Isolation and Functional Characterization of the Promoters of Miltiradiene Synthase Genes, TwTPS27a and TwTPS27b, and Interaction Analysis with the Transcription Factor TwTGA1 from Tripterygium wilfordii
Abstract
1. Introduction
2. Results
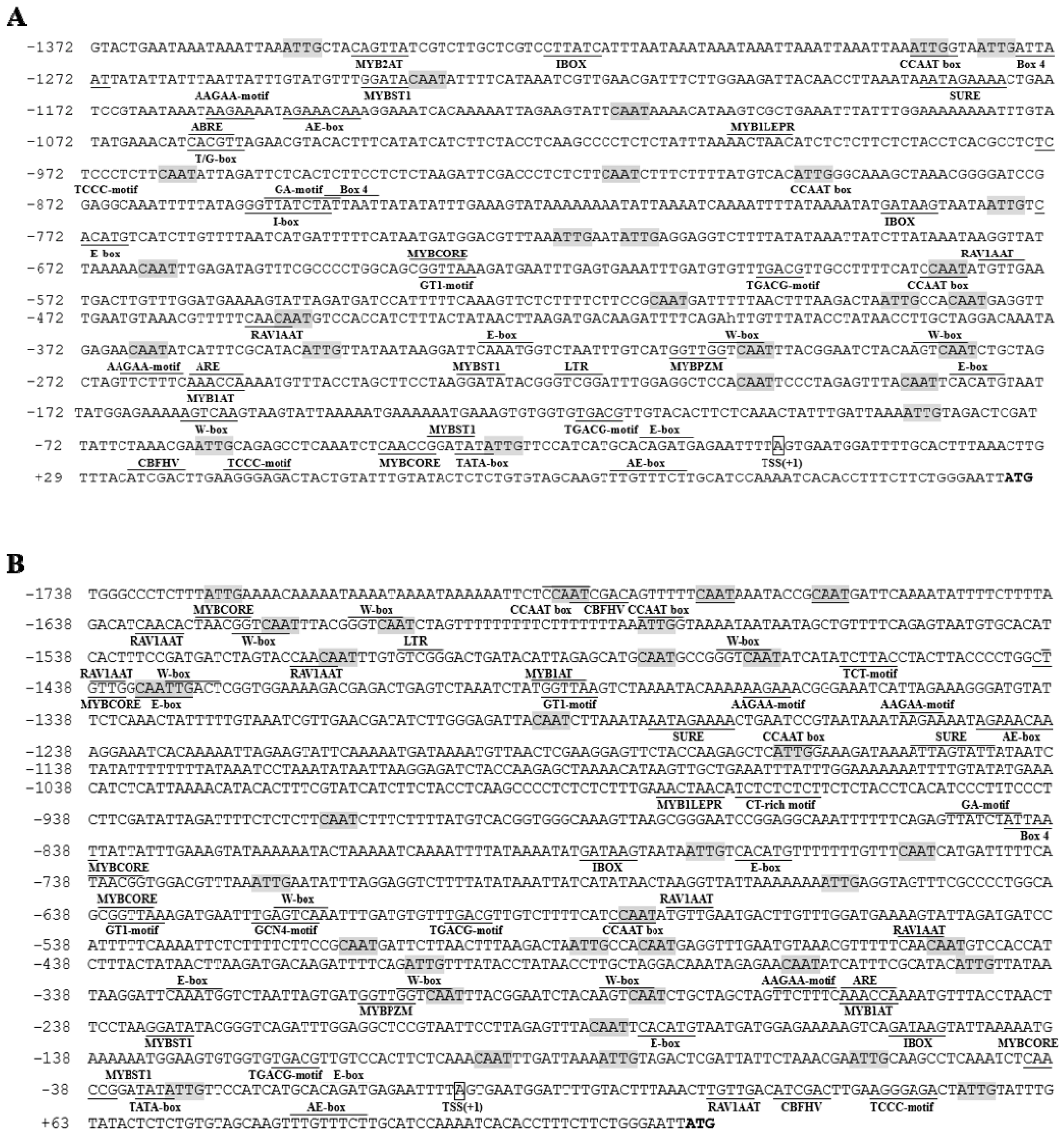
2.1. Isolation of Promoters and Prediction of Cis‑Regulatory Elements
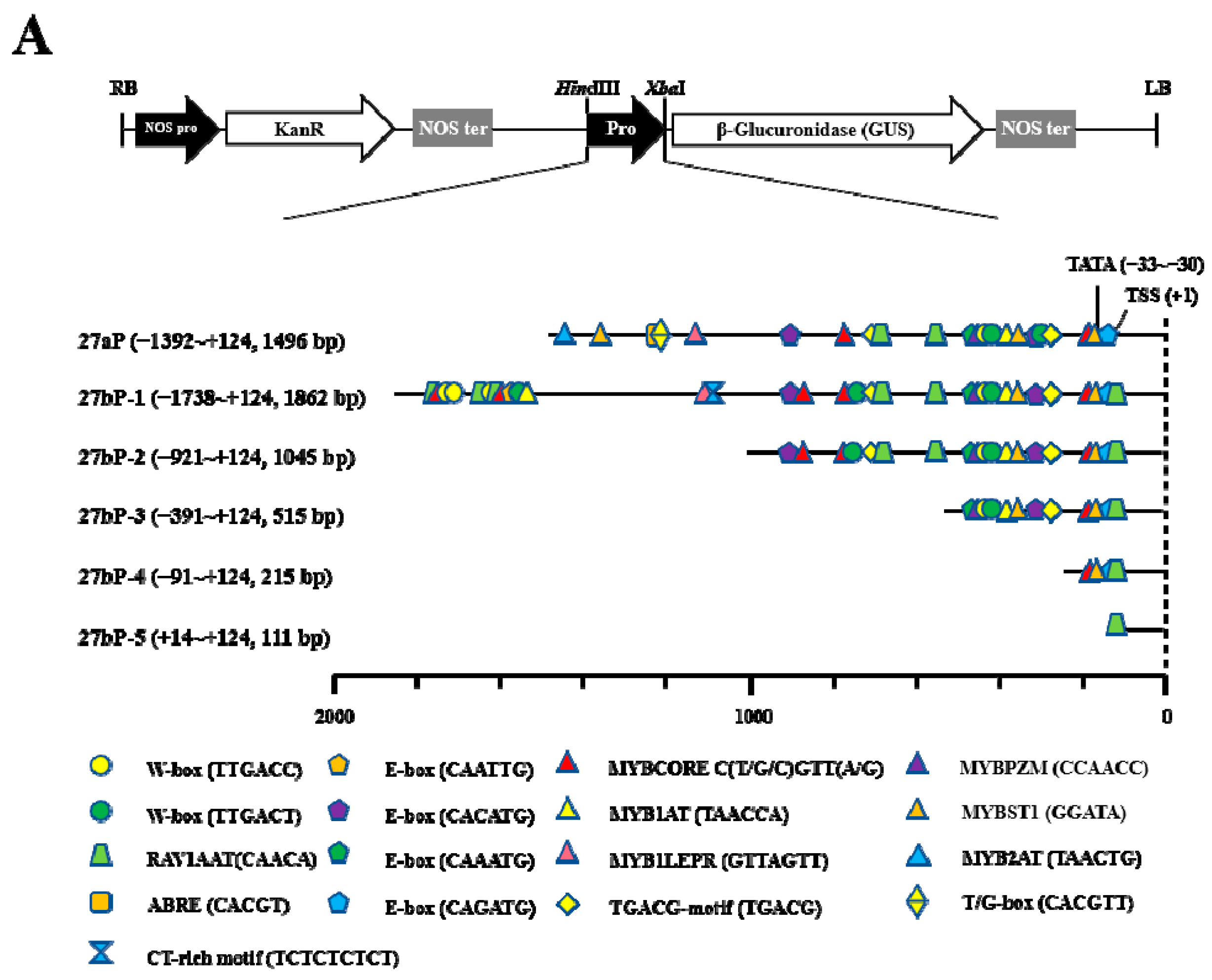
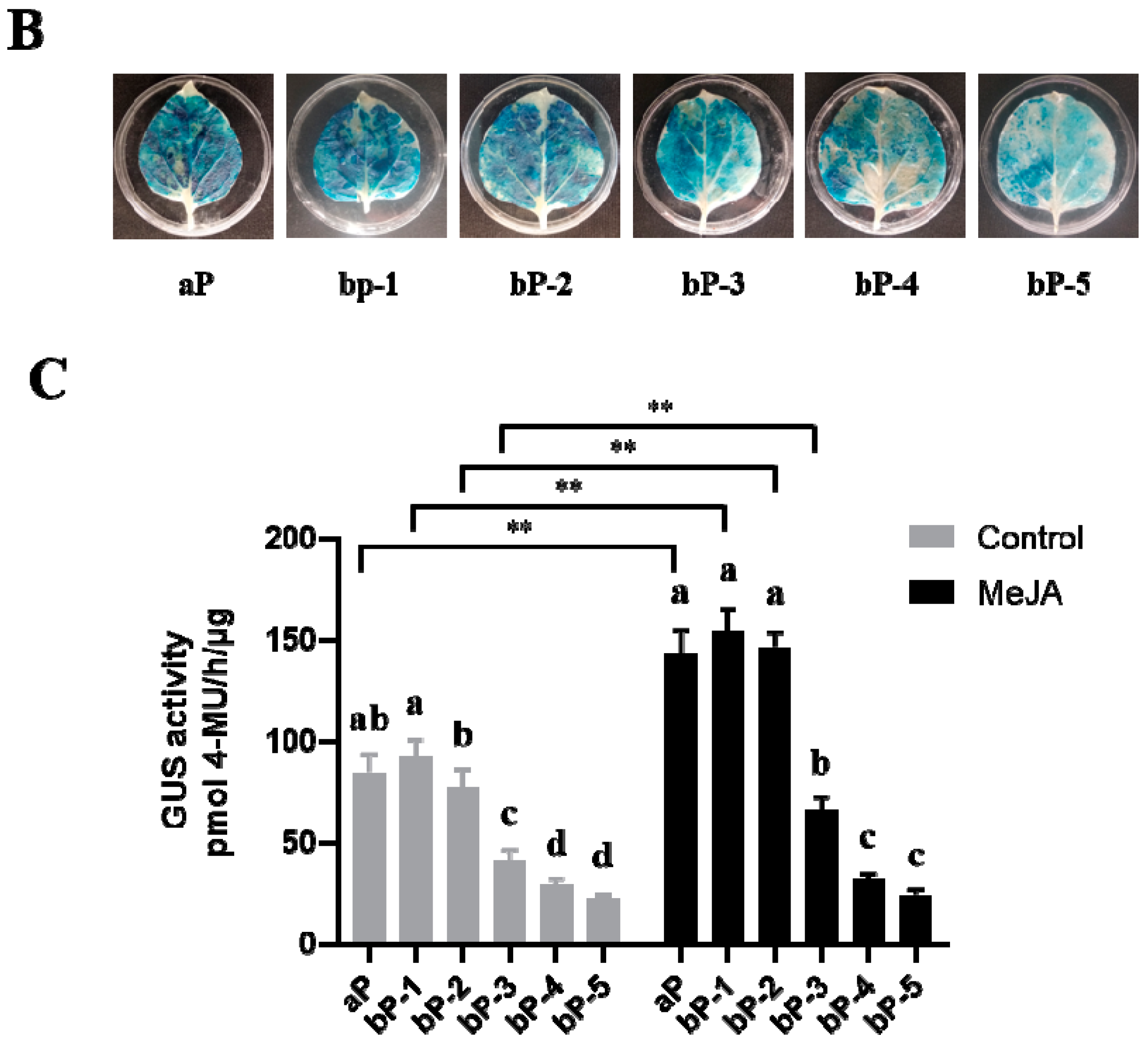
2.2. Promoter Deletion Analysis under Normal and MeJA Treatment Conditions
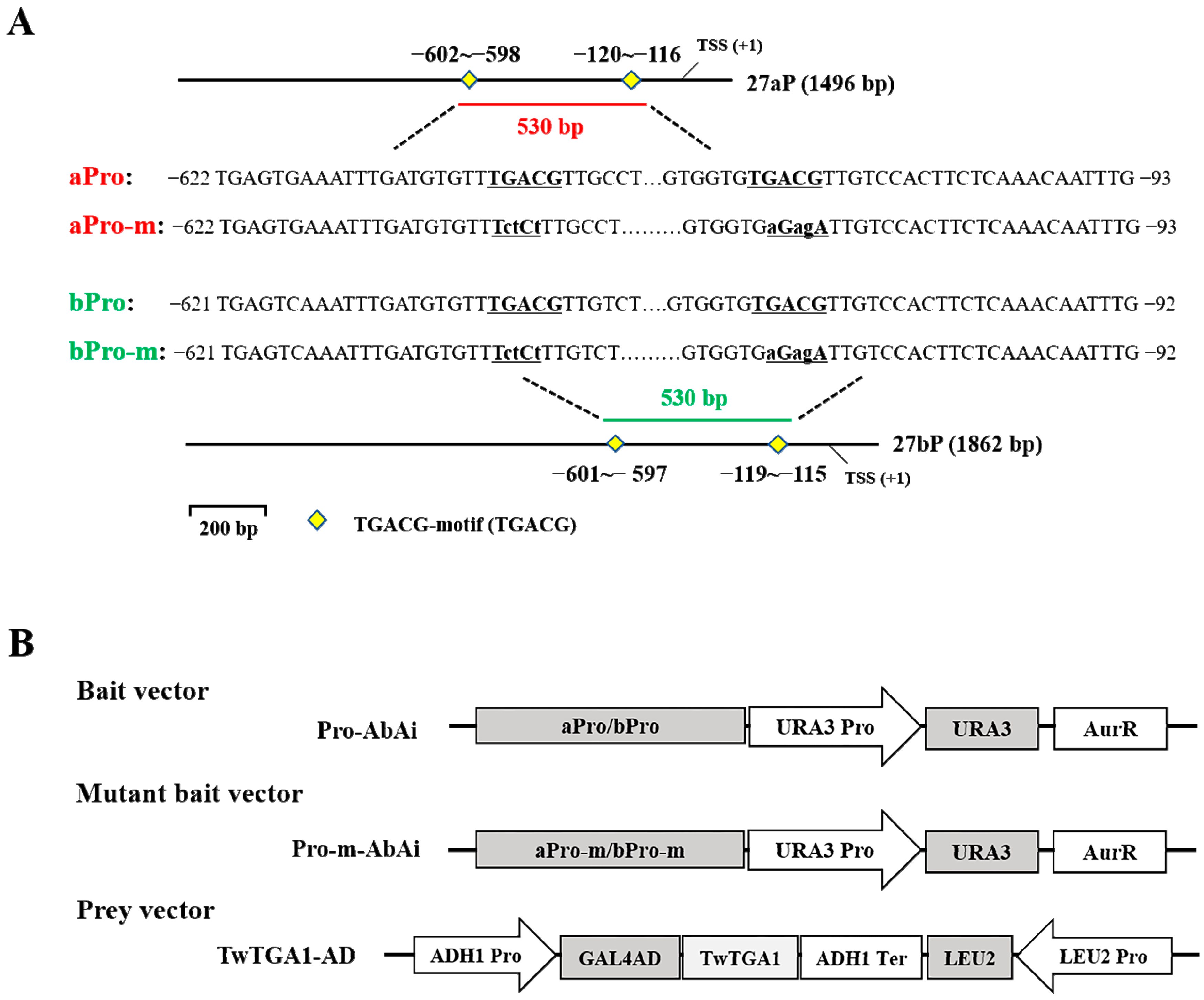
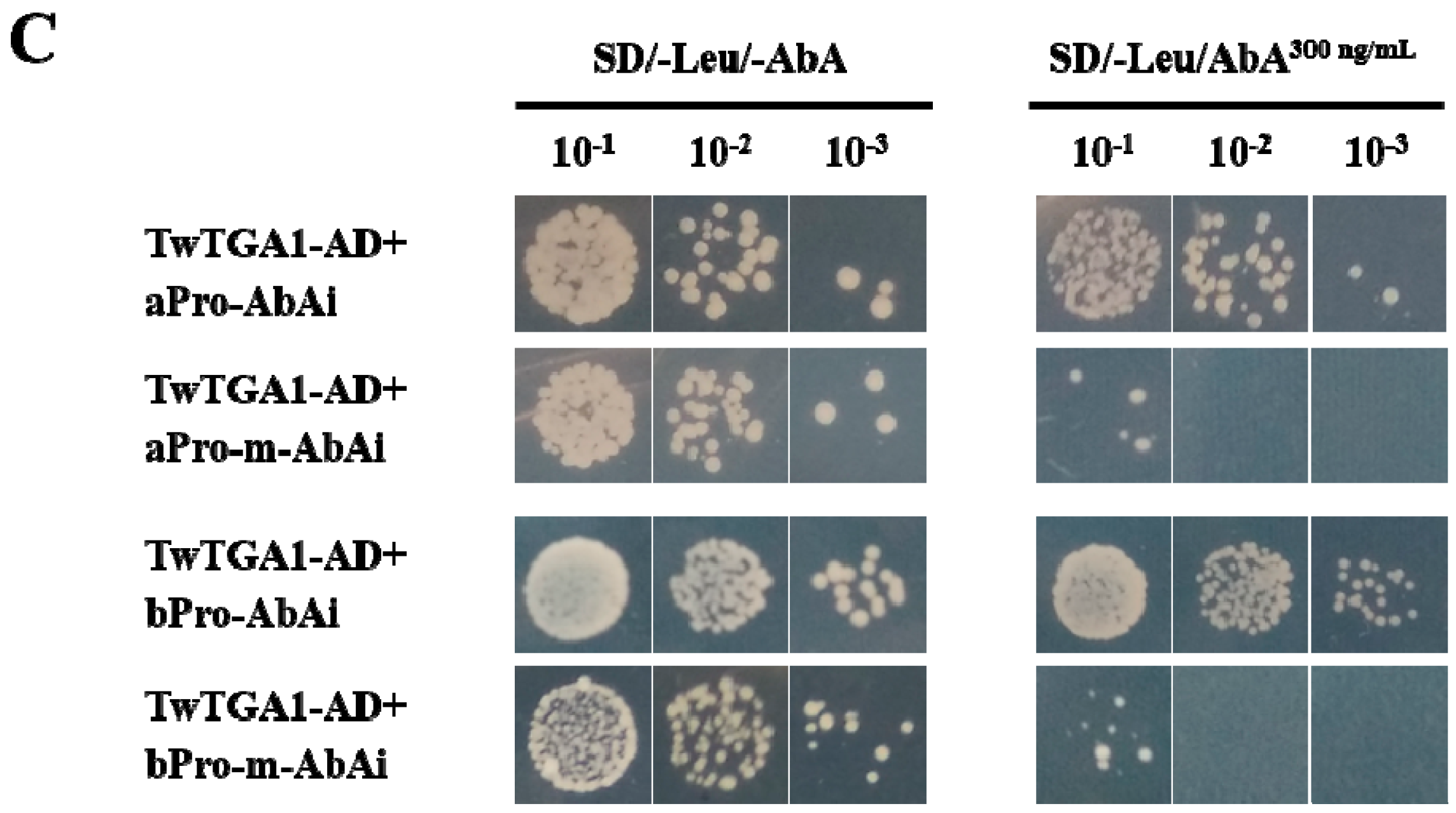
2.3. TwTGA1 Specifically Binds to the TGACG-motif in the TwTPS27a/b Promoters
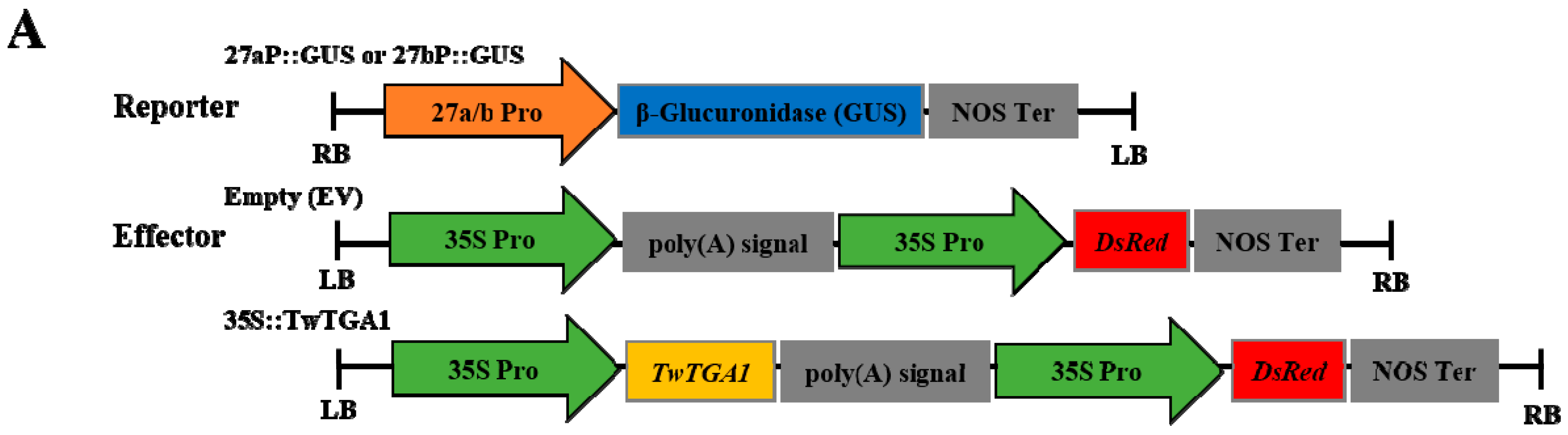
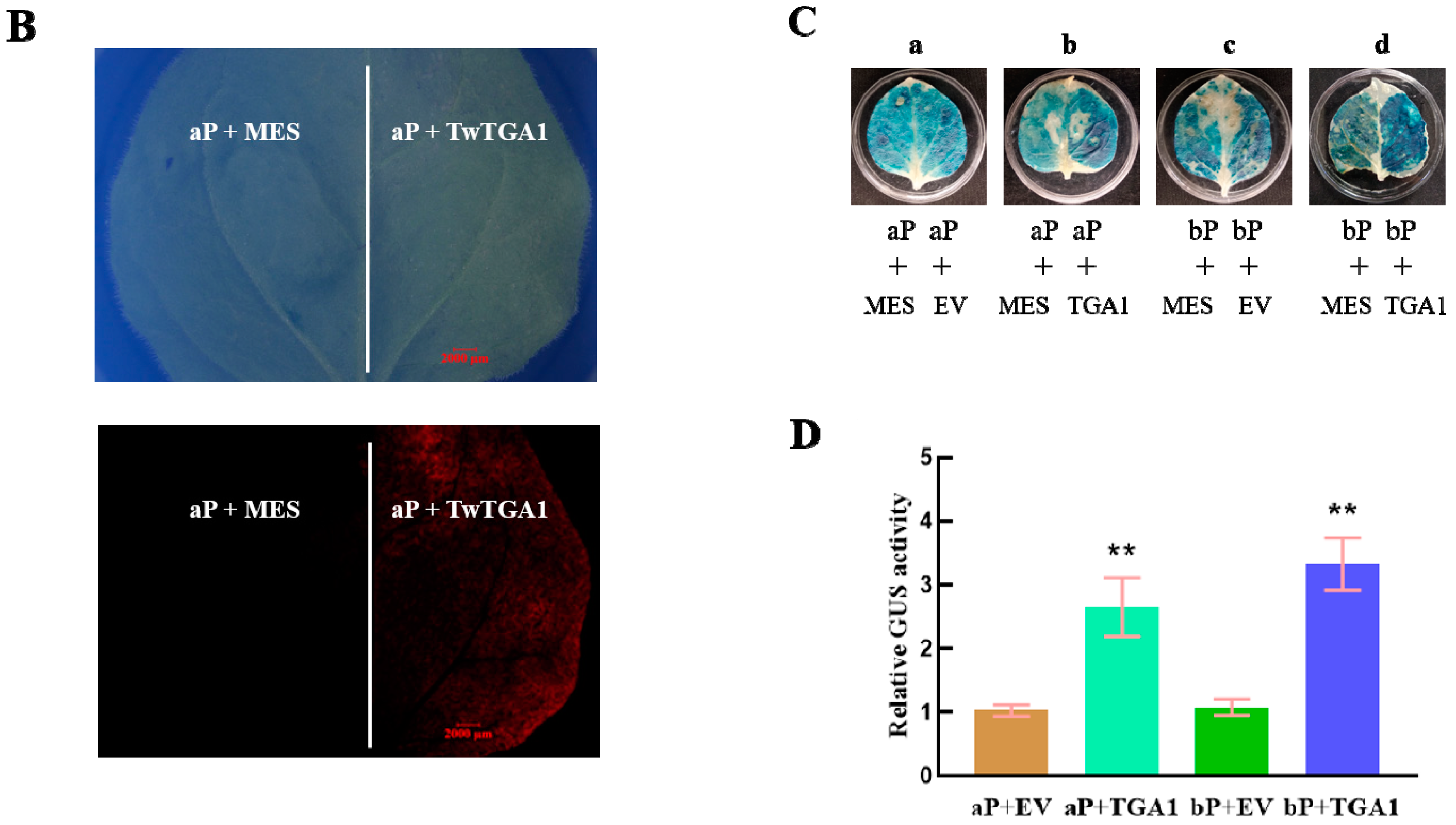
2.4. TwTGA1 Can Activate the TwTPS27a/b Promoters
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions
4.2. Isolation of the TwTPS27a/b Genes
4.3. Cloning and Analysis of the TwTPS27a/b Promoter Sequences
4.4. Construction of a Series of Promoters::GUS Vectors
4.5. Agrobacterium-Mediated Transient Expression in Tobacco Leaves and MeJA Treatment
4.6. GUS Staining and GUS Fluorimetric Assays
4.7. The Binding of TwTGA1 to the TGACG-motif Present in the TwTPS27a/b Promoters
4.8. Activation of the TwTPS7a/b Promoters by TwTGA1 in Tobacco Leaves
4.9. Statistical Analysis
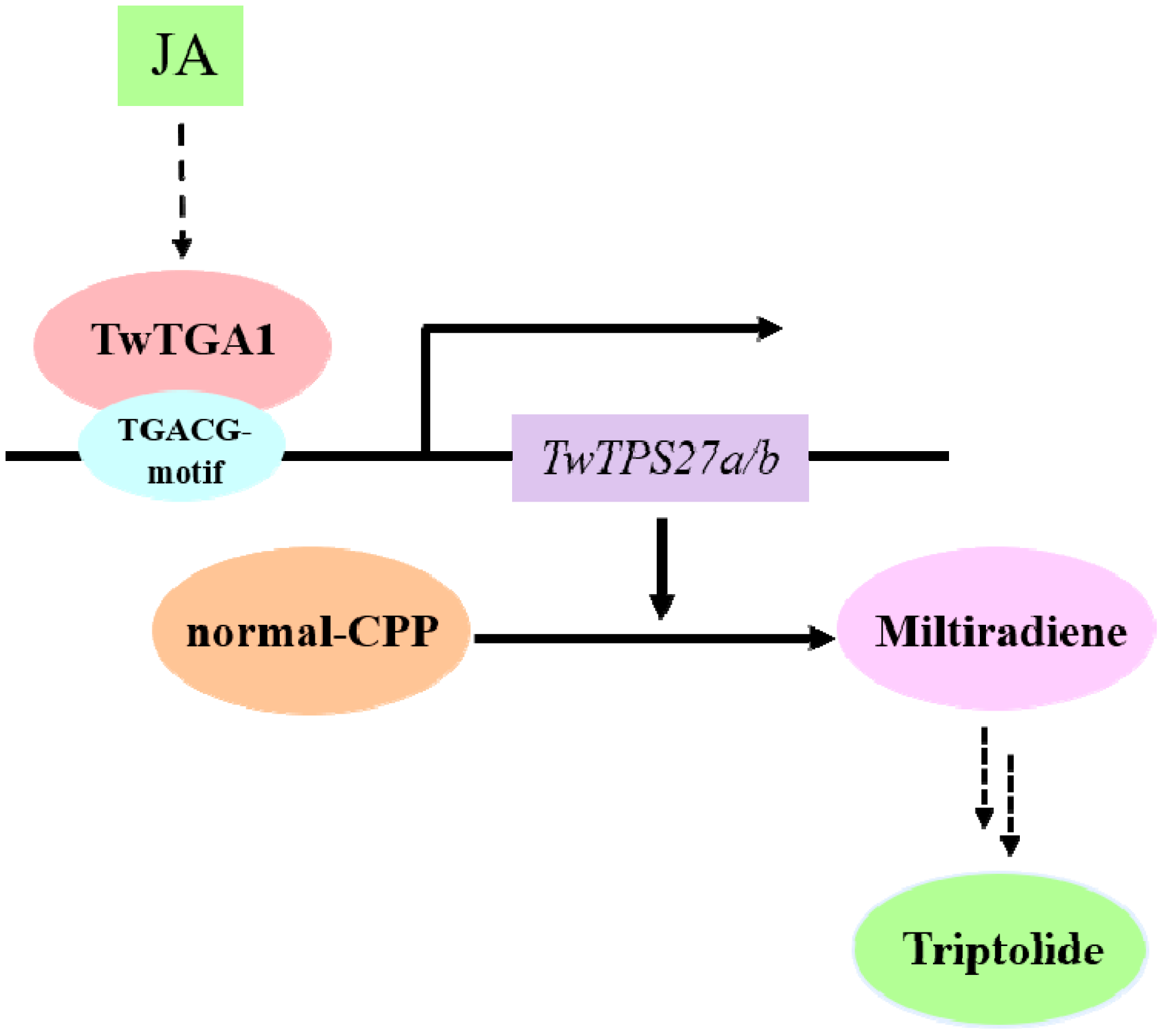
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sylvester, J.; Liacini, A.; Li, W.Q.; Dehnade, F.; Zafarullah, M. Tripterygium Wilfordii Hook F Extract Suppresses Proinflammatory Cytokine-Induced Expression of Matrix Metalloproteinase Genes in Articular Chondrocytes by Inhibiting Activating Protein-1 and Nuclear Factor-KappaB Activities. Mol. Pharm. 2001, 59, 1196–1205. [Google Scholar] [CrossRef]
- Ma, J.; Dey, M.; Yang, H.; Poulev, A.; Pouleva, R.; Dorn, R.; Lipsky, P.E.; Kennelly, E.J.; Raskin, I. Anti-Inflammatory and Immunosuppressive Compounds from Tripterygium Wilfordii. Phytochemistry 2007, 68, 1172–1178. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, C.; Ma, J.; Ni, L.; Huang, J.; Li, L.; Lin, M.; Hou, Q.; Zhang, D. Diterpenoids and Lignans from the Leaves of Tripterygium Wilfordii. Fitoterapia 2018, 129, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Shen, S.-X.; Liu, L.-X.; Xu, J.-H.; Ma, X.-H.; Shi, D.-M.; Zhang, Z.-Q. A New Demethyl Abietane Diterpenoid from the Roots of Tripterygium Wilfordii. Nat. Prod. Res. 2020, 34, 3094–3100. [Google Scholar] [CrossRef]
