Abstract
Drought is the most prevalent unfavorable condition that impairs plant growth and development by altering morphological, physiological, and biochemical functions, thereby impeding plant biomass production. To survive the adverse effects, water limiting condition triggers a sophisticated adjustment mechanism orchestrated mainly by hormones that directly protect plants via the stimulation of several signaling cascades. Predominantly, water deficit signals cause the increase in the level of endogenous ABA, which elicits signaling pathways involving transcription factors that enhance resistance mechanisms to combat drought-stimulated damage in plants. These responses mainly include stomatal closure, seed dormancy, cuticular wax deposition, leaf senescence, and alteration of the shoot and root growth. Unraveling how plants adjust to drought could provide valuable information, and a comprehensive understanding of the resistance mechanisms will help researchers design ways to improve crop performance under water limiting conditions. This review deals with the past and recent updates of ABA-mediated molecular mechanisms that plants can implement to cope with the challenges of drought stress.
1. Introduction
As sessile organisms, plants are exposed to an ever-changing environment during their entire lifespan. Plants have acquired adaptive physiological and molecular mechanisms by diverse mitigating strategies in the course of evolution to cope with unfavorable conditions. Predominantly, plants are exposed to both abiotic and biotic stress at various stages of their development. Biotic stress includes the attack by pests and pathogens, e.g., bacteria, viruses, fungi, and nematodes, whereas abiotic stress includes drought, flooding, temperature, salinity, nutrient deficiency, heavy metals, and ultraviolet radiation [1,2]. Consequently, they adversely affect the productivity of the crop, which is a major obstacle to attaining global food security essential for the continuously growing world population. Among the abiotic stresses, drought has become a major plague as a result of climate-change scenarios around the world and a certain percentage of developing countries will face water scarcities by 2030 (FAO 2003) [3]. In the least developed countries (LDCs) and low to middle-income countries (LMICs), over 34% of crop and livestock production was reduced by drought from 2008-2018 (FAO 2021) [4]. Among the world’s major crops, rice (>50% yield reduction) was more sensitive towards drought compared to maize (39.3% yield reduction) and wheat (20.6% yield reduction) under comparable water reduction (approximately 40%) [5,6]. Under drought severity growing from moderate to exceptional, the yield loss risk is anticipated to increase by 9%–12%, 5.6%–6.3%, and 18.1%–19.4% in wheat, maize, and rice, respectively [7].
Water scarcity in the soil directly constrains physiological functions in plants, such as leaf growth, photosynthetic capacity, nutrient uptake, stomatal conductance etc. [8]. The limited water availability initiates when the transpiration rate is higher than the amount of water absorbed by plant roots [9]. Typically, plants need water from seedling to the reproductive stage to transport nutrients, for photosynthesis, and to maintain the turgidity of cell walls and various cellular processes [10]. Not having adequate water poses a severe threat to seed germination and seedling growth in plants with decreasing osmotic potential [11,12,13,14,15,16]. Prolonged drought condition reduces uptake of nutrients by root, leaf water potential, transpiration rate, water-use efficiency and stomatal conductance essential for both vegetative and reproductive growth [11]. Under drought, the interruption of water flow between xylem and adjacent cells negatively affects turgor pressure, which in turn decreases cell elongation, consequently resulting in reduced plant height, leaf area, and crop yield [17]. A decline in stomatal conductance is responsible for a net reduction in photosynthesis due to inhibited CO2 assimilation [18]. These physiological changes lead to the production of reactive oxygen species (ROS), which promotes oxidative stress via the impairment of cell membranes, nucleic acids, and proteins to destabilize cellular functions [19,20]. To abate the effects of drought stress, plants respond in very complex ways, bringing morphological and physiological adaptation. Such processes include closing stomata, increased root growth, decreased stem and leaf expansion, cuticular wax biosynthesis, and shortening life cycle [17,21]. In addition, plants have evolved efficient antioxidant machinery, including enzymatic and non-enzymatic systems to attenuate the effect of ROS [19]. The promoter of catalase gene Cat2 in wheat contains ARE (antioxidant responsive element), which is induced by H2O2 during drought [22]. Besides, plants also regulate osmotic adjustment via the accumulation of soluble sugars, free amino acid, and proline required for normal cellular homeostasis [23]. To counter drought stress by triggering different mitigating strategies is directly controlled by a well synchronization of gene expression, such as RD22, RD29B, RD20A, Gly (glyoxalase I family) etc., predominantly modulated by multiple hormonal signaling [24].
Along with many fundamental processes plant hormone abscisic acid (ABA), a key regulator of abiotic stress resistance, plays an important role in mediating drought stress responses in coordination with other plant hormones. Drought stress enhances cellular calcium level, which leads to a calcium-dependent phosphorylation cascade to activate the essential genes required for ABA biosynthesis, such as zeaxanthin oxidase (ZEP), 9-cis-epoxycarotenoid dioxygenase (NCED), ABA-aldehyde oxidase (AAO), and molybdenum cofactor sulphurase (MCSU) [25]. Synthesized active ABA is predominantly accumulated in the vascular tissue of leaves and transported to sites of action for stress response [26,27]. ABA is also stored in vacuole in biologically inactive form by conjugated with glucose ester (ABA-GE) [28]. In addition to the core biosynthetic pathway, endogenous ABA levels increase in response to drought stress through the hydrolyzation of an inactive form of ABA, ABA-GE, to active form [29]. The enhancement of active ABA concentration can cause both repression and increased expression of ABA-responsive genes via ABA signaling machinery.
Forward genetics screening of pyrabactin resistant mutants has identified pyrabactin resistance 1 (PYR1)/PYR1-Like (PYL)/regulatory component of ABA receptor (RCAR) genes as ABA receptors [30,31,32]. There are two major steps to ABA signaling that include PYR1/PYL/RCAR receptor activation by ABA to negatively regulate PP2Cs (Protein phosphatase 2C) and concurrent activation of SNF1-related protein kinase 2 (SnRK2s) to modulate downstream genetic circuits. PYR/PYL/RCAR receptors are homologous to the START (steroidogenic acute regulatory) domain superfamily, which contains a conserved helix-grip motif to generate a central hydrophobic ligand-binding pocket essential for lipids and hormones binding [33]. After binding to the central hydrophobic ligand-binding pocket, ABA creates conformational change in the receptor by forming a gate-latch interface by closing the gate. This structural alteration facilitates binding ABA bound receptor to the PP2C active site to further lock the receptor, thereby forming a receptor-ABA-PP2C complex via gate-latch-lock mechanism [34,35].
Among the other plant phosphatase, PP2Cs, an evolutionarily conserved serine (Ser)/threonine (Thr)-specific phosphatases (STPs) or metal-dependent protein phosphatases (PPMs), act as major negative regulators of ABA signal transduction pathways. The genetic screen for ABA-insensitive mutants and sequence similarity identified multiple PP2Cs in Arabidopsis, such as ABA-INSENSTIVE1 (ABI1), ABI2, ABA-HYPERSENSITIVE GERMINATION1 (AHG1), AHG3/AtPP2CA, HOMOLOGY TO ABI1 (HAB1), and HAB2 [36,37,38,39,40]. PP2Cs are Mg2+/Mn2+–dependent monomeric enzymes, which predominantly inhibit activation of the ABA-responsive transcription factors (TFs) by dephosphorylation of SnRK2s. When endogenous ABA levels are upregulated by developmental or environmental cues, ABA bound PYR/PYL/RCAR receptors interact with PP2Cs to inhibit its protein phosphatase activity, resulting in the release of active SnRK2s [34,41,42,43].
Protein phosphorylation of Snf1-Related Kinases2 (SnRK2s) is one of the major cellular events in ABA signaling. Among the three subclasses: I, II, and III, Subclass III SnRK2s act as positive regulators of ABA signaling as they are rapidly activated by ABA [44]. In Arabidopsis, three members of Subclass III SnRK2s (SRK2D/SnRK2.2, SRK2I/SnRK2.3, and SRK2E/OST1/SnRK2.6) are activated by ABA within 30 min. Generally, SnRK2s are plant-specific Ser/Thr protein kinases, either auto-phosphorylated or trans-phosphorylated by other kinases when ABA receptors specifically sequester PP2Cs, thereby facilitating SnRK2s activation. The active SnRK2s by their well-conserved kinase catalytic domain positively regulate downstream ABA-responsive genes via the phosphorylation of TFs, which include bZIP transcription factors like ABRE (ABA-responsive element)-binding (AREB) proteins or ABRE-binding factors (ABFs) [45,46,47]. SnRK2s mediated AREB/ABFs phosphorylation is a crucial ABA-dependent regulation. These phosphorylated AREBs or ABFs bind to conserved ABA-responsive elements (ABRE) present on the promoter of ABA-regulated genes to up-regulate several downstream genes, for example, RD29B [24]. In addition, several other TFs, including MYC (myelocytomatosis), MYB (myeloblastosis), DREB2 (drought-responsive element binding), NAC (NAM, ATAF1,2, and CUC), AP2/ERF (apetala 2/ethylene responsive factor), basic leucine zipper, and HD-ZIP (homeodomain leucine zipper), greatly influence plant abiotic stress resistance via ABA-dependent and ABA-independent signal transduction pathways by binding with specific cis-acting elements present in promoter regions of several stress-induced genes [48,49,50]. For instance, the RD29 (response to dessication) genes are regulated by AREBs and DREBs TFs via both ABA-dependent and independent signal transduction pathways [51]. MYC (myelocytomatosis) and MYB (myeloblastosis) family’s TFs are involved in the ABA-dependent pathway for the up-regulation of abiotic stress-responsive genes like RD22 [52,53], which is involved in drought stress response via stomatal regulation [54]. In contrast, DREB proteins that bind to DRE cis-elements induce an ABA-independent stress-responsive gene expression, leading to stress resistance ability via the accumulation of osmoprotectants like proline, sucrose [34,55,56]. For example, DREB triggers the expression of RD29A gene during drought stress without involvement of ABA [56,57]. In addition, heterotrimeric G-proteins regulate the ABA signaling pathway and drought resistance by manipulating downstream signaling cascades [58,59,60]. For example, the two subunits of G-protein in rice, qPE9-1 (Gγ subunit) and RGB1 (Gβ subunit), show contrast regulation of ABA dependent stress responses acting as negative and positive regulators, respectively [61].
2. General Aspects of Plant Drought Stress
ABA is the major phytohormone that accumulates in the presence of drought stress to modulate an array of biochemical and physiological changes for acclimatization against stress conditions via ABA-mediated a wide variety of gene expression [62]. Various morpho-physiological alterations were induced by ABA during short-term and prolonged exposure of plants in drought [63]. Here, this review deals with the molecular adaptive mechanisms that plants can implement to combat various drought stress challenges.
2.1. Stomata Closure
Stomata are specialized structures constituting a pair of guard cells enclosing a central aperture through which CO2 gas enters the leaf interior for photosynthesis and concomitant loss of water vapor by transpiration [64]. Under water-deficit conditions, plants can decrease the stomatal pore size to control gas exchange and transpiration rates via reducing guard cell turgor pressure and stomatal conductance [65,66]. Predominantly, enhanced ABA synthesis in guard cells permits stomatal closure under drought. Besides enzymatic activation of ABA biosynthesis enzymes, an inert ABA conjugate activation by β-glucosidase is also responsible for ABA accumulation in leaf guard cells [67,68].
