, 177, 652–670. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Chang, C. Concerto on Chromatin: Interplays of Different Epigenetic Mechanisms in Plant Development and Environmental Adaptation. Plants 2021, 10, 2766. https://doi.org/10.3390/plants10122766
Liu J, Chang C. Concerto on Chromatin: Interplays of Different Epigenetic Mechanisms in Plant Development and Environmental Adaptation. Plants. 2021; 10(12):2766. https://doi.org/10.3390/plants10122766
Chicago/Turabian StyleLiu, Jiao, and Cheng Chang. 2021. "Concerto on Chromatin: Interplays of Different Epigenetic Mechanisms in Plant Development and Environmental Adaptation" Plants 10, no. 12: 2766. https://doi.org/10.3390/plants10122766
APA StyleLiu, J., & Chang, C. (2021). Concerto on Chromatin: Interplays of Different Epigenetic Mechanisms in Plant Development and Environmental Adaptation. Plants, 10(12), 2766. https://doi.org/10.3390/plants10122766






