Deciphering the Proteotoxic Stress Responses Triggered by the Perturbed Thylakoid Proteostasis in Arabidopsis
Abstract
:1. Introduction
2. Results
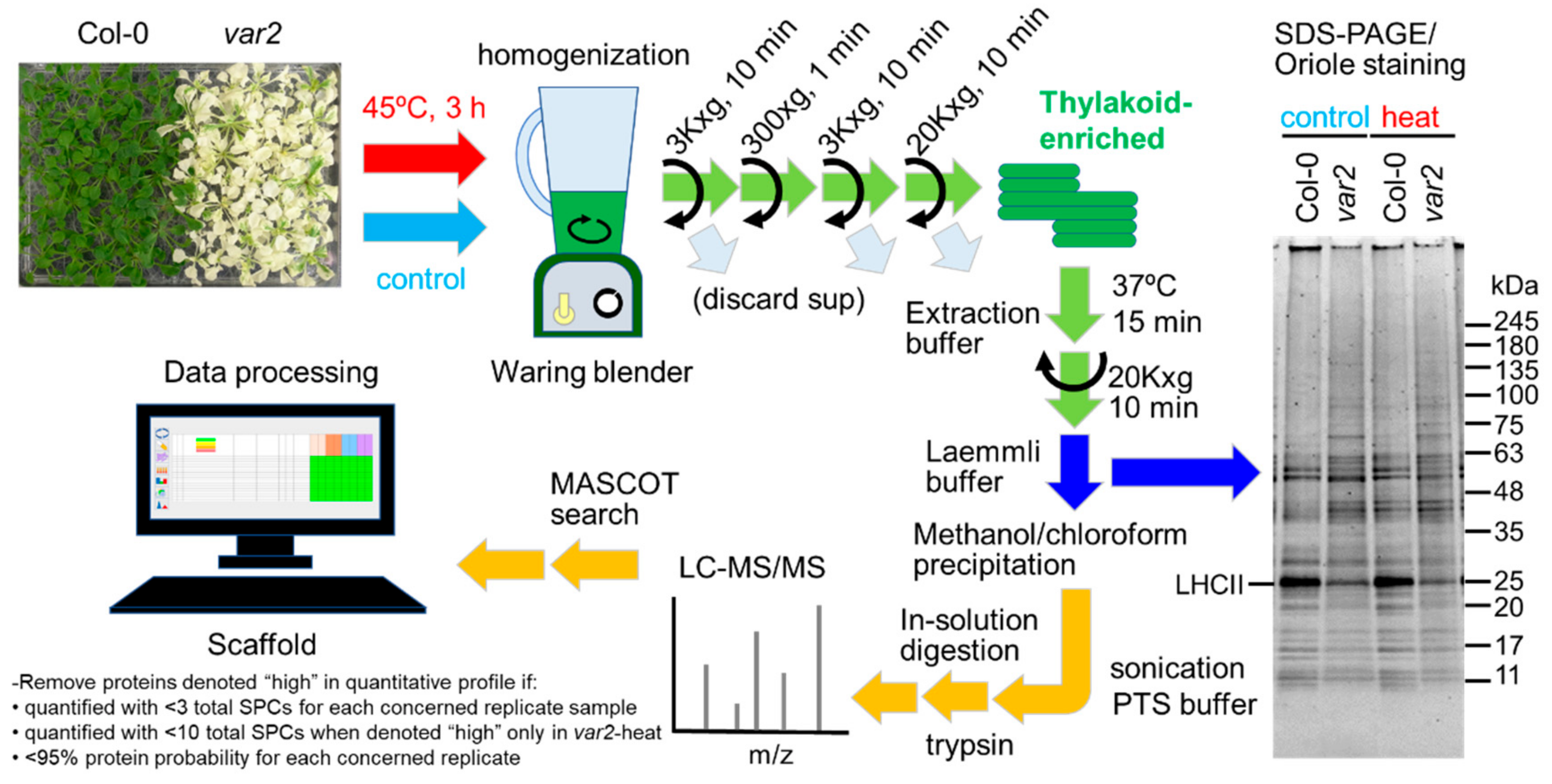
2.1. Comparing Thylakoid-Enriched Proteomes of var2 versus Wild-Type Plants in Response to Heat Exposure