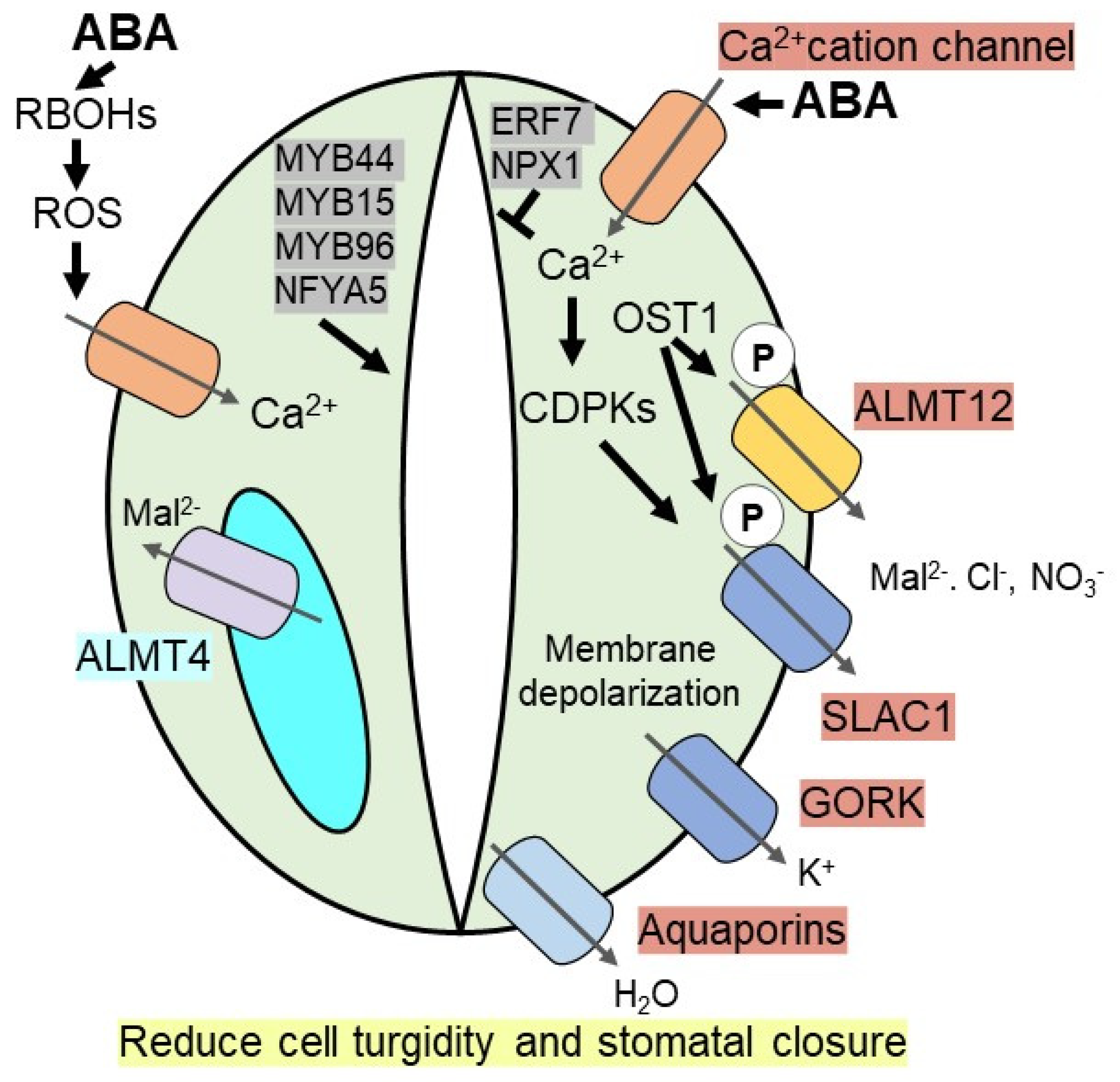
Elevated ABA in guard cells under drought stress induces the level of calcium in the cytosol by triggering Ca2+ influx via non-selective Ca2+ cation channels or hyperpolarization-activated Ca2+ channels. Increased levels of cytosolic calcium activate calcium-dependent protein kinases (CDPKs), which induces SLAC1 (Slow Anion Channel-Associated 1), a S-type anion channel, via phosphorylation [69,70]. For example, activation of SLAC1 is attenuated in CDPK mutants (cpk3cpk6) [71]. In addition, OST1 (open stomata 1), a calcium-independent SnRK2-type kinase, phosphorylate SLAC1 and also R-type anion channels, like aluminium-activated malate transporter 12/quickly activating anion channel 1 (ALMT12/QUAC1) in Arabidopsis [65,72,73]. Phosphorylated anion channels efflux anions (malate (Mal2−), Cl− and NO3−) out of the guard cells, which subsequently promote plasma membrane depolarization to drive K+ efflux through the voltage-dependent outward K+ (K+out) channel like GORK (guard cell outwardly rectifying K+ channel). The constant efflux of ions from the guard cells promotes the efflux of water out of the cell via aquaporins to reduce cell turgidity, thereby facilitating stomatal closure [28,64,65,67] (Figure 1).
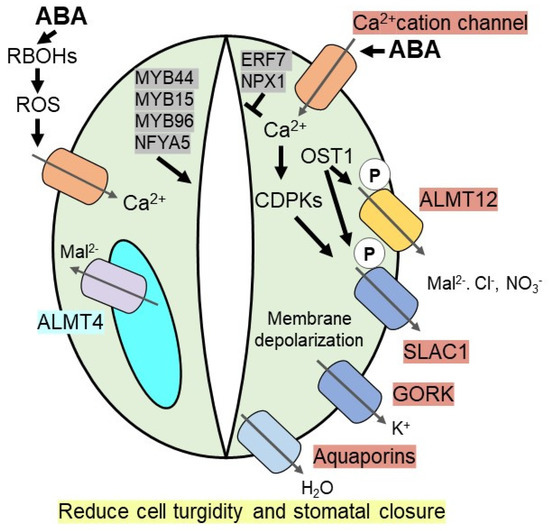
Figure 1.
ABA induces stomatal closure under drought stress. During drought, accumulated ABA inside guard cells induces Ca2+ level, which activates CDPKs (calcium-dependent protein kinases), thereby triggering SLAC1 (slow anion channel-associated 1) channel. Ca2+ independent kinase, OST1 (open stomata 1), phosphorylates SLAC1 anion channel as well as R-type ALMT12 (aluminium-activated malate transporter 12) anion channel to efflux the anions such as malate (Mal2−), Cl− and NO3−. It promotes plasma membrane depolarization to efflux K+ ion via GORK (guard cell outwardly rectifying K+ channel) and water via aquaporins. ABA induces RBOHs (NADPH oxidase/respiratory burst oxidase homolog) on guard cells membrane to generate ROS (reactive oxygen species), which promotes Ca2+ level. A vacuolar anion channel, ALMT4 triggers Mal2− ion outside of vacuole required for stomatal closure. Several transcription factors [MYB44, MYB15, MYB96, NFYA5 (nuclear transcription factor Y subunit A-5), ERF7 (ethylene responsive factor), NPX1 (Nuclear Protein X1)] either positively or negatively promote ABA induced stomatal closure to enhance drought resistance.
Apart from plasma membrane-bound anion channels, the ABA-mediated unknown dephosphorylation mechanism activates vacuolar anion channels, subsequently inducing stomatal closure or impeding stomatal opening [74]. Recently discovered ALMT4 (aluminum activated malate transporter 4), a vacuolar anion channel in Arabidopsis guard cells, is essential for ABA-mediated stomatal closure as knockout mutants of almt4 are unable to control stomatal movement in response to ABA or drought stress. During drought, stress-induced ABA-mediated stomatal closure, ALMT4 is involved in malate (Mal2−) efflux from the vacuole [75]. In addition, ABA induces two NADPH oxidases, AtrbohD and AtrbohF, to generate reactive oxygen species (ROS), like oxygen radicals and H2O2, which act as a positive regulator for stomatal closure by increasing influx of Ca2+ through the Ca2+ channel [76,77]. Increased cytosolic Ca2+ governs multiple Ca2+ dependent kinases to regulate ion channels as well as ROS producing enzymes such as RBOH (NADPH oxidase/respiratory burst oxidase homolog) required for stomatal closure (Figure 1).
Transcriptional regulation plays an essential role in ABA-mediated stomatal closure. Recent studies show that several R2R3 MYB TFs are involved in the modulation of guard cells in the ABA-dependent pathway. For instance, in Arabidopsis both AtMYB44 and AtMYB15 overexpression lines are more sensitive to ABA-induced stomatal closure compared to wild-type plants. Therefore, these transgenic lines exhibit remarkably improved resistance to drought stress [78,79]. During water deficit conditions, contrasting stomatal aperture is observed in MYB96 overexpressing and knockout mutant plants. Overexpression of MYB96 results in increased resistance to drought stress as it triggers stomatal closure, whereas its knockout mutants show a lesser extent of decrease in stomatal aperture under drought, indicating that MYB96 plays a role in controlling stomatal opening [80]. Besides MYB TFs, several other ABA inducible TFs regulate stomatal movement. For example, AtERF7, an APETALA2/ethylene-responsive element-binding protein (AP2/EREBP) family of TFs, acts as a negative regulator of stomatal closure. Thus, aterf7 RNAi lines show increased ABA sensitivity and enhanced survival compared to wild-type in Arabidopsis [81]. NFYA5 (Nuclear transcription factor Y subunit A-5) TF belongs to the Arabidopsis NF-YA family and is critically important in stomatal movement. Overexpression of NFYA5 significantly enhances stomatal closure and increases plant survival under drought stress by positively regulating other drought-responsive genes via binding at CCAAT box cis-element [82]. NPX1 (Nuclear Protein X1) TF represses genes involved in ABA synthesis and ABA signaling; thus, npx1 null mutant shows higher ABA-induced stomatal closure and water deficit resistance than wild-type [83] (Figure 1). In rice, ABA inducible SNAC1 (STRESS RESPONSIVE NAC1) promotes ABA-induced stomatal closure and enhanced drought resistance [84,85]. Apart from TFs, E3 Ub ligase genes in Arabidopsis, such as AtPUB18 and AtPUB19, are negative regulators of ABA mediated stomatal closure as double mutants show enhanced ABA sensitivity and drought tolerance [86]. Metabolites like trehalose also affect stomatal movement. For example, the overexpression of AtTRE1 gene encoding trehalase shows sensitivity towards the ABA-dependent stomatal closure [87].
2.2. Seed Dormancy
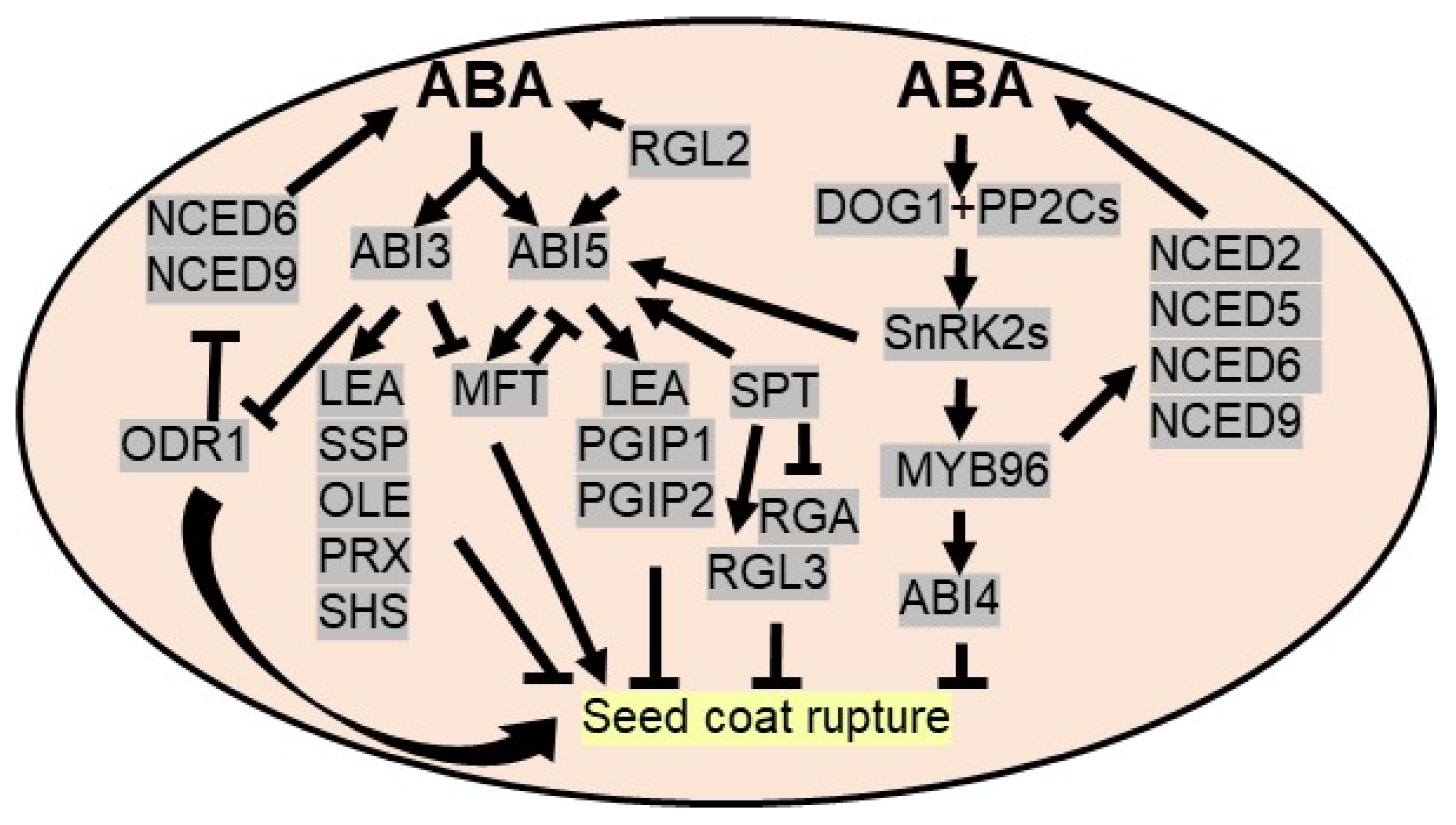
Seed dormancy, a temporary quiescent state, is an adaptive mechanism to prevent viable seed germination under unfavorable growth conditions. To avoid the harsh and challenging growing season or drought, seeds in the dormant stage suppress metabolic processes and eventually germinate under favorable conditions. A seed can maintain its viability at a dormant stage for long duration [88], widely dictated by environmental factors and plant hormones. A dynamic balance between abscisic acid (ABA) and gibberellins (GAs) regulates seed dormancy and germination. ABA promotes and maintains seed dormancy, whereas GAs break seed dormancy. The levels of ABA upsurge during embryonic development and remains high in mature seed. ABA inhibits the cell wall loosening of embryos and prevents water imbibition required for germination. Several genetic analyses demonstrated the involvement of ABA-mediated signaling in seed dormancy. For example, abscisic acid insensitive 3 (ABI3), a major downstream component of ABA signaling, is a pivotal regulator of seed dormancy and desiccation tolerance during embryogenesis by affecting ABA signaling and ABA biosynthesis. ABI3 activates the expression of ABA-inducible genes, like seed storage proteins (SSPs), oleosin (OLE), late embryogenesis abundant (LEA) proteins, peroxiredoxin-like proteins (PRXs), and small heat shock proteins (SHSs) [89]. In addition, ABI3 transcriptionally represses ODR1 (reversal of reduced dormancy5 1 or RDO51) by binding to its promoter at the proximal RY motif. ODR1 also interacts with bHLH57 to negatively regulate the expression of ABA biosynthetic genes, like 9-cis-epoxycarotenoid dioxygenase6 (NCED6) and NCED9, required for seed dormancy [90,91]. ABI5, another major downstream component of ABA signaling, is activated by SnRK2s via phosphorylation before binding to various promoters consisting ABRE/G-box elements like LEA genes [92,93]. ABI5 also interacts physically with ABI3 to synergistically regulate promoters of many ABA-induced genes [94]. Predominantly, ABI5 inhibits embryo development by stimulating a group of LEA genes to combat drought stress. ABI5 also induces PGIP (POLYGALACTURONASE INHIBITING PROTEIN) and PGIP2 encoding polygalacturonase inhibitors to inhibit seed germination in Arabidopsis [95] (Figure 2).