- Titov, D.V.; Gilman, B.; He, Q.-L.; Bhat, S.; Low, W.-K.; Dang, Y.; Smeaton, M.; Demain, A.L.; Miller, P.S.; Kugel, J.F.; et al. XPB, a Subunit of TFIIH, Is a Target of the Natural Product Triptolide. Nat. Chem. Biol. 2011, 7, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.-L.; Yang, Y.-X.; Ding, J.; Li, Y.-C.; Miao, Z.-H. Triptolide: Structural Modifications, Structure-Activity Relationships, Bioactivities, Clinical Development and Mechanisms. Nat. Prod. Rep. 2012, 29, 457–475. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, Y.; Wang, C.; Liu, L.; Wang, H.; Zhang, Y.; Long, C.; Sun, X. Triptolide Inhibits Epstein-Barr Nuclear Antigen 1 Expression by Increasing Sensitivity of Mitochondria Apoptosis of Nasopharyngeal Carcinoma Cells. J. Exp. Clin. Cancer Res. 2018, 37, 192. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.-F.; Liu, L.; He, L.-L.; Ye, J.; Lin, Z.-J.; Yuan, D.-L.; Deng, M.-M.; Fang, Z.-H.; Carter, B.Z.; Xu, B. Combining Triptolide with ABT-199 Is Effective against Acute Myeloid Leukemia through Reciprocal Regulation of Bcl-2 Family Proteins and Activation of the Intrinsic Apoptotic Pathway. Cell Death Dis. 2020, 11, 555. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-J.; Zhao, Y.-J.; Zhang, M.; Su, P.; Wang, X.-J.; Zhang, X.-N.; Gao, W.; Huang, L.-Q. Cloning and Characterisation of the Gene Encoding 3-Hydroxy-3-Methylglutaryl-CoA Synthase in Tripterygium Wilfordii. Molecules 2014, 19, 19696–19707. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Su, P.; Zhao, Y.; Zhang, M.; Wang, X.; Liu, Y.; Zhang, X.; Gao, W.; Huang, L. Molecular Cloning and Characterization of DXS and DXR Genes in the Terpenoid Biosynthetic Pathway of Tripterygium Wilfordii. Int. J. Mol. Sci. 2015, 16, 25516–25535. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, Y.; Wang, J.; Hu, T.; Tong, Y.; Zhou, J.; Gao, J.; Huang, L.; Gao, W. The Expression of TwDXS in the MEP Pathway Specifically Affects the Accumulation of Triptolide. Physiol. Plant 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, Y.; Wang, J.; Hu, T.; Tong, Y.; Zhou, J.; Song, Y.; Gao, W.; Huang, L. Overexpression and RNA Interference of TwDXR Regulate the Accumulation of Terpenoid