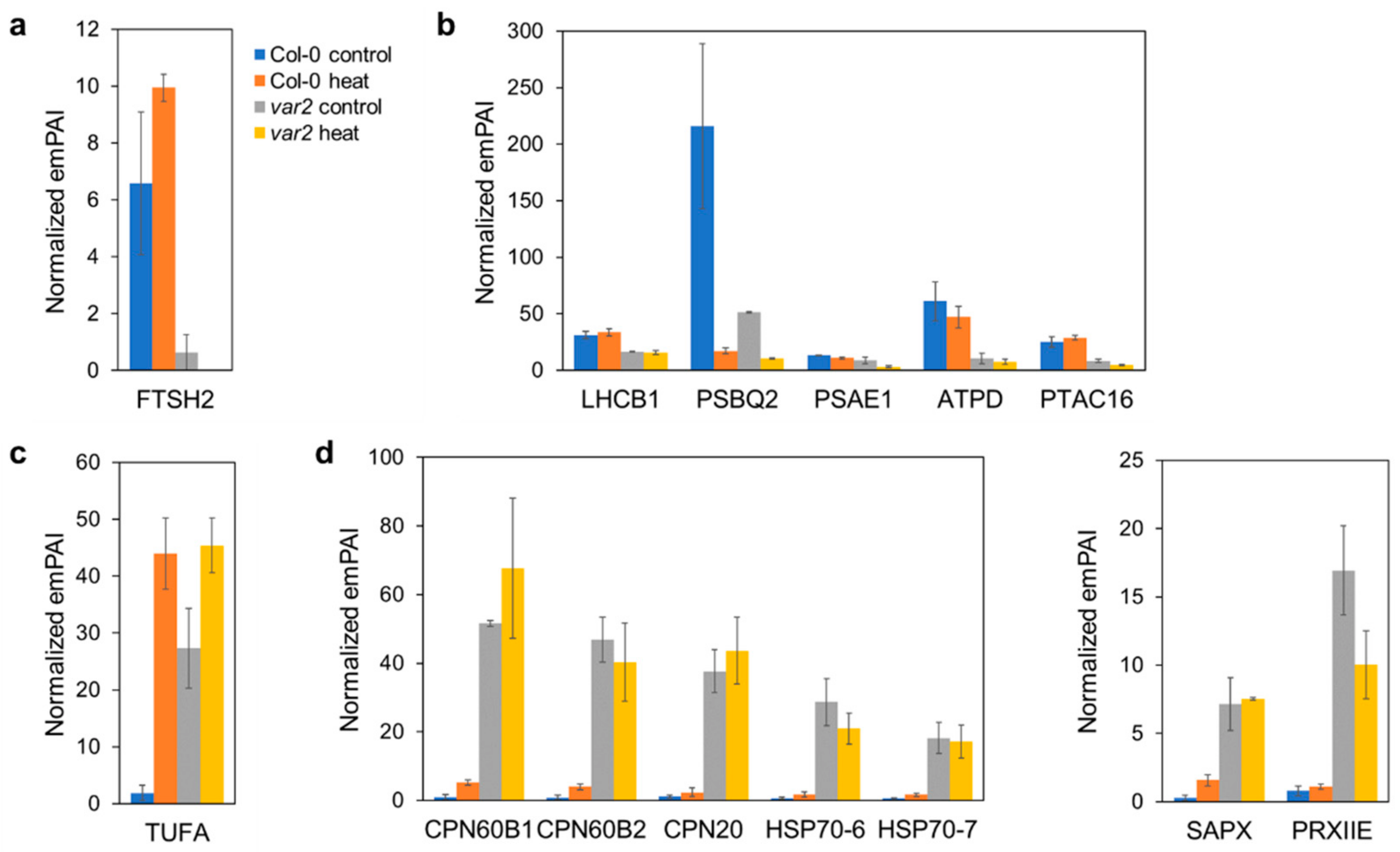
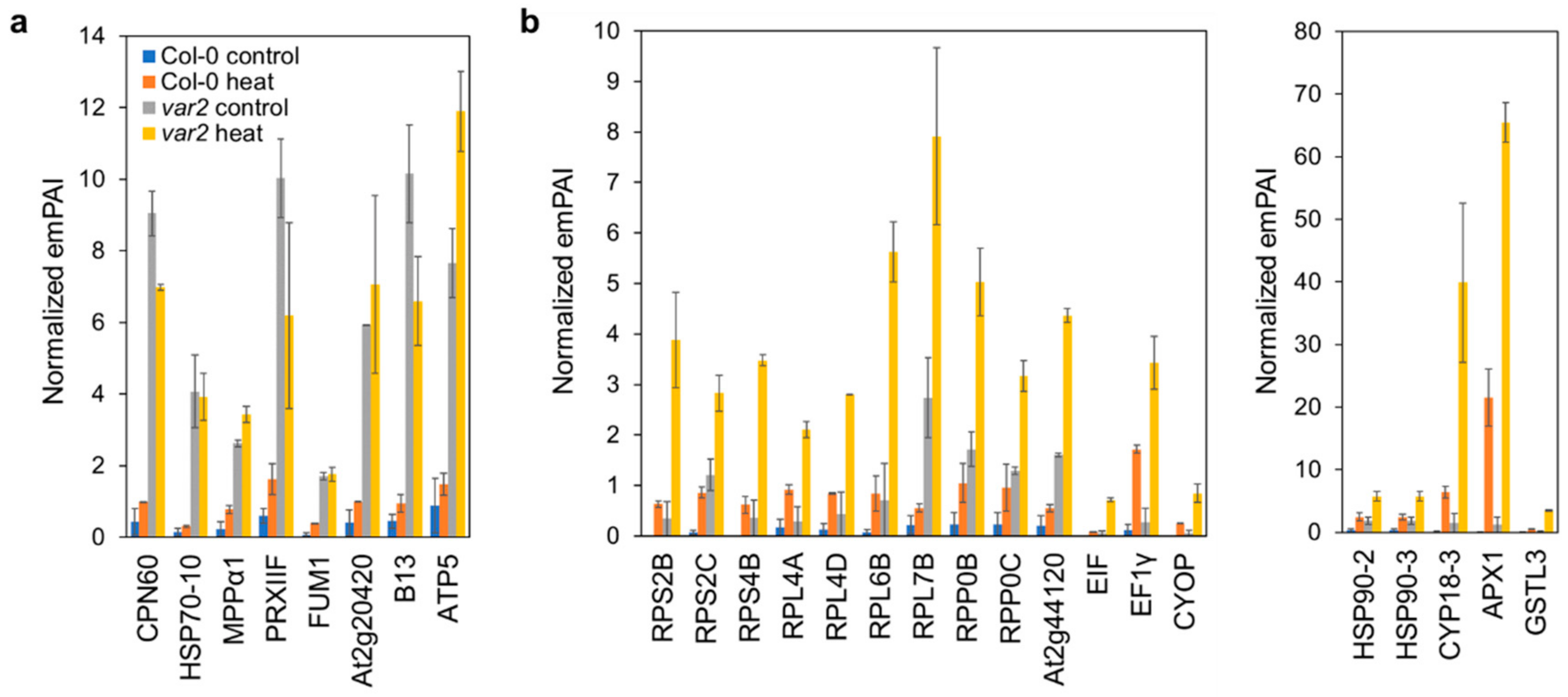
2.2. Verifying Thylakoid Proteostasis