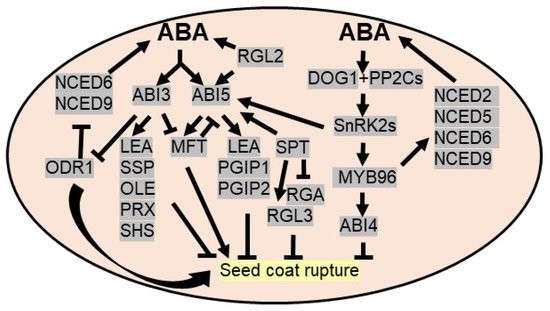
Figure 2.
ABA triggers seed dormancy to avoid drought stress. ABI3 (abscisic acid insensitive 3), a major downstream component of ABA signaling, triggers SSP (seed storage protein), OLE (Oleosin), LEA (late embryogenesis abundant), PRX (peroxiredoxin-like proteins) and SHS (small heat shock proteins) to promote seed dormancy. DELLA protein RGA-LIKE2 (RGL2), a negative regulator of GA signaling, induces ABI5 TFs to upregulate LEA and PGIP1/PGIP2 (POLYGALACTURONASE INHIBITING PROTEIN) genes and triggers endogenous ABA concentration to promote seed dormancy. ABI3 downregulates ODR1 (reversal of reduced dormancy5 1 or RDO51) as it inhibits ABA biosynthesis enzymes, NCED6 (9-cis-epoxycarotenoid dioxygenase6) and NCED9. ABI3 and ABI5 regulate MFT (MOTHER OF FT AND TFL1) expression and MFT via negative feedback regulation represses ABI5. DOG1 (delay of germination-1), a master regulator of dormancy, is induced by ABA/drought. DOG1 binds to PP2Cs to derepress ABA signaling by SnRK2. MYB96 TF activates ABA biosynthesis enzymes to increase the endogenous ABA concentration and induces ABI4 to inhibit seed germination. SPT (SPATULA) regulates the expression of ABI5, RGA and RGL3 to promote seed dormancy.
DELLA proteins are a GRAS family of transcription factors and serve as a convergence point of ABA and GA signaling pathways. DELLA inhibits the GA responses concomitantly, activating ABA to stimulate seed dormancy to escape from the water deficit condition. In Arabidopsis, out of five DELLA proteins (GA INSENSITIVE (GAI), REPRESSOR OF GA1-3 (RGA), RGA-LIKE1 (RGL1), RGL2 and RGL3), RGL2 acts as a major repressor of seed germination as it induces ABI5 expression as well as endogenous ABA concentration when GA levels are low. Similar to ABI5, the expression of RGL2 is stimulated by exogenous ABA [92]. The expression of MFT (MOTHER OF FT AND TFL1), a phosphatidylethanolamine-binding protein, is positively and negatively regulated by ABI5 and ABI3, respectively, to deploy ABA signaling pathways by suppressing ABI5 during seed germination via a negative feedback regulation [96]. Another TF, SPATULA (SPT), acts as both dormancy promoter and repressor by regulating the expression of ABI5, RGA and RGL3 in Arabidopsis [97,98]. One of the R2R3 TFs, MYB96, transcriptionally regulates ABA biosynthetic genes like NCED2, NCED5, NCED6, and NCED9 as well as GA biosynthetic genes like GA3ox1 and GA20ox1 to sustain a balance between ABA and GA, thereby regulating seed dormancy [99]. Catabolism of embryonic lipid reserves (triacylglycerol) assists the seed germination event via acting as energy source. ABI4, another positive regulator of ABA signaling, which is essential for inhibiting seed germination by lipid breakdown, is controlled by MYB96 [100]. In addition, WRKY2 TF acts as a negative regulator of ABA-mediated seed dormancy in Arabidopsis [101] (Figure 2).
Apart from these, a heme-binding protein, i.e., delay of germination-1 (DOG1), acts as a crucial regulator of dormancy as it binds to PP2Cs (ABA-hypersensitive germination 1/AHG1, AHG2) to positively regulate ABA signaling. Thus, dog1 mutant seeds display a non-dormancy phenotype [102] (Figure 2). Under abiotic stresses like drought, bZIP67 TF, epigenetic regulation and alternative splicing regulate the expression levels of DOG1 to control seed dormancy [103]. Homologues of DOG1 in wheat (TaDOG1L4) promote seed dormancy as confirmed by overexpression and RNA interference studies [104]. Like Arabidopsis, TaDOG1L4 interacts with TaPP2C-a10 to modulate the ABA signaling mechanism in wheat seed dormancy [105].
2.3. Cuticular Wax Biosynthesis
The plant cuticle is an extracellular, thick, waxy layer that remains outside part of the epidermis to protect against a dehydrating environment, UV radiation, pathogen entry, and other abiotic stresses. The primary constituent of the plant cuticle is a macromolecular scaffold of cutin and waxes. These waxes are organic solvent-soluble lipids, typically derived from very-long-chain fatty acids (C20–C34) [106]. During the transition from an aquatic to a land lifestyle, plants were exposed to a set of challenges in the terrestrial environment, including drought, high temperature, exposure to UV radiation etc. In order to sustain under such a challenging environment, plants would have necessitated some morphological and physiological features. Establishing a hydrophobic surface layer or cuticle was one of the adaptive milestones to retain water inside plant cells under dehydrating conditions [107]. Apart from reducing leaf transpiration and maintaining stomatal conductance, cuticular wax can act as a photoprotective layer of PS II complex under drought stress in wheat [108].
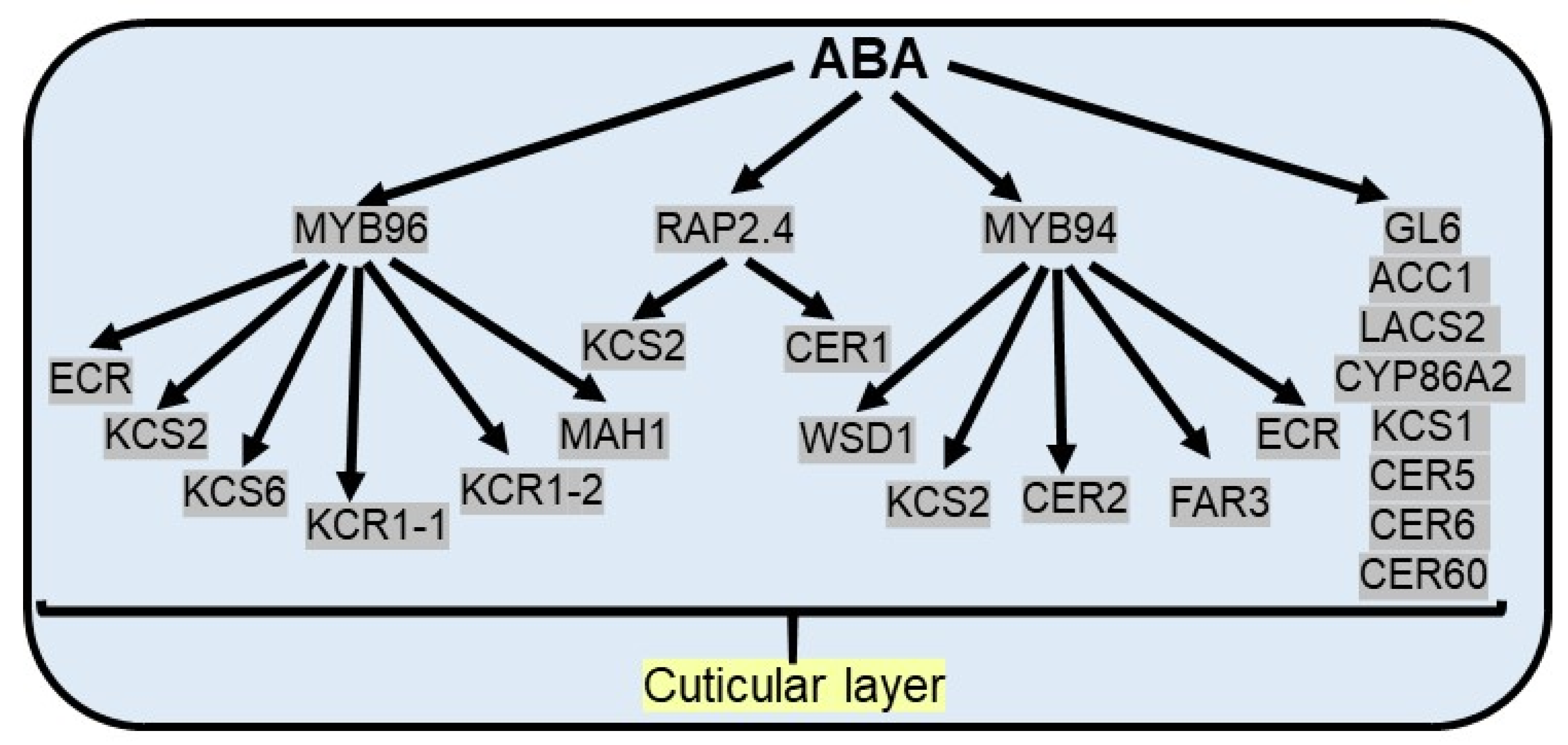
Cuticular wax composition can vary considerably within the same plant during drought conditions, viz. an increased percentage of alkane in total wax is observed under water deficit conditions [107,109,110]. In addition, wax load per unit area and cuticle thickness can substantially increase in a dehydrating environment. The wax ester synthase (WSD1) gene is upregulated in water deficit conditions, resulting in an increased cuticular wax load in leaves and stems of Arabidopsis [111]. Similarly, up-regulation of some genes in the aliphatic wax biosynthetic pathway enhances cuticular wax load, including wax esters in grape berries under drought [112]. Altering cuticular wax accumulation by intracellular trafficking and augmented expression of candidate genes in the fatty acid biosynthesis pathway is regulated by the glossy gene (GL6), causing slower water losses to survive in water deficit conditions [113] (Figure 3).
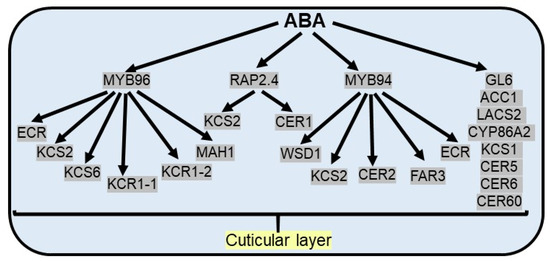
Figure 3.
ABA promotes the cuticular wax accumulation under drought stress. ABA responsive MYB96 TF activates cuticular wax biosynthesis by upregulation of ECR (trans-2-enoyl-CoA reductases), KCS2/6 (3-ketoacyl-CoA synthetases), KCR1-1/1-2 (3-ketoacyl-CoA reductases) and MAH1 (mid-chain alkane hydroxylase 1) genes. ABA responsive MYB94 TF activates cuticular wax biosynthesis by upregulation of WSD1 (wax synthase/acyl-CoA: diacylglycerol acyltransferase), KCS2, CER2 (eceriferums), FAR3 (fatty acyl-CoA reductase) and ECR genes. Apart from MYB TF, ABA responsive RAP2.4 TF activates cuticular wax biosynthesis by upregulation of KCS2 and CER1. In addition, several cuticular wax biosynthetic genes and cuticle-associated genes [GL6 (glossy gene), ACC1 (acetyl-CoA carboxylase 1), LACS2 (long-chain acyl-CoA synthetase 2), KCS1, CYP86A2 (cytochrome P450), CER5, CER6, CER60] regulate ABA induced cuticular wax biosynthesis.
ABA has been established as an important regulator, which leads to the increase in cuticular wax biosynthetic genes and cuticle-associated genes, including acetyl-CoA carboxylase 1 (ACC1), long-chain acyl-CoA synthetase 2 (LACS2), 3-ketoacyl-CoA synthase (KCS1), cytochrome P450 (CYP86A2), and eceriferums (CER1, CER2, CER5, CER6, CER60) [107,114]. In Arabidopsis, ABA treatment induces the expression of the CER6, which causes an increase in surface wax accumulation in Arabidopsis [115]. BnKCS1-1, BnKCS1-2, and BnCER1-2 promote cuticular wax production in Brassica napus and thereby increase resistance to water deficit conditions [116]. Apart from stomatal closure and the regulation of seed dormancy, ABA-responsive MYB96 TF plays a substantial role in the biosynthesis of cuticular wax by binding with the promoter of fatty acid elongating enzymes like 3-ketoacyl-CoA synthetases (KCS), 3-ketoacyl-CoA reductases (KCR), 3-hydroxyacyl-CoA dehydratases, and trans-2-enoyl-CoA reductases (ECR), essential for cuticular wax biosynthesis. The same is validated from the contrasting phenotypes observed, like enhanced levels of epicuticular wax crystals on the leaf surface in MYB96 overexpression lines and reduced levels of cuticular wax in myb96-1 mutants [54]. The expression of multiple wax biosynthetic genes, like KCS2, KCS6, KCR1-1, KCR1-2, ECR, and MAH1 (mid-chain alkane hydroxylase 1), is significantly enhanced as a result of the overexpression of MYB96 in Camelina sativa [117]. Under drought stress conditions, another R2R3 TF MYB94 along with MYB96 additively upregulate the expression of wax biosynthetic genes to prevent the loss of water from aerial organs as double mutants (myb96myb94) show an additional reduction in wax load and transcript level of wax biosynthetic genes than single mutants [118]. MYB94 regulates wax biosynthesis genes via direct binding to the promoter of the WSD1 (wax synthase/acyl-CoA: diacylglycerol acyltransferase), KCS2/DAISY (β-ketoacyl-CoA synthase), CER2, FAR3 (fatty acyl-CoA reductase), and ECR (enoyl-CoA reductase) genes in Arabidopsis [119]. In maize, ZmFDL1/MYB94 has been reported to function as a positive regulator in cuticular wax biosynthesis under drought conditions [120]. Apart from MYB TFs, AP2/ERF TF family members activate wax biosynthetic genes to enhance drought resistance. For example, ABA and drought inducible RAP2.4 TF increase cuticular wax biosynthesis via direct interaction with CCGAC or GCC consensus motifs in promoters of KCS2 and CER1 genes in Arabidopsis [121] (Figure 3).