Active Ingredients in Tripterygium Wilfordii. Biotechnol. Lett. 2018, 40, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Tong, Y.; Wang, Z.; Su, P.; Gao, W.; Huang, L. Molecular Cloning and Functional Identification of a CDNA Encoding 4-Hydroxy-3-Methylbut-2-Enyl Diphosphate Reductase from Tripterygium Wilfordii. Acta. Pharm. Sin. B 2017, 7, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Zhang, M.; Su, P.; Zhao, Y.; Wang, X.; Zhang, X.; Gao, W.; Huang, L. Cloning and Functional Characterization of an Isopentenyl Diphosphate Isomerase Gene from Tripterygium Wilfordii. Biotechnol. Appl. Biochem. 2016, 63, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, Y.; Zhang, Y.; Su, P.; Hu, T.; Lu, Y.; Zhang, R.; Zhou, J.; Ma, B.; Gao, W.; et al. Overexpression and RNAi-Mediated Downregulation of TwIDI Regulates Triptolide and Celastrol Accumulation in Tripterygium Wilfordii. Gene 2018, 679, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Su, P.; Zhou, Y.-J.J.; Wang, X.-J.J.; Zhao, Y.-J.J.; Liu, Y.-J.J.; Tong, Y.-R.R.; Hu, T.-Y.Y.; Huang, L.-Q.Q.; Gao, W. Identification of Geranylgeranyl Diphosphate Synthase Genes from Tripterygium Wilfordii. Plant Cell Rep. 2015, 34, 2179–2188. [Google Scholar] [CrossRef] [PubMed]
- Su, P.; Gao, L.; Tong, Y.; Guan, H.; Liu, S.; Zhang, Y.; Zhao, Y.; Wang, J.; Hu, T.; Tu, L.; et al. Analysis of the Role of Geranylgeranyl Diphosphate Synthase 8 from Tripterygium Wilfordii in Diterpenoids Biosynthesis. Plant Sci. 2019, 285, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Hansen, N.L.; Heskes, A.M.; Hamberger, B.; Olsen, C.E.; Hallström, B.M.; Andersen-Ranberg, J.; Hamberger, B. The Terpene Synthase Gene Family in Tripterygium Wilfordii Harbors a Labdane-Type Diterpene Synthase among the Monoterpene Synthase TPS-b Subfamily. Plant J. 2017, 89, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Su, P.; Guan, H.; Zhao, Y.; Tong, Y.; Xu, M.; Zhang, Y.; Hu, T.; Yang, J.; Cheng, Q.; Gao, L.; et al. Identification and Functional Characterization of Diterpene Synthases for Triptolide Biosynthesis from Tripterygium Wilfordii. Plant J. 2018, 93, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Tu, L.; Su, P.; Zhang, Z.; Gao, L.; Wang, J.; Hu, T.; Zhou, J.; Zhang, Y.; Zhao, Y.; Liu, Y.; et al. Genome of Tripterygium Wilfordii and Identification of Cytochrome P450 Involved in Triptolide Biosynthesis. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Zhou, J.; Tong, Y.; Su, P.; Li, X.; Liu, Y.; Liu, N.; Wu, X.; Zhang, Y.; Wang, J.; et al. Engineering Chimeric Diterpene Synthases and Isoprenoid Biosynthetic Pathways Enables High-Level Production of Miltiradiene in Yeast. Metab. Eng. 2020, 60, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Sun, K.; Mackaluso, J.D.; Seddon, A.E.; Jin, R.; Thomashow, M.F.; Shiu, S.-H. Cis-Regulatory Code of Stress-Responsive Transcription in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 14992–14997. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and Validation of Promoters and Cis-Acting Regulatory Elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Jin, B.; Sheng, Z.; Muhammad, I.; Chen, J.; Yang, H. Cloning and Functional Analysis of the Promoter of a Stress-Inducible Gene (Zmap) in Maize. PLoS ONE 2019, 14. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Xiong, Y.; Teixeira da Silva, J.A.; Pang, J.; Zhang, T.; Yu, X.; Zhang, X.; Niu, M.; Ma, G. Molecular Cloning and Functional Characterization of Bisabolene Synthetase (SaBS) Promoter from Santalum Album. Forests 2020, 11, 85. [Google Scholar] [CrossRef]
- Xing, B.; Yang, D.; Yu, H.; Zhang, B.; Yan, K.; Zhang, X.; Han, R.; Liang, Z. Overexpression of SmbHLH10 Enhances Tanshinones Biosynthesis in Salvia Miltiorrhiza Hairy Roots. Plant Sci. 2018, 276, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Li, Y.; Yang, S.; Yang, Z.; Qiu, M.; Gao, R.; Han, T.; Meng, X.; Xu, Z.; Wang, L.; et al. The Spatio-Temporal Biosynthesis of Floral Flavonols Is Controlled by Differential Phylogenetic MYB Regulators in Freesia Hybrida. New Phytol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Wu, C.; Zhao, L.; Wang, Y.; Wang, C.; Zhou, W.; Ming, Y.; Kai, G. WRKY Transcription Factor OpWRKY1 Acts as a Negative Regulator of Camptothecin Biosynthesis in Ophiorrhiza Pumila Hairy Roots. Plant Cell Tiss. Organ Cult. 2020, 142, 69–78. [Google Scholar] [CrossRef]