Impairment and Proteotoxic Stress Responses
2.3. Oxidative Stress-Dependent Protein Accumulation
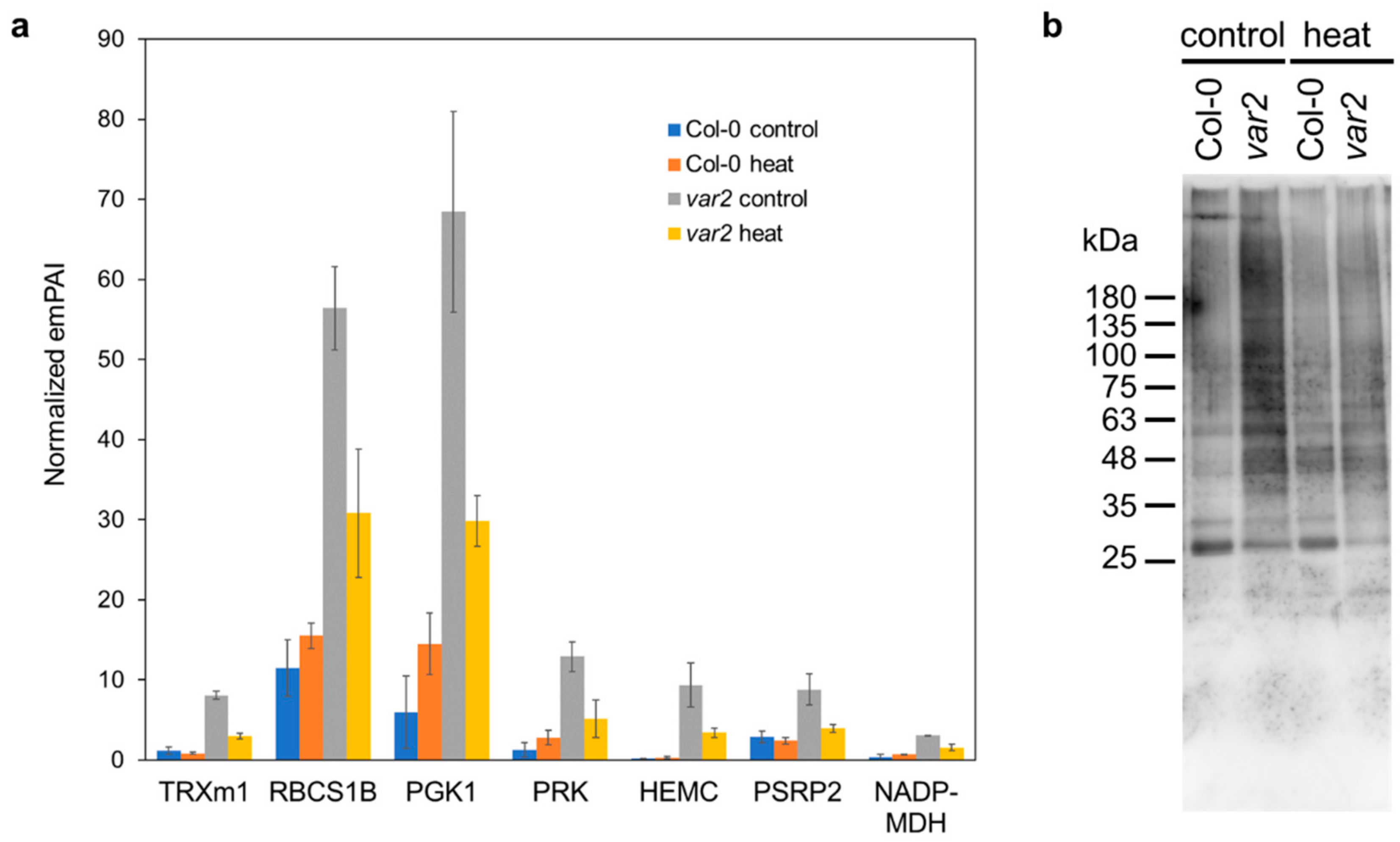