2.4. Leaf Senescence
Organ senescence causes programmed cell death regulating the development of all living organisms. Leaf senescence in plants is not only age-related, but also acts as the long-term adaptive mechanism under drought conditions facilitating minimal water loss for survival and completion of their life cycle. Leaf senescence and abscission, involving the termination of photosynthesis, increase of reactive oxygen species (ROS), accumulation of exhausted materials to dying cells, and remobilization of nutrients from senescent leaves to young leaves, meristem, or storage organs, is predominantly regulated by various factors modulated by various phytohormones, viz. ABA, ethylene, jasmonic acid, salicylic acid, and strigolactones [122,123,124]. Among these, ABA is a critical phytohormone that mediates leaf senescence. Accumulation of ABA by overexpression of OsNCED5 accelerates senescence in transgenic rice and contrasting phenotype has been detected in nced5 mutant [125]. ABA is involved in the biosynthesis of ethylene by inducing 1-aminocyclopropane-1-carboxylic acid (ACC) synthase to promote senescence as ethylene is reported to induce organized cell disassembly and nutrient mobilization from senescent leaves to young organs [126,127].
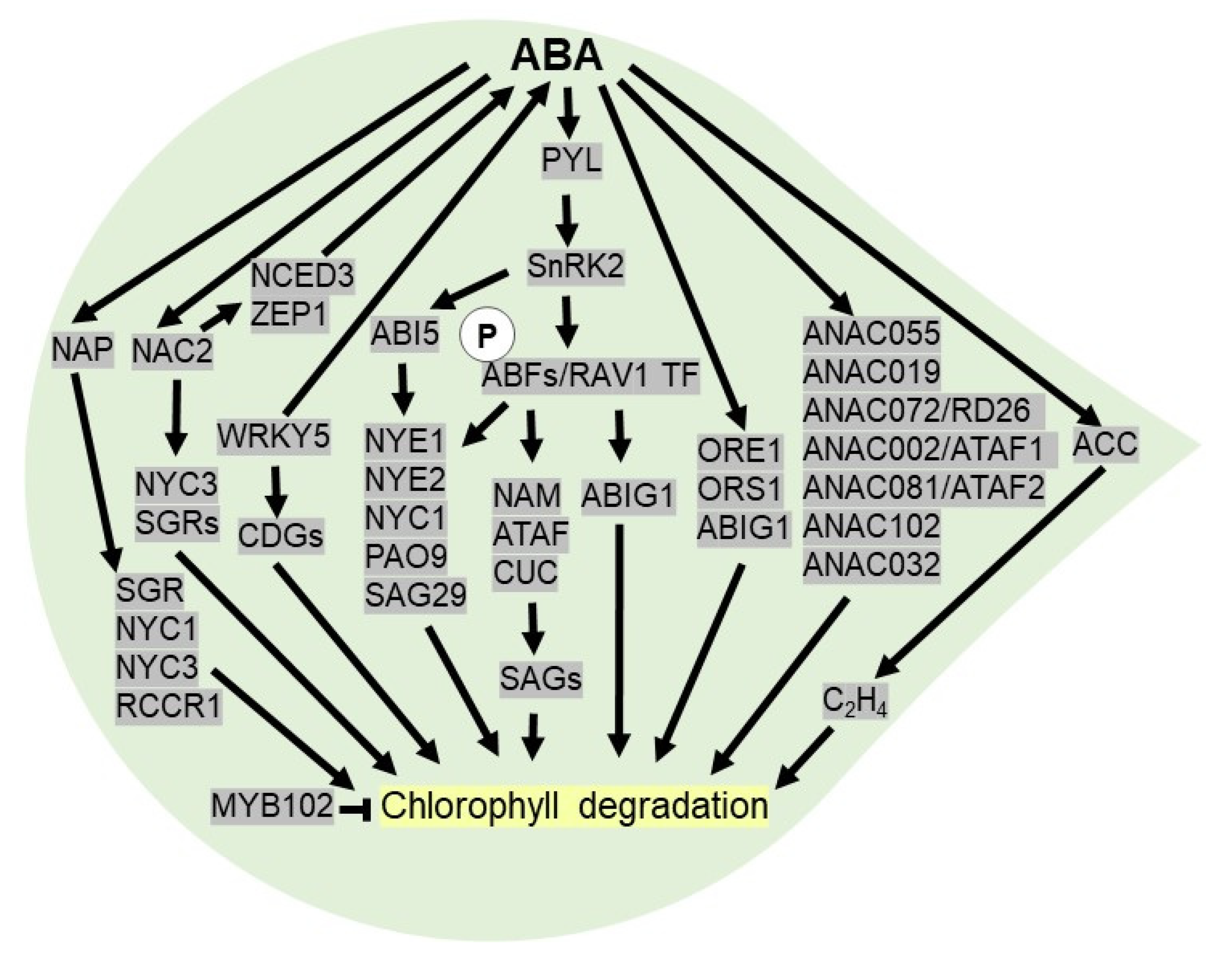
In ethylene-independent manner, core ABA signaling components like PYL receptors, PP2Cs phosphatase and protein kinases like SnRK2 play a crucial role in regulating leaf senescence. For example, the overexpression of PYL9 under stress-inducible promoter in Arabidopsis increases ABA sensitivity and drought resistance by promoting leaf senescence, thereby facilitating water transport to developing tissues [128]. Accumulation of ABA under drought conditions activates SnRK2 mediated phosphorylation of ABFs (ABA-responsive element-binding factors) and RAV1 (ABA-Insensitive 3/VP1) TFs via ABA signaling. The phosphorylated ABFs and RAV1 bind to ABRE motif elements in the promoter of NAC (NAM, ATAF, and CUC) TFs, which are likely to act as crucial regulators in mediating ABA-triggered leaf senescence by modulating downstream SAGs (senescence-associated genes). Precocious leaf senescence has been observed in Arabidopsis after overexpression of ABA inducible NAC TFs, like NAP, ORESARA1 (ORE1), and Oresara 1 sister 1 (ORS1) [128,129,130]. In Arabidopsis, an ABA-inducible group of stress-responsive NAC TFs, SNAC-As, including ANAC055, ANAC019, ANAC072/RD26, ANAC002/ATAF1, ANAC081/ATAF2, ANAC102, and ANAC032, triggers leaf senescence by activating a set of ABA-inducible genes independent of AREB/ABFs [131]. In rice, OsNAC2 plays a fundamental role in leaf senescence as it transcriptionally activates OsNYC3 (non-yellow coloring1) and OsSGR (STAY-GREEN) genes (Figure 4). Additionally, OsNAC2 modulates ABA biosynthetic (OsNCED3 and OsZEP1) and catabolic genes (OsABA8ox1) to increase ABA levels. Thus, leaf senescence is significantly delayed in OsNAC2-RNAi lines, whereas the overexpression of OsNAC2 accelerates senescence in transgenic rice plants [132].
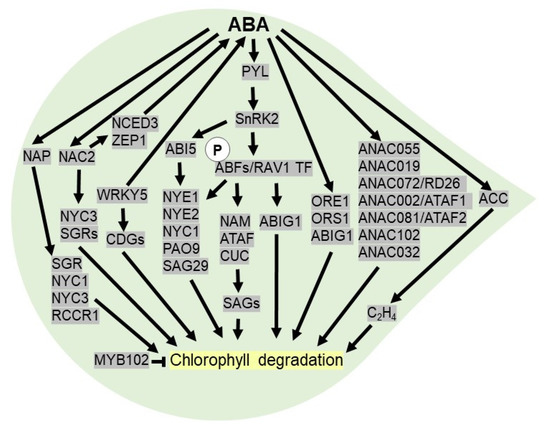
Figure 4.
ABA activates leaf senescence under drought stress. After perception of ABA, PYL receptor triggers SnRK2 to phosphorylates ABFs (ABA-responsive element-binding factors) and RAV1 (ABA-insensitive 3/VP1) TF, which binds to promoter of NAC (NAM, ATAF and CUC) TFs to regulate downstream SAGs (senescence-associated genes) genes required for chlorophyll degradation. ABFs and ABI5 induce NYE1 (non-yellowing 1), NYE2, NYC1 (Non-Yellow Coloring1) and PAO9 (pheophorbiden a oxygenase) to induce leaf senescence. ABA inducible NAC TFs (NAP, ORE1, ORS1, ANAC055, ANAC019, ANAC072/RD26, ANAC002/ATAF1, ANAC081/ATAF2, ANAC102 and ANAC032) accelerate chlorophyll degradation. ABA promotes leaf senescence by stimulating ACC (1-aminocyclopropane-1-carboxylic acid) synthase essential for ethylene biosynthesis. In rice, WRKY TF modulates leaf senescence by triggering ABA biosynthetic genes and CDGs (chlorophyll degradation genes). ABA inducible NAP TF triggers SGR (STAY-GREEN), NYC1, NYC3 and RCCR1 (red chlorophyll catabolite reductase 1) to promote chlorophyll degradation. ABA inducible NAC2 TF triggers NYC3, SGRs and ABA biosynthesis by activating NCED3 and ZEP1 (zeaxanthin oxidase) to promote chlorophyll degradation.
To date, there are pieces of evidence indicating that chlorophyll breakdown is a marker of leaf senescence and ABA can promote the degradation of chlorophyll under drought [133]. The ABA-responsive element (ABRE)-binding TFs, viz ABI5 or EEL (ENHANCED EM LEVEL), ABF2, ABF3, and ABF4, which are activated by PYLs-PP2C-SnRK2 core-sensing system, trigger NYE1 (NON-YELLOWING 1) or SGR1, NYE2, NYC1, and PAO9 (pheophorbiden a oxygenase) to accelerate chlorophyll degradation. In addition, ABF can directly activate the SAG gene, like SAG29 essential for leaf senescence [134]. In rice, OsNAP is a positive regulator of early leaf senescence as it induces chlorophyll degradation genes (CDGs), such as SGR, NYC1, NYC3, and RCCR1 (red chlorophyll catabolite reductase 1) [135] (Figure 4). A WRKY TF, OsWRKY5, promotes ABA biosynthesis and chlorophyll degradation genes, leading to early leaf senescence [136]. In addition, the transcript levels of ABIG1 (ABA insensitive growth 1) or HAT22 (homeobox from Arabidopsis thaliana 22) is increased in the presence of ABA. In Arabidopsis, ABIG1, a part of an ABA signaling pathway, accelerates leaf senescence by activating multiple pathways in drought conditions [137]. On the other hand, OsMYB102 delays the leaf senescence in rice as it acts as a negative regulator of ABA accumulation and signaling [138]. Similarly, a WRKY TF in cotton, GhWRKY91, acts as a negative regulator of ABA- and drought-induced leaf senescence [139].
2.5. Root and Shoot Length
Roots are essential for plant growth and development as they utilize soil resources via the uptake of water and nutrients. Under water-limited conditions, plant sustainability, as well as productivity, depends on root traits like root length, root diameter, root angle, root density, lateral root number, root hair density etc. To support existing shoots in water deficit condition, plants produce considerably longer roots with decreased diameter, which is vital to acquire the available water at depths in the soil and thus, maintenance of root elongation at low water potentials (ψw) is an essential adaptive feature under dry conditions [140]. Typically, drought stress activates ABA to accumulate in the roots, and enhanced drought duration increases the level of ABA in the root apex to trigger the adaptive morphological changes, including root tip swelling and root apical meristem premature (RAM) differentiation [141,142]. Accumulation of ABA also regulates the architecture of the root system and hydraulic conductivity or unit length root conductance [143,144]. Predominantly, root water uptake in plants is influenced by ABA inducible water channel proteins named aquaporins that alter cell water permeability to maintain cellular water and osmotic homeostasis [145,146].