- Li, B.; Liu, Y.; Cui, X.-Y.; Fu, J.-D.; Zhou, Y.-B.; Zheng, W.-J.; Lan, J.-H.; Jin, L.-G.; Chen, M.; Ma, Y.-Z.; et al. Genome-Wide Characterization and Expression Analysis of Soybean TGA Transcription Factors Identified a Novel TGA Gene Involved in Drought and Salt Tolerance. Front. Plant Sci. 2019, 10, 549. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Salasini, B.C.; Khan, M.; Devi, B.; Bush, M.; Subramaniam, R.; Hepworth, S.R. Clade I TGACG-Motif Binding Basic Leucine Zipper Transcription Factors Mediate BLADE-ON-PETIOLE-Dependent Regulation of Development. Plant Physiol. 2019, 180, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Budimir, J.; Treffon, K.; Nair, A.; Thurow, C.; Gatz, C. Redox-Active Cysteines in TGACG-BINDING FACTOR 1 (TGA1) Do Not Play a Role in Salicylic Acid or Pathogen-Induced Expression of TGA1-Regulated Target Genes in Arabidopsis Thaliana. New Phytol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Busta, L.; Zhang, Q.; Ding, P.; Jetter, R.; Zhang, Y. TGACG-BINDING FACTOR 1 (TGA1) and TGA4 Regulate Salicylic Acid and Pipecolic Acid Biosynthesis by Modulating the Expression of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol. 2018, 217, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Gatz, C. From Pioneers to Team Players: TGA Transcription Factors Provide a Molecular Link between Different Stress Pathways. Mol. Plant-Microbe Interact. MPMI 2013, 26, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Guo, Z.; Zhang, L.; Zhang, F.; Jiang, W.; Shen, Q.; Fu, X.; Yan, T.; Shi, P.; Hao, X.; et al. Interaction of BZIP Transcription Factor TGA6 with Salicylic Acid Signaling Modulates Artemisinin Biosynthesis in Artemisia Annua. J. Exp. Bot. 2019, 70, 3969–3979. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Miyamoto, K.; Yamane, H.; Nishizawa, Y.; Minami, E.; Nojiri, H.; Okada, K. OsTGAP1 Is Responsible for JA-Inducible Diterpenoid Phytoalexin Biosynthesis in Rice Roots with Biological Impacts on Allelopathic Interaction. Physiol. Plant 2017, 161, 532–544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, Z.; Ji, A.; Luo, H.; Song, J. Genomic Survey of BZIP Transcription Factor Genes Related to Tanshinone Biosynthesis in Salvia Miltiorrhiza. Acta Pharm. Sin. B 2018, 8, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Liu, H.; Wang, S.; Wang, C.; Miao, G. A Class I TGA Transcription Factor from Tripterygium Wilfordii Hook.f. Modulates the Biosynthesis of Secondary Metabolites in Both Native and Heterologous Hosts. Plant Sci. 2020, 290, 110293. [Google Scholar] [CrossRef]