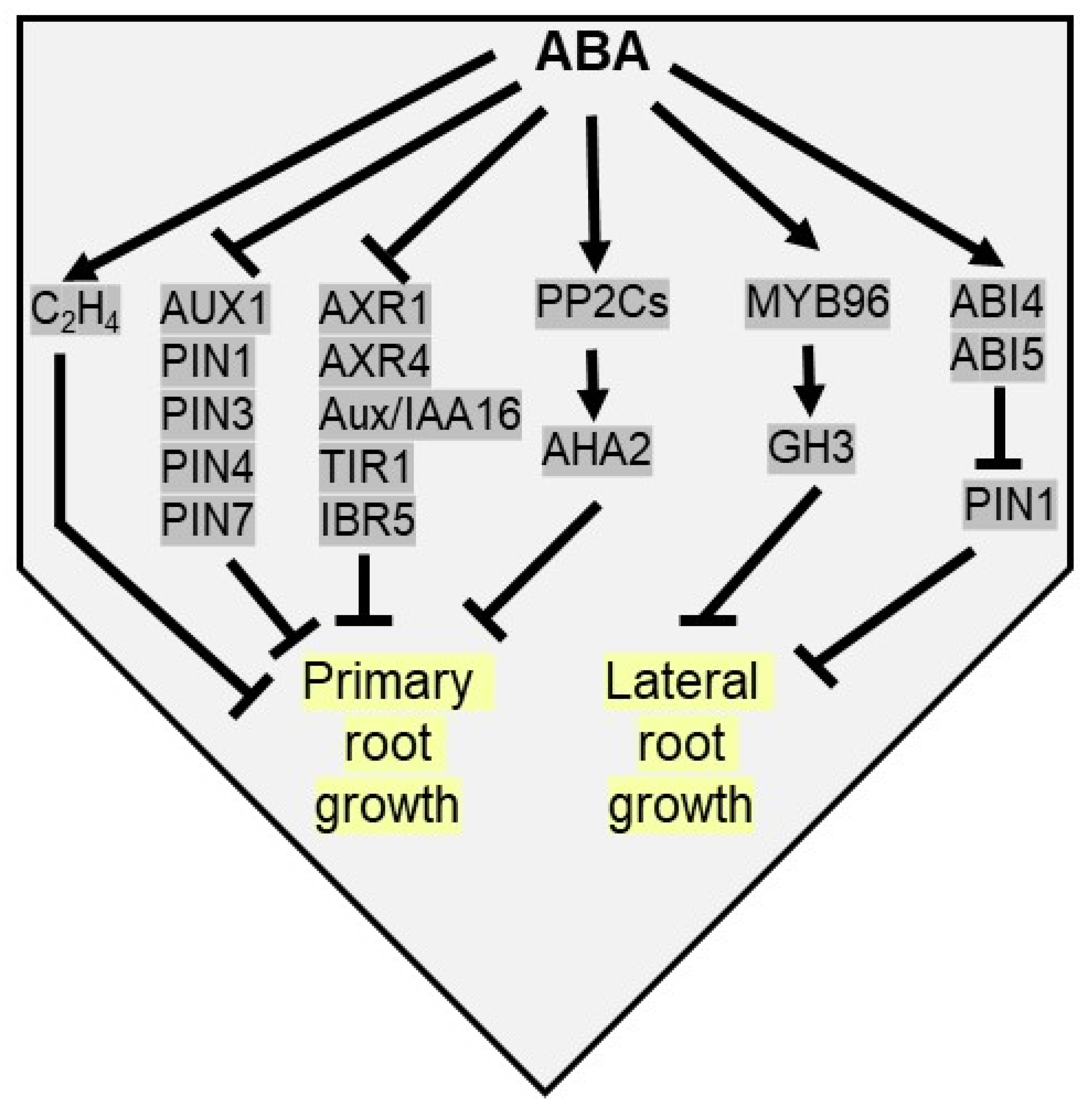
The underlying mechanisms that allow morpho-physiological effects on root growth by ABA are intricate as they connect with diverse hormonal regulatory networks. For example, in ABA-deficient seedlings, increased ethylene accumulation inhibits root growth, and therefore ABA maintains root growth under low water conditions by inhibiting ethylene production [147]. To promote root stem cell maintenance, a low concentration of ABA inhibits quiescent centre (QC) division and differentiation of stem cells and their daughter cells in primary root meristem [148,149]. Typically, moderate water stress or low concentration of ABA positively modulates root growth by manipulating auxin levels via auxin transport and auxin signaling [150]. Root-specific NF-Yb21 (NUCLEAR FACTOR-Y) TF interacts with FUS3 (FUSCA3) to promote ABA biosynthesis via activation of NCED3, which in turn promotes auxin transport leading to root growth and drought tolerance in populus [151]. On the other hand, high concentrations of ABA inhibit root growth by reducing the expression of auxin transport genes in Arabidopsis roots, viz. AUX1, PIN1 (PIN-FORMED 1), PIN3, PIN4, and PIN7 [152,153]. As a result, decreased sensitivity to ABA is detected in aux1 (auxin resistant 1), axr4, and pin2 mutants. Apart from auxin transport genes, several signaling components of auxin like AXR1 (Auxin resistant 1), AXR4, Aux/IAA16 (Aux/Indole-3-acetic acid), TIR1 (transport inhibitor response 1), and IBR5 (IBA response 5) are downregulated by ABA, resulting in the suppression of primary root growth [154]. Apart from these, a recent study depicts a model of ABA concentration-dependent root growth modulation by H+ extrusion across the plasma membrane. High ABA concentration upregulates PP2Cs, which dephosphorylate Thr947 of H+-dependent adenosine triphosphatase 2 (AHA2) after binding its C-terminal R domain, resulting in primary root growth arrest of Arabidopsis by inhibition of apoplastic H+ efflux. Whereas, low ABA concentration positively regulates root growth by derepressing AHA2 and H+ extrusion via ABA receptor-mediated inhibition of PP2C activity [155] (Figure 5).
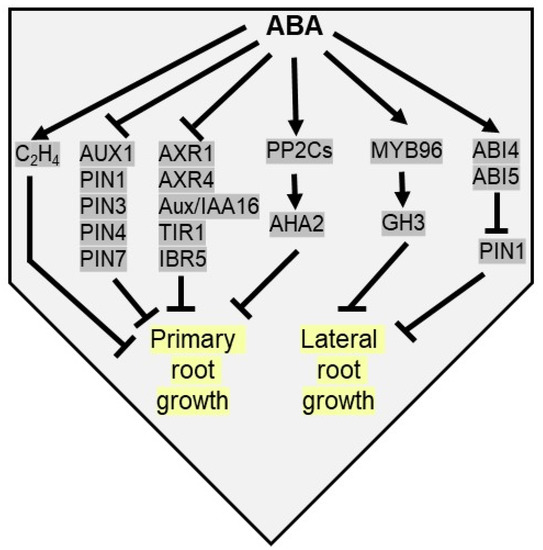
Figure 5.
ABA inhibits both primary and lateral root elongation under drought stress. ABA activates ethylene biosynthesis to inhibit primary root growth. ABA inhibits auxin transport [AUX1, PIN1 (pin-formed 1), PIN3, PIN4 and PIN7) and auxin signaling genes [AXR1 (auxin resistant 1), AXR4, Aux/IAA16, TIR1 (transport inhibitor response 1) and IBR5 (IBA response 5)] to arrest primary root growth. ABA upregulates PP2Cs to dephosphorylate AHA2 (H+-dependent adenosine triphosphatase 2) to inhibit primary root growth. ABA inducible MYB96 TF upregulates GH3 (gretchen hagen 3) to inhibit lateral root elongation. ABA restricts lateral root growth by suppressing PIN1 proteins via ABI4 and ABI5.
The inhibition of lateral root (LR) growth is another adaptive phenotypic response of plants against drought stress. Plants adapt to drought by restricting the horizontal spread of lateral growth and utilizing energy in primary root elongation to acquire water in water deficit soil [156]. Thus, despite a similar number of lateral root primordia in both wild type and ABA related mutants in Arabidopsis, root primordia fail to elongate in mutants [156,157]. Predominantly, lateral root initiation is promoted by auxin-dependent cell cycle-related genes. In Arabidopsis, ABA-induced MYB96 TF enhances the expression of GH3, encoding an auxin-amido synthetase, which inhibits lateral root elongation by inactivating endogenous auxin pool [80]. An auxin efflux carrier gene PIN1 modulates polar auxin transport from the shoot to root apices, affecting lateral root development. ABA negatively regulates PIN1 expression by its downstream signaling components, viz ABI4 and ABI5 (Figure 5). Thus, LR initiation and the elongation of emerged LRs are inhibited in ABI4 overexpression lines of Arabidopsis [154,158,159,160].
Although different sensitivity is observed between shoot and root tissues under water-limited conditions, drought-induced ABA typically inhibits shoot growth. Water deficit and ABA cause prevention of shoot growth of maize, but it is derepressed in fluridone-treated seedlings as it targets carotenoid biosynthetic pathway to reduce endogenous ABA pool [161]. Similarly, ABA accumulation under low water potential has been reported to prevent the shoot growth of soybean [162]. Predominantly, endogenous ABA restricts ethylene production under water deficit conditions to maintain shoot growth in the early and late developmental stages [143,163,164]. Apart from that, it has been reported that OsbZIP23 TF plays a key role in conferring drought tolerance by enhancing the expression of many stress related genes, like LEA, RD22, etc., in rice [165].
3. Conclusions and Future Perspectives
Plants are exposed to a changing environment throughout their existence. Drought is one of the most prevalent global problems that negatively affect agricultural production, reducing net photosynthesis by altering plant carbon allocation and metabolism. To cope with drought, plants can elicit physiological and biochemical responses aimed at enhancing resistance. Phenotypic plasticity, including modifications of vegetative and reproductive architecture, is essential for resistance to water deficiency. The developmental plasticity of the plant organs is regulated by endogenous ABA level, which precisely regulates numerous signaling proteins, TFs, and even ABA biosynthetic genes. Extensive cross-talk among those proteins leads to the formation of complex signaling networks (Figure 6). Here, we provided considerable phenotypic and genetic evidence for the ABA-mediated drought stress resistance in plants through various molecular mechanisms, which is critically important to understanding the fundamental biology that underscores the stress resistance phenotype. This detailed knowledge about how plants modify when challenged by drought is essential to enhance drought-stress resistance in different crop plants.
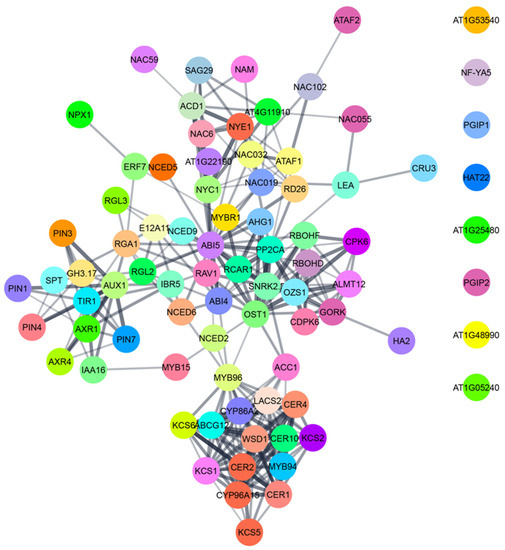
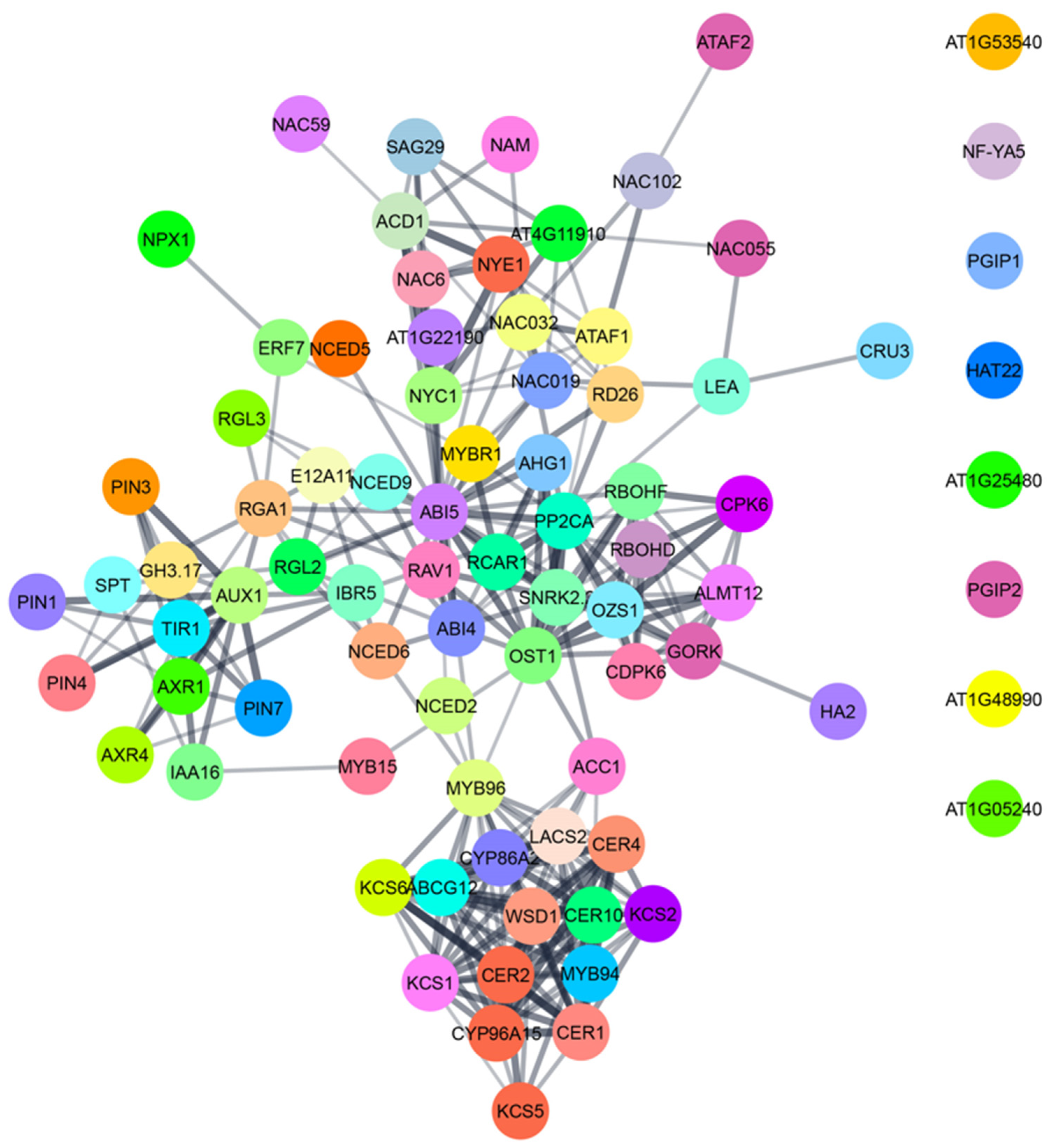
Figure 6.
A network of proteins involved in ABA-mediated physiological adaptations like stomatal closure, seed dormancy, cuticular wax biosynthesis, leaf senescence and alteration of root and shoot growth in Arabidopsis thaliana. The data for AGI locus code was collected from TAIR—(arabidopsis.org (accessed on 16 November 2021)). The interaction network was prepared by using STRING (string-db.org (accessed on 16 November 2021)) and represented by using Cytoscape software.
Drought stress responses are coordinated by complex signaling networks. It is implausible that a single gene alone regulates plant drought tolerance. QTL mapping facilitated the establishment of relationships between drought stress tolerance and agronomic and physiological traits by identifying genomic regions linked with traits of interest. Recent genomics tools, molecular techniques, and precise phenotype analysis detected several candidate genes for crop drought tolerance. Multiple cross-talk among different regulatory networks during drought stress makes it a challenge to discriminate those interactions that most effect under water deficit conditions. Therefore, it is critically important to recognize convergent points in the drought stress response circuitry essential for translational research. In future, a systems biology approach, including transcriptomics, proteomics, and metabolomics, high-throughput phenotyping, functional characterization of novel regulatory candidate genes, and detailed study of epigenetics, will be needed to precisely manipulate physiological processes for developing drought-tolerant crops.
Author Contributions
A.A. and S.R.C. were involved in writing and editing the review. S.R.C. secured the funding. All authors have read and agreed to the published version of the manuscript.
Funding
Research in the author’s lab is supported by Indian Institute of Science Education and Research (IISER), Tirupati, SERB Start-up Research Grant (SRG/2019/000901) of S.R.C., DBT-Ramalingaswami Re- entry Fellowship of S.R.C. (BT/RLF/Re-entry/01/2018) and STARS Research Grant of S.R.C. (MoE/STARS-1/508) and A.A. acknowledges the Council of Scientific and Industrial Research (CSIR), India for JRF fellowship.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We apologize to all authors whose work was not cited due to the length limitations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Iqbal, Z.; Iqbal, M.S.; Hashem, A.; Abd_Allah, E.F.; Ansari, M.I. Plant Defense Responses to Biotic Stress and Its Interplay With Fluctuating Dark/Light Conditions. Front. Plant Sci. 2021, 12, 631810. [Google Scholar] [CrossRef] [PubMed]
- He, M.; He, C.-Q.; Ding, N.-Z. Abiotic Stresses: General Defenses of Land Plants and Chances for Engineering Multistress Tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef] [PubMed]
- FAO. More Crop Per Drop. Available online: https://www.fao.org/english/newsroom/focus/2003/water.htm (accessed on 1 November 2021).
- FAO. Disasters Take Heavy Toll on Agri-Food Systems as New Threats Emerge. Available online: https://www.fao.org/news/story/en/item/1381672/icode/ (accessed on 1 November 2021).
- Daryanto, S.; Wang, L.; Jacinthe, P.-A. Global Synthesis of Drought Effects on Maize and Wheat Production. PLoS ONE 2016, 11, e0156362. [Google Scholar] [CrossRef]
- Daryanto, S.; Wang, L.; Jacinthe, P.-A. Global synthesis of drought effects on cereal, legume, tuber and root crops production: A review. Agric. Water Manag. 2017, 179, 18–33. [Google Scholar] [CrossRef]
- Leng, G.; Hall, J. Crop yield sensitivity of global major agricultural countries to droughts and the projected changes in the future. Sci. Total Environ. 2019, 654, 811–821. [Google Scholar] [CrossRef]
- Fathi, A.; Tari, D.B. Effect of Drought Stress and its Mechanism in Plants. Int. J. Life Sci. 2016, 10, 1–6. [Google Scholar] [CrossRef]
- Kapoor, D.; Bhardwaj, S.; Landi, M.; Sharma, A.; Ramakrishnan, M.; Sharma, A. The Impact of Drought in Plant Metabolism: How to Exploit Tolerance Mechanisms to Increase Crop Production. Appl. Sci. 2020, 10, 5692. [Google Scholar] [CrossRef]
- Blatt, M.R.; Chaumont, F.; Farquhar, G. Focus on Water. Plant Physiol. 2014, 164, 1553–1555. [Google Scholar] [CrossRef]
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, S.A.; Men, S.; Wang, L. Chilling and Drought Stresses in Crop Plants: Implications, Cross Talk, and Potential Management Opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef]
- Bernau, V.M.; Jardón Barbolla, L.; McHale, L.K.; Mercer, K.L. Germination Response of Diverse Wild and Landrace Chile Peppers (Capsicum spp.) under Drought Stress Simulated with Polyethylene Glycol. PLoS ONE 2020, 15, e0236001. [Google Scholar] [CrossRef]
- Mukami, A.; Ngetich, A.; Mweu, C.; Oduor, R.O.; Muthangya, M.; Mbinda, W.M. Differential characterization of physiological and biochemical responses during drought stress in finger millet varieties. Physiol. Mol. Biol. Plants 2019, 25, 837–846. [Google Scholar] [CrossRef]
- Islam, M.M.; Kayesh, E.; Zaman, E.; Urmi, T.A.; Haque, M.M. Evaluation of Rice (Oryza sativa L.) Genotypes for Drought Tolerance at Germination and Early Seedling Stage. Agriculturists 2018, 16, 44–54. [Google Scholar] [CrossRef]
- Razzaq, H.; Tahir, M.H.N.; Sadaqat, H.A.; Sadia, B. Screening of sunflower (Helianthus annus L.) accessions under drought stress conditions, an experimental assay. J. Soil Sci. Plant Nutr. 2017, 17, 662–671. [Google Scholar] [CrossRef]
- Okçu, G. Mehmet Kaya, and Mehmet Atak. Effects of Salt and Drought Stresses on Germination and Seedling Growth of Pea (Pisum sativum L.). Turk J. Agric. For. 2005, 29, 237–242. [Google Scholar]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Wang, Z.; Li, G.; Sun, H.; Ma, L.; Guo, Y.; Zhao, Z.; Gao, H.; Mei, L. Effects of drought stress on photosynthesis and photosynthetic electron transport chain in young apple tree leaves. Biol. Open 2018, 7, bio035279. [Google Scholar] [CrossRef] [PubMed]
- Abid, M.; Ali, S.; Qi, L.K.; Zahoor, R.; Tian, Z.; Jiang, D.; Snider, J.L.; Dai, T. Physiological and biochemical changes during drought and recovery periods at tillering and jointing stages in wheat (Triticum aestivum L.). Sci. Rep. 2018, 8, 4615. [Google Scholar] [CrossRef] [PubMed]
- Auten, R.L.; Davis, J.M. Oxygen Toxicity and Reactive Oxygen Species: The Devil Is in the Details. Pediatr. Res. 2009, 66, 121–127. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to Environmental Stresses. Plant Cell 1995, 7, 1099. [Google Scholar] [CrossRef]
- Pastori, G.M.; Foyer, C. Common Components, Networks, and Pathways of Cross-Tolerance to Stress. The Central Role of “Redox” and Abscisic Acid-Mediated Controls. Plant Physiol. 2002, 129, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shahzad, B.; Kumar, V.; Kohli, S.K.; Sidhu, G.P.S.; Bali, A.S.; Handa, N.; Kapoor, D.; Bhardwaj, R.; Zheng, B. Phytohormones Regulate Accumulation of Osmolytes Under Abiotic Stress. Biomolecules 2019, 9, 285. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene Networks Involved in Drought Stress Response and Tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Tuteja, N. Abscisic Acid and Abiotic Stress Signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef]
- Efetova, M.; Zeier, J.; Riederer, M.; Lee, C.-W.; Stingl, N.; Mueller, M.; Hartung, W.; Hedrich, R.; Deeken, R. A Central Role of Abscisic Acid in Drought Stress Protection of Agrobacterium-Induced Tumors on Arabidopsis. Plant Physiol. 2007, 145, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, K.; Okamoto, M.; Seo, M.; Koshiba, T. Activation of abscisic acid biosynthesis in the leaves of Arabidopsis thaliana in response to water deficit. J. Plant Res. 2008, 122, 235–243. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.I.; Nambara, E. A Quick Release Mechanism for Abscisic Acid. Cell 2006, 126, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.M.; Melcher, K.; Teh, B.T.; Xu, H.E. Abscisic acid perception and signaling: Structural mechanisms and applications. Acta Pharmacol. Sin. 2014, 35, 567–584. [Google Scholar] [CrossRef]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Klingler, J.P.; Batelli, G.; Zhu, J.-K. ABA receptors: The START of a new paradigm in phytohormone signalling. J. Exp. Bot. 2010, 61, 3199–3210. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Weiner, J.J.; Peterson, F.C.; Volkman, B.F.; Cutler, S.R. Structural and functional insights into core ABA signaling. Curr. Opin. Plant Biol. 2010, 13, 495–502. [Google Scholar] [CrossRef]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K. Molecular Basis of the Core Regulatory Network in ABA Responses: Sensing, Signaling and Transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef]
- Gosti, F.; Beaudoin, N.; Serizet, C.; Webb, A.A.R.; Vartanian, N.; Giraudat, J. ABI1 Protein Phosphatase 2C Is a Negative Regulator of Abscisic Acid Signaling. Plant Cell 1999, 11, 1897. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Yoshida, T.; Kitahata, N.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007, 50, 935–949. [Google Scholar] [CrossRef]
- Saez, A.; Apostolova, N.; Gonzalez-Guzman, M.; Gonzalez-Garcia, M.P.; Nicolas, C.; Lorenzo, O.; Rodriguez, P.L. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 2004, 37, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-Hypersensitive Germination3 Encodes a Protein Phosphatase 2C (AtPP2CA) That Strongly Regulates Abscisic Acid Signaling during Germination among Arabidopsis Protein Phosphatase 2Cs. Plant Physiol. 2006, 140, 115–126. [Google Scholar] [CrossRef]
- Komatsu, K.; Suzuki, N.; Kuwamura, M.; Nishikawa, Y.; Nakatani, M.; Ohtawa, H.; Takezawa, D.; Seki, M.; Tanaka, M.; Taji, T.; et al. Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat. Commun. 2013, 4, 2219. [Google Scholar] [CrossRef]
- Hirayama, T.; Umezawa, T. The Pp2c-Snrk2 Complex: The Central Regulator of an Abscisic Acid Signaling Pathway. Plant Signal Behav. 2010, 5, 160–163. [Google Scholar] [CrossRef]
- Lin, Z.; Li, Y.; Wang, Y.; Liu, X.; Ma, L.; Zhang, Z.; Mu, C.; Zhang, Y.; Peng, L.; Xie, S.; et al. Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 2021, 12, 2456. [Google Scholar] [CrossRef]
- Kamiyama, Y.; Katagiri, S.; Umezawa, T. Growth Promotion or Osmotic Stress Response: How SNF1-Related Protein Kinase 2 (SnRK2) Kinases Are Activated and Manage Intracellular Signaling in Plants. Plants 2021, 10, 1443. [Google Scholar] [CrossRef]
- Kulik, A.; Wawer, I.; Krzywinska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 Protein Kinases—Key Regulators of Plant Response to Abiotic Stresses. OMICS J. Integr. Biol. 2011, 15, 859–872. [Google Scholar] [CrossRef]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 Protein Kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, Involved in ABA Signaling are Essential for the Control of Seed Development and Dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [PubMed]
- Maszkowska, J.; Szymańska, K.P.; Kasztelan, A.; Krzywińska, E.; Sztatelman, O.; Dobrowolska, G. The Multifaceted Regulation of SnRK2 Kinases. Cells 2021, 10, 2180. [Google Scholar] [CrossRef] [PubMed]
- Hoang, X.L.T.; Nhi, D.N.H.; Thu, N.B.A.; Thao, N.P.; Tran, L.-S.P. Transcription Factors and Their Roles in Signal Transduction in Plants under Abiotic Stresses. Curr. Genom. 2017, 18, 483–497. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Laxmi, A. Transcriptional regulation of drought response: A tortuous network of transcriptional factors. Front. Plant Sci. 2015, 6, 895. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. Ap2/Erf Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, S.; Ruan, M.; Wang, Y.; Wang, C. Analysis and application of RD29 genes in abiotic stress response. Acta Physiol. Plant. 2012, 34, 1239–1250. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Jha, B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol. Plant. 2010, 54, 201–212. [Google Scholar] [CrossRef]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.-J.; Go, Y.S.; Park, C.-M. The MYB96 Transcription Factor Regulates Cuticular Wax Biosynthesis under Drought Conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef]