- Mason, H.S.; DeWald, D.B.; Mullet, J.E. Identification of a Methyl Jasmonate-Responsive Domain in the Soybean VspB Promoter. Plant Cell 1993, 5, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Rouster, J.; Leah, R.; Mundy, J.; Cameron-Mills, V. Identification of a Methyl Jasmonate-Responsive Region in the Promoter of a Lipoxygenase 1 Gene Expressed in Barley Grain. Plant J. 1997, 11, 513–523. [Google Scholar] [CrossRef]
- Xiong, J.; Liu, L.; Ma, X.; Li, F.; Tang, C.; Li, Z.; Lü, B.; Zhou, T.; Lian, X.; Chang, Y.; et al. Characterization of PtAOS1 Promoter and Three Novel Interacting Proteins Responding to Drought in Poncirus Trifoliata. Int. J. Mol. Sci. 2020, 21, 4705. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, L.; Cui, L.; Lei, J.; Zhang, X. Effects of elicitors on growth of adventitious roots and contents of secondary metabolites in Tripterygium wilfordii Hook. f. Chin. J. Biotechnol. 2015, 31, 734–743. [Google Scholar]
- Zerbe, P.; Hamberger, B.; Yuen, M.M.S.; Chiang, A.; Sandhu, H.K.; Madilao, L.L.; Nguyen, A.; Hamberger, B.; Bach, S.S.; Bohlmann, J. Gene Discovery of Modular Diterpene Metabolism in Nonmodel Systems. Plant Physiol. 2013, 162, 1073–1091. [Google Scholar] [CrossRef] [PubMed]
- Atchley, W.R.; Fitch, W.M. A Natural Classification of the Basic Helix-Loop-Helix Class of Transcription Factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-Wide Classification and Evolutionary Analysis of the BHLH Family of Transcription Factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.-G.; Yu, J.-Q.; Han, P.-L.; Xie, X.-B.; Sun, C.-H.; Zhang, Q.-Y.; Wang, J.-H.; Hao, Y.-J. The Regulatory Module MdPUB29-MdbHLH3 Connects Ethylene Biosynthesis with Fruit Quality in Apple. New Phytol. 2019, 221, 1966–1982. [Google Scholar] [CrossRef]
- Shen, Q.; Lu, X.; Yan, T.; Fu, X.; Lv, Z.; Zhang, F.; Pan, Q.; Wang, G.; Sun, X.; Tang, K. The Jasmonate-Responsive AaMYC2 Transcription Factor Positively Regulates Artemisinin Biosynthesis in Artemisia Annua. New Phytol. 2016, 210, 1269–1281. [Google Scholar] [CrossRef]