- Finkelstein, R. Abscisic Acid Synthesis and Response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Lata, C.; Prasad, M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef]
- Riera, M.; Valon, C.; Fenzi, F.; Giraudat, J.; Leung, J. The genetics of adaptive responses to drought stress: Abscisic acid-dependent and abscisic acid-independent signalling components. Physiol. Plant 2005, 123, 111–119. [Google Scholar] [CrossRef]
- Jose, J.; Choudhury, S.R. Heterotrimeric G-proteins mediated hormonal responses in plants. Cell. Signal 2020, 76, 109799. [Google Scholar] [CrossRef]
- Kaur, J.; Choudhury, S.R.; Vijayakumar, A.; Hovis, L.; Rhodes, Z.; Polzin, R.; Blumenthal, D.; Pandey, S. Arabidopsis Type III Gγ Protein AGG3 Is a Positive Regulator of Yield and Stress Responses in the Model Monocot Setaria viridis. Front. Plant Sci. 2018, 9, 109. [Google Scholar] [CrossRef]
- Choudhury, S.R.; Riesselman, A.J.; Pandey, S. Constitutive or seed-specific overexpression of ArabidopsisG-protein γ subunit 3(AGG3) results in increased seed and oil production and improved stress tolerance in Camelina sativa. Plant Biotechnol. J. 2013, 12, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.P.; Zhou, Y.; Yin, J.F.; Yan, X.J.; Lin, S.; Xu, W.F.; Baluška, F.; Wang, Y.P.; Xia, Y.J.; Liang, G.H.; et al. Rice G-Protein Subunits Qpe9-1 and Rgb1 Play Distinct Roles in Abscisic Acid Responses and Drought Adaptation. J. Exp. Bot. 2015, 66, 6371–6384. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic Acid Signaling and Abiotic Stress Tolerance in Plants: A Review on Current Knowledge and Future Prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, A.A.; Gori, A.; Da-Silva, C.J.; Brunetti, C. Abscisic Acid Biosynthesis and Signaling in Plants: Key Targets to Improve Water Use Efficiency and Drought Tolerance. Appl. Sci. 2020, 10, 6322. [Google Scholar] [CrossRef]
- Kim, T.-H.; Böhmer, M.; Hu, H.; Nishimura, N.; Schroeder, J.I. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and CA2+ signaling. Annu. Rev. Plant Biol. 2010, 61, 561–591. [Google Scholar] [CrossRef]
- Munemasa, S.; Hauser, F.; Park, J.; Waadt, R.; Brandt, B.; Schroeder, J.I. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 2015, 28, 154–162. [Google Scholar] [CrossRef]
- Pirasteh-Anosheh, H.; Saed-Moucheshi, A.; Pakniyat, H.; Pessarakli, M. Stomatal responses to drought stress. In Water Stress and Crop Plants; Wiley: Hoboken, NJ, USA, 2016; pp. 24–40. [Google Scholar]
- Bauer, H.; Ache, P.; Lautner, S.; Fromm, J.; Hartung, W.; Al-Rasheid, K.A.; Sonnewald, S.; Sonnewald, U.; Kneitz, S.; Lachmann, N.; et al. The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol. 2013, 23, 53–57. [Google Scholar] [CrossRef]
- Lee, K.H.; Piao, H.L.; Kim, H.-Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.-J.; Hwang, I. Activation of Glucosidase via Stress-Induced Polymerization Rapidly Increases Active Pools of Abscisic Acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Jose, J.; Ghantasala, S.; Choudhury, S.R. Arabidopsis Transmembrane Receptor-Like Kinases (RLKs): A Bridge between Extracellular Signal and Intracellular Regulatory Machinery. Int. J. Mol. Sci. 2020, 21, 4000. [Google Scholar] [CrossRef]
- Geiger, D.; Scherzer, S.; Mumm, P.; Marten, I.; Ache, P.; Matschi, S.; Liese, A.; Wellmann, C.; Al-Rasheid, K.A.S.; Grill, E.; et al. Guard Cell Anion Channel Slac1 Is Regulated by Cdpk Protein Kinases with Distinct Ca2+ Affinities. Proc. Natl. Acad. Sci. USA 2010, 107, 8023–8028. [Google Scholar] [CrossRef]
- Mori, I.C.; Murata, Y.; Yang, Y.; Munemasa, S.; Wang, Y.-F.; Andreoli, S.; Tiriac, H.; Alonso, J.; Harper, J.F.; Ecker, J.; et al. CDPKs CPK6 and CPK3 Function in ABA Regulation of Guard Cell S-Type Anion- and Ca2+-Permeable Channels and Stomatal Closure. PLoS Biol. 2006, 4, e327. [Google Scholar] [CrossRef] [PubMed]
- Imes, D.; Mumm, P.; Böhm, J.; Al-Rasheid, K.A.S.; Marten, I.; Geiger, D.; Hedrich, R. Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 2013, 74, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.; Mumm, P.; Imes, D.; Endler, A.; Weder, B.; Al-Rasheid, K.A.; Geiger, D.; Marten, I.; Martinoia, E.; Hedrich, R. Atalmt12 Represents an R-Type Anion Channel Required for Stomatal Movement in Arabidopsis Guard Cells. Plant J. 2010, 63, 54–62. [Google Scholar] [CrossRef]
- Eisenach, C.; De Angeli, A. Ion Transport at the Vacuole during Stomatal Movements. Plant Physiol. 2017, 174, 520–530. [Google Scholar] [CrossRef]
- Eisenach, C.; Baetz, U.; Huck, N.V.; Zhang, J.; De Angeli, A.; Beckers, G.J.; Martinoia, E. ABA-Induced Stomatal Closure Involves ALMT4, a Phosphorylation-Dependent Vacuolar Anion Channel of Arabidopsis. Plant Cell 2017, 29, 2552–2569. [Google Scholar] [CrossRef]
- Kwak, J.M.; Mori, I.C.; Pei, Z.-M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.; Schroeder, J. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Sierla, M.; Waszczak, C.; Vahisalu, T.; Kangasjärvi, J. Reactive Oxygen Species in the Regulation of Stomatal Movements. Plant Physiol. 2016, 171, 1569–1580. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.-J. Overexpression of AtMYB44 Enhances Stomatal Closure to Confer Abiotic Stress Tolerance in Transgenic Arabidopsis. Plant Physiol. 2008, 146, 323–324. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Li, S.; An, X.; Liu, X.; Qin, H.; Wang, D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genom. 2009, 36, 17–29. [Google Scholar] [CrossRef]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.-Y.; Na Lee, Y.; Kim, S.-G.; Lee, Y.-H.; Park, W.J.; Park, C.-M. The MYB96 Transcription Factor Mediates Abscisic Acid Signaling during Drought Stress Response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef]
- Song, C.P.; Agarwal, M.; Ohta, M.; Guo, Y.; Halfter, U.; Wang, P.; Zhu, J.K. Role of an Arabidopsis Ap2/Erebp-Type Transcriptional Repressor in Abscisic Acid and Drought Stress Responses. Plant Cell 2005, 17, 2384–2396. [Google Scholar] [CrossRef]
- Li, W.-X.; Oono, Y.; Zhu, J.; He, X.-J.; Wu, J.; Iida, K.; Lu, X.-Y.; Cui, X.; Jin, H.; Zhu, J.-K. The Arabidopsis NFYA5 Transcription Factor Is Regulated Transcriptionally and Posttranscriptionally to Promote Drought Resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef]
- Kim, M.J.; Shin, R.; Schachtman, D. A Nuclear Factor Regulates Abscisic Acid Responses in Arabidopsis. Plant Physiol. 2009, 151, 1433–1445. [Google Scholar] [CrossRef]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef]
- Cominelli, E.; Galbiati, M.; Tonelli, C. Transcription factors controlling stomatal movements and drought tolerance. Transcr. 2010, 1, 41–45. [Google Scholar] [CrossRef]
- Seo, D.H.; Ryu, M.Y.; Jammes, F.; Hwang, J.H.; Turek, M.; Kang, B.G.; Kwak, J.M.; Kim, W.T. Roles of Four Arabidopsis U-Box E3 Ubiquitin Ligases in Negative Regulation of Abscisic Acid-Mediated Drought Stress Responses. Plant Physiol. 2012, 160, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Van Houtte, H.; Vandesteene, L.; López-Galvis, L.; Lemmens, L.; Kissel, E.; Carpentier, S.; Feil, R.; Avonce, N.; Beeckman, T.; Lunn, J.E.; et al. Overexpression of the Trehalase Gene AtTRE1 Leads to Increased Drought Stress Tolerance in Arabidopsis and Is Involved in Abscisic Acid-Induced Stomatal Closure. Plant Physiol. 2013, 161, 1158–1171. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Gao, J.; Kim, J.I.; Chen, K.; Bressan, R.A.; Zhu, J.-K. Control of Plant Water Use by ABA Induction of Senescence and Dormancy: An Overlooked Lesson from Evolution. Plant Cell Physiol. 2017, 58, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Kermode, A.R. Role of Abscisic Acid in Seed Dormancy. J. Plant Growth Regul. 2005, 24, 319–344. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, H.; Ding, L.; Soppe, W.J.; Xiang, Y. REVERSAL OF RDO5 1, a Homolog of Rice Seed Dormancy4, Interacts with bHLH57 and Controls ABA Biosynthesis and Seed Dormancy in Arabidopsis. Plant Cell 2020, 32, 1933–1948. [Google Scholar] [CrossRef]
- Sano, N.; Marion-Poll, A. ABA Metabolism and Homeostasis in Seed Dormancy and Germination. Int. J. Mol. Sci. 2021, 22, 5069. [Google Scholar] [CrossRef]
- Piskurewicz, U.; Jikumaru, Y.; Kinoshita, N.; Nambara, E.; Kamiya, Y.; Lopez-Molina, L. The Gibberellic Acid Signaling Repressor RGL2 Inhibits Arabidopsis Seed Germination by Stimulating Abscisic Acid Synthesis and ABI5 Activity. Plant Cell 2008, 20, 2729–2745. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, Y.-N.; Xue, L.-J.; Zou, M.-J.; Liu, J.-Y.; Chen, F.; Xue, H.-W. Rice ABI5-Like1 Regulates Abscisic Acid and Auxin Responses by Affecting the Expression of ABRE-Containing Genes. Plant Physiol. 2011, 156, 1397–1409. [Google Scholar] [CrossRef]
- Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and Role of the Arabidopsis Abscisic Acid-Insensitive 5 Gene in Abscisic Acid, Sugar, and Stress Response. Plant Physiol. 2002, 129, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Kanai, M.; Nishimura, M.; Hayashi, M. A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J. 2010, 62, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1Regulates Seed Germination through a Negative Feedback Loop Modulating ABA Signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Vaistij, F.E.; Gan, Y.; Penfield, S.; Gilday, A.D.; Dave, A.; He, Z.; Josse, E.M.; Choi, G.; Halliday, K.J.; Graham, I.A. Differential Control of Seed Primary Dormancy in Arabidopsis Ecotypes by the Transcription Factor Spatula. Proc. Natl. Acad. Sci. USA 2013, 110, 10866–10871. [Google Scholar] [CrossRef]
- Gil Lee, H.; Lee, K.; Seo, P.J. The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Biol. 2015, 87, 371–381. [Google Scholar] [CrossRef]
- Lee, K.; Gil Lee, H.; Yoon, S.; Kim, H.U.; Seo, P.J. The Arabidopsis MYB96 Transcription Factor Is a Positive Regulator of ABSCISIC ACID-INSENSITIVE4 in the Control of Seed Germination. Plant Physiol. 2015, 168, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. Wrky Transcription Factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Née, G.; Kramer, K.; Nakabayashi, K.; Yuan, B.; Xiang, Y.; Miatton, E.; Finkemeier, I.; Soppe, W.J.J. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 2017, 8, 72. [Google Scholar] [CrossRef]
- Carrillo-Barral, N.; Rodríguez-Gacio, M.D.C.; Matilla, A.J. Delay of Germination-1 (DOG1): A Key to Understanding Seed Dormancy. Plants 2020, 9, 480. [Google Scholar] [CrossRef]
- Ashikawa, I.; Mori, M.; Nakamura, S.; Abe, F. A transgenic approach to controlling wheat seed dormancy level by using Triticeae DOG1-like genes. Transgenic Res. 2014, 23, 621–629. [Google Scholar] [CrossRef]
- Yu, X.; Han, J.; Li, L.; Zhang, Q.; Yang, G.; He, G. Wheat PP2C-a10 regulates seed germination and drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 2020, 39, 635–651. [Google Scholar] [CrossRef]
- Armando, C.L.; Yahia, E.M. Chapter 6—Morphology and Anatomy. In Postharvest Physiology and Biochemistry of Fruits and Vegetables; Yahia, E.M., Ed.; Woodhead Publishing: Sawston, UK, 2019; pp. 113–130. [Google Scholar]
- Xue, D.; Zhang, X.; Lu, X.; Chen, G.; Chen, Z.-H. Molecular and Evolutionary Mechanisms of Cuticular Wax for Plant Drought Tolerance. Front. Plant Sci. 2017, 8, 621. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Xu, W.; Yu, X.; Shen, H.; Li, H.; Cheng, D.; Liu, A.; Liu, J.; Liu, C.; Zhao, S.; et al. Cuticular Wax Accumulation Is Associated with Drought Tolerance in Wheat Near-Isogenic Lines. Front. Plant Sci. 2016, 7, 1809. [Google Scholar] [CrossRef]
- Chen, M.; Zhu, X.; Zhang, Y.; Du, Z.; Chen, X.; Kong, X.; Sun, W.; Chen, C. Drought stress modify cuticle of tender tea leaf and mature leaf for transpiration barrier enhancement through common and distinct modes. Sci. Rep. 2020, 10, 6696. [Google Scholar] [CrossRef]
- Post-Beittenmiller, D. BIOCHEMISTRY AND MOLECULAR BIOLOGY OF WAX PRODUCTION IN PLANTS. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 405–430. [Google Scholar] [CrossRef]
- Patwari, P.; Salewski, V.; Gutbrod, K.; Kreszies, T.; Dresen-Scholz, B.; Peisker, H.; Steiner, U.; Meyer, A.J.; Schreiber, L.; Dörmann, P. Surface wax esters contribute to drought tolerance in Arabidopsis. Plant J. 2019, 98, 727–744. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulos, N.; Tindjau, R.; Wong, D.C.J.; Matzat, T.; Haslam, T.; Song, C.; Gambetta, G.A.; Kunst, L.; Castellarin, S.D. Drought stress modulates cuticular wax composition of the grape berry. J. Exp. Bot. 2020, 71, 3126–3141. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Du, Y.; He, C.; Dietrich, C.R.; Li, J.; Ma, X.; Wang, R.; Liu, Q.; Liu, S.; Wang, G.; et al. Maize glossy6 is involved in cuticular wax deposition and drought tolerance. J. Exp. Bot. 2019, 70, 3089–3099. [Google Scholar] [CrossRef]
- Kosma, D.K.; Jenks, M.A. Eco-physiological and molecular-genetic determinants of plant cuticle function in drought and salt stress tolerance. In Advances in Molecular Breeding towards Drought and Salt Tolerant Crops; Jenks, M.A., Hasegawa, P.M., Jain, S.M., Eds.; Springer: Dortrecht, The Netherlands, 2007; pp. 91–120. [Google Scholar]
- Hooker, T.S.; Millar, A.A.; Kunst, L. Significance of the Expression of the CER6 Condensing Enzyme for Cuticular Wax Production in Arabidopsis. Plant Physiol. 2002, 129, 1568–1580. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, S.; Xu, Y.; Li, S.; Zhang, S.; Yuan, Z.; Li, J.; Ni, Y. Overexpression of BnKCS1-1, BnKCS1-2, and BnCER1-2 promotes cuticular wax production and increases drought tolerance in Brassica napus. Crop. J. 2019, 8, 26–37. [Google Scholar] [CrossRef]
- Lee, S.B.; Kim, H.; Kim, R.J.; Suh, M.C. Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Rep. 2014, 33, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Kim, H.U.; Suh, M.C. MYB94 and MYB96 Additively Activate Cuticular Wax Biosynthesis in Arabidopsis. Plant Cell Physiol. 2016, 57, 2300–2311. [Google Scholar] [CrossRef]
- Lee, S.B.; Suh, M.C. Cuticular Wax Biosynthesis is Up-Regulated by the MYB94 Transcription Factor in Arabidopsis. Plant Cell Physiol. 2014, 56, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Castorina, G.; Domergue, F.; Chiara, M.; Zilio, M.; Persico, M.; Ricciardi, V.; Horner, D.S.; Consonni, G. Drought-Responsive Zmfdl1/Myb94 Regulates Cuticle Biosynthesis and Cuticle-Dependent Leaf Permeability. Plant Physiol. 2020, 184, 266–282. [Google Scholar] [CrossRef]
- Yang, S.U.; Kim, H.; Kim, R.J.; Kim, J.; Suh, M.C. AP2/DREB Transcription Factor RAP2.4 Activates Cuticular Wax Biosynthesis in Arabidopsis Leaves Under Drought. Front. Plant Sci. 2020, 11, 895. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Alegre, L. Die and let live: Leaf senescence contributes to plant survival under drought stress. Funct. Plant Biol. 2004, 31, 203–216. [Google Scholar] [CrossRef]
- Zhang, Z.; Guo, Y. Hormone Treatments in Studying Leaf Senescence. In Springer Protocols Handbooks; Springer: Singapore, 2018; Volume 1744, pp. 125–132. [Google Scholar]
- Tiwari, S.; Lata, C.; Chauhan, P.S.; Prasad, V.; Prasad, M. A Functional Genomic Perspective on Drought Signalling and its Crosstalk with Phytohormone-mediated Signalling Pathways in Plants. Curr. Genom. 2017, 18, 469–482. [Google Scholar] [CrossRef]
- Huang, Y.; Jiao, Y.; Xie, N.; Guo, Y.; Zhang, F.; Xiang, Z.; Wang, R.; Wang, F.; Gao, Q.; Tian, L.; et al. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 2019, 287, 110188. [Google Scholar] [CrossRef] [PubMed]
- Riov, J.; Dagan, E.; Goren, R.; Yang, S.F. Characterization of Abscisic Acid-Induced Ethylene Production in Citrus Leaf and Tomato Fruit Tissues. Plant Physiol. 1990, 92, 48–53. [Google Scholar] [CrossRef]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Kwasniewski, M.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Xue, G.P.; Mueller-Roeber, B. Ors1, an H₂O₂-Responsive Nac Transcription Factor, Controls Senescence in Arabidopsis Thaliana. Mol. Plant 2011, 4, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Snac-as, Stress-Responsive Nac Transcription Factors, Mediate Aba-Inducible Leaf Senescence. Plant J. 2015, 84, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Worley, E.; Udvardi, M. A NAP-AAO3 Regulatory Module Promotes Chlorophyll Degradation via ABA Biosynthesis in Arabidopsis Leaves. Plant Cell 2014, 26, 4862–4874. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Gao, J.; Zhu, X.; Song, Y.; Li, Z.; Ren, G.; Zhou, X.; Kuai, B. ABF2, ABF3, and ABF4 Promote ABA-Mediated Chlorophyll Degradation and Leaf Senescence by Transcriptional Activation of Chlorophyll Catabolic Genes and Senescence-Associated Genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef]
- Kim, T.; Kang, K.; An, G.; Paek, N.-C. OsWRKY5 Promotes Rice Leaf Senescence via Senescence-Associated NAC and Abscisic Acid Biosynthesis Pathway. Int. J. Mol. Sci. 2019, 20, 4437. [Google Scholar] [CrossRef]
- Liu, T.; Longhurst, A.D.; Talavera-Rauh, F.; Hokin, S.A.; Barton, M.K. The Arabidopsis transcription factor ABIG1 relays ABA signaled growth inhibition and drought induced senescence. eLife 2016, 5, e13768. [Google Scholar] [CrossRef]
- Piao, W.; Kim, S.-H.; Lee, B.-D.; An, G.; Sakuraba, Y.; Paek, N.-C. Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 2019, 70, 2699–2715. [Google Scholar] [CrossRef]
- Gu, L.; Ma, Q.; Zhang, C.; Wang, C.; Wei, H.; Wang, H.; Yu, S. The Cotton GhWRKY91 Transcription Factor Mediates Leaf Senescence and Responses to Drought Stress in Transgenic Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1352. [Google Scholar] [CrossRef]
- Spollen, W.G.; Sharp, R.E.; Saab, I.N.; Wu, Y. Water Deficits: Plant Responses from Cell to Community; Smith, J.A.C., Griffiths, H., Eds.; Bios Scientific Publishers: Oxford, UK, 1993; pp. 37–52. [Google Scholar]
- Robertson, J.M.; Pharis, R.P.; Huang, Y.Y.; Reid, D.M.; Yeung, E.C. Drought-Induced Increases in Abscisic Acid Levels in the Root Apex of Sunflower. Plant Physiol. 1985, 79, 1086–1089. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Li, X. ABA mediates PEG-mediated premature differentiation of root apical meristem in plants. Plant Signal. Behav. 2014, 9, e977720. [Google Scholar] [CrossRef][Green Version]
- Sharp, R.E. Interaction with ethylene: Changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant Cell Environ. 2002, 25, 211–222. [Google Scholar] [CrossRef]
- Maurel, C.; Verdoucq, L.; Luu, D.T.; Santoni, V. Plant Aquaporins: Membrane Channels with Multiple Integrated Functions. Annu. Rev. Plant Biol. 2008, 59, 595–624. [Google Scholar] [CrossRef] [PubMed]
- Parent, B.; Hachez, C.; Redondo, E.; Simonneau, T.; Chaumont, F.; Tardieu, F. Drought and Abscisic Acid Effects on Aquaporin Content Translate into Changes in Hydraulic Conductivity and Leaf Growth Rate: A Trans-Scale Approach. Plant Physiol. 2009, 149, 2000–2012. [Google Scholar] [CrossRef]
- Patel, J.; Mishra, A. Plant aquaporins alleviate drought tolerance in plants by modulating cellular biochemistry, root-architecture, and photosynthesis. Physiol. Plant. 2021, 172, 1030–1044. [Google Scholar] [CrossRef] [PubMed]
- Spollen, W.G.; Lenoble, M.E.; Samuels, T.D.; Bernstein, N.; Sharp, R.E. Abscisic Acid Accumulation Maintains Maize Primary Root Elongation at Low Water Potentials by Restricting Ethylene Production. Plant Physiol. 2000, 122, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Han, W.; De Smet, I.; Talboys, P.; Loya, R.; Hassan, A.; Rong, H.; Jürgens, G.; Knox, J.P.; Wang, M.-H. ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 2010, 64, 764–774. [Google Scholar] [CrossRef]
- Takatsuka, H.; Umeda, M. Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 2014, 65, 2633–2643. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Jia, L.; Shi, W.; Liang, J.; Zhou, F.; Li, Q.; Zhang, J. Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytol. 2013, 197, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.; Wang, X.; Han, X.; An, Y.; Lin, S.; Shen, C.; Wen, J.; Liu, C.; Yin, W.; et al. Root-Specific Nf-Y Family Transcription Factor, Pdnf-Yb21, Positively Regulates Root Growth and Drought Resistance by Abscisic Acid-Mediated Indoylacetic Acid Transport in Populus. New Phytol. 2020, 227, 407–426. [Google Scholar] [CrossRef]
- Thole, J.M.; Beisner, E.R.; Liu, J.; Venkova, S.V.; Strader, L.C. Abscisic Acid Regulates Root Elongation Through the Activities of Auxin and Ethylene in Arabidopsis thaliana. G3 Genes Genomes Genet. 2014, 4, 1259–1274. [Google Scholar] [CrossRef]
- Rowe, J.; Topping, J.F.; Liu, J.; Lindsey, K. Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 2016, 211, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.R.; Wang, Y.B.; He, S.B.; Hao, F.S. Mechanisms for Abscisic Acid Inhibition of Primary Root Growth. Plant Signal Behav. 2018, 13, e1500069. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Yuan, W.; Wang, Y.; Garcia-Maquilon, I.; Dang, X.; Li, Y.; Zhang, J.; Zhu, Y.; Rodriguez, P.L.; Xu, W. Low ABA concentration promotes root growth and hydrotropism through relief of ABA INSENSITIVE 1-mediated inhibition of plasma membrane H+-ATPase 2. Sci. Adv. 2021, 7, eabd4113. [Google Scholar] [CrossRef]
- Xiong, L.; Wang, R.-G.; Mao, G.; Koczan, J.M. Identification of Drought Tolerance Determinants by Genetic Analysis of Root Response to Drought Stress and Abscisic Acid. Plant Physiol. 2006, 142, 1065–1074. [Google Scholar] [CrossRef]
- Deak, K.I.; Malamy, J. Osmotic regulation of root system architecture. Plant J. 2005, 43, 17–28. [Google Scholar] [CrossRef]
- Shkolnik-Inbar, D.; Bar-Zvi, D. Expression of Abscisic Acid Insensitive 4 (Abi4) in Developing Arabidopsis Seedlings. Plant Signal Behav. 2011, 6, 694–696. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.-T.; Xu, H.-H.; Zhang, K.-X.; Guo, T.-T.; Lu, Y.-T. Glucose inhibits root meristem growth viaABA INSENSITIVE 5, which represses PIN1 accumulation and auxin activity in Arabidopsis. Plant Cell Environ. 2013, 37, 1338–1350. [Google Scholar] [CrossRef]
- Shkolnik-Inbar, D.; Bar-Zvi, D. ABI4 Mediates Abscisic Acid and Cytokinin Inhibition of Lateral Root Formation by Reducing Polar Auxin Transport in Arabidopsis. Plant Cell 2010, 22, 3560–3573. [Google Scholar] [CrossRef]
- Saab, I.N.; Sharp, R.E.; Pritchard, J.; Voetberg, G.S. Increased Endogenous Abscisic Acid Maintains Primary Root Growth and Inhibits Shoot Growth of Maize Seedlings at Low Water Potentials. Plant Physiol. 1990, 93, 1329–1336. [Google Scholar] [CrossRef] [PubMed]
- Creelman, R.A.; Mason, H.S.; Bensen, R.J.; Boyer, J.S.; Mullet, J.E. Water Deficit and Abscisic Acid Cause Differential Inhibition of Shoot versus Root Growth in Soybean Seedlings: Analysis of Growth, Sugar Accumulation, and Gene Expression. Plant Physiol. 1990, 92, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Sharp, R.E.; LeNoble, M.E. ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 2002, 53, 33–37. [Google Scholar] [CrossRef]
- Lenoble, M.E.; Spollen, W.G.; Sharp, R.E. Maintenance of shoot growth by endogenous ABA: Genetic assessment of the involvement of ethylene suppression. J. Exp. Bot. 2003, 55, 237–245. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a Key Player of the Basic Leucine Zipper Transcription Factor Family for Conferring Abscisic Acid Sensitivity and Salinity and Drought Tolerance in Rice. Plant Physiol. 2008, 148, 1938–1952. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).