- Li, T.; Xu, Y.; Zhang, L.; Ji, Y.; Tan, D.; Yuan, H.; Wang, A. The Jasmonate-Activated Transcription Factor MdMYC2 Regulates ETHYLENE RESPONSE FACTOR and Ethylene Biosynthetic Genes to Promote Ethylene Biosynthesis during Apple Fruit Ripening. Plant Cell 2017, 29, 1316–1334. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Y.; Gao, F.; Jin, W.; Li, S.; Kimani, S.; Yang, S.; Bao, T.; Gao, X.; Wang, L. MYB21 Interacts with MYC2 to Control the Expression of Terpene Synthase Genes in Flowers of F. Hybrida and A. Thaliana. J. Exp. Bot. 2020. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Luo, X.; Zhang, C.; Xu, X.; Huang, J.; Chen, Y.; Feng, S.; Zhan, X.; Zhang, L.; Yuan, H.; et al. Tissue-Specific Study across the Stem of Taxus Media Identifies a Phloem-Specific TmMYB3 Involved in the Transcriptional Regulation of Paclitaxel Biosynthesis. Plant J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yan, T.; Shen, Q.; Lu, X.; Pan, Q.; Huang, Y.; Tang, Y.; Fu, X.; Liu, M.; Jiang, W.; et al. GLANDULAR TRICHOME-SPECIFIC WRKY 1 Promotes Artemisinin Biosynthesis in Artemisia Annua. New Phytol. 2017, 214, 304–316. [Google Scholar] [CrossRef]
- Sun, P.-W.; Xu, Y.-H.; Yu, C.-C.; Lv, F.-F.; Tang, X.-L.; Gao, Z.-H.; Zhang, Z.; Wang, H.; Liu, Y.; Wei, J.-H. WRKY44 Represses Expression of the Wound-Induced Sesquiterpene Biosynthetic Gene ASS1 in Aquilaria Sinensis. J. Exp. Bot. 2020, 71, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Xiao, L.; Gao, S.; Li, Q.; Chen, J.; Xiao, Y.; Ji, Q.; Chen, R.; Chen, W.; Zhang, L. TRICHOME AND ARTEMISININ REGULATOR 1 Is Required for Trichome Development and Artemisinin Biosynthesis in Artemisia Annua. Mol. Plant 2015, 8, 1396–1411. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Sun, M.; Yuan, T.; Wang, Y.; Shi, M.; Lu, S.; Tang, B.; Pan, J.; Wang, Y.; Kai, G. The AP2/ERF Transcription Factor SmERF1L1 Regulates the Biosynthesis of Tanshinones and Phenolic Acids in Salvia Miltiorrhiza. Food Chem. 2019, 274, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Miao, G.; Guo, J.; Huo, Y.; Zhang, X.; Xie, J.; Feng, J. Establishment of Tripterygium Wilfordii Hook. f. Hairy Root Culture and Optimization of Its Culture Conditions for the Production of Triptolide and Wilforine. J. Microbiol. Biotechnol. 2014, 24, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.; Gao, Y.; Shi, M.; Zhang, X.; He, S.; Chen, Z.; An, C. SiteFinding-PCR: A Simple and Efficient PCR Method for Chromosome Walking. Nucleic Acids Res. 2005, 33, e122. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ye, S.; Li, J.; Zheng, B.; Bao, M.; Ning, G. Fusion Primer and Nested Integrated PCR (FPNI-PCR): A New High-Efficiency Strategy for Rapid Chromosome Walking or Flanking Sequence Cloning. BMC Biotechnol 2011, 11, 109. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant Cis-Acting Regulatory DNA Elements (PLACE) Database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Li, H.-L.; Tang, X.; Peng, S.-Q. Molecular and Functional Characterization of the HbSRPP Promoter in Response to Hormones and Abiotic Stresses. Transgenic Res. 2014, 23, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yu, D.; Yu, J.; Zhu, D.; Zhao, Q. Functional Analysis of Maize Silk-Specific ZmbZIP25 Promoter. Int. J. Mol. Sci. 2018, 19, 822. [Google Scholar] [CrossRef] [PubMed]
- Trishla, V.S.; Marriboina, S.; Boyidi, P.; Kirti, P.B. GUS-Reporter Based Analysis of the Promoter Activity of Gossypium Hirsutum NAC Transcription Factor, GhNAC4 That Is Induced by Phytohormones and Environmental Stresses. Plant Cell Tiss. Organ Cult. 2020, 141, 643–654. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huo, Y.; Zhang, B.; Chen, L.; Zhang, J.; Zhang, X.; Zhu, C. Isolation and Functional Characterization of the Promoters of Miltiradiene Synthase Genes, TwTPS27a and TwTPS27b, and Interaction Analysis with the Transcription Factor TwTGA1 from Tripterygium wilfordii. Plants 2021, 10, 418. https://doi.org/10.3390/plants10020418
Huo Y, Zhang B, Chen L, Zhang J, Zhang X, Zhu C. Isolation and Functional Characterization of the Promoters of Miltiradiene Synthase Genes, TwTPS27a and TwTPS27b, and Interaction Analysis with the Transcription Factor TwTGA1 from Tripterygium wilfordii. Plants. 2021; 10(2):418. https://doi.org/10.3390/plants10020418
Chicago/Turabian StyleHuo, Yanbo, Bin Zhang, Ling Chen, Jing Zhang, Xing Zhang, and Chuanshu Zhu. 2021. "Isolation and Functional Characterization of the Promoters of Miltiradiene Synthase Genes, TwTPS27a and TwTPS27b, and Interaction Analysis with the Transcription Factor TwTGA1 from Tripterygium wilfordii" Plants 10, no. 2: 418. https://doi.org/10.3390/plants10020418
APA StyleHuo, Y., Zhang, B., Chen, L., Zhang, J., Zhang, X., & Zhu, C. (2021). Isolation and Functional Characterization of the Promoters of Miltiradiene Synthase Genes, TwTPS27a and TwTPS27b, and Interaction Analysis with the Transcription Factor TwTGA1 from Tripterygium wilfordii. Plants, 10(2), 418. https://doi.org/10.3390/plants10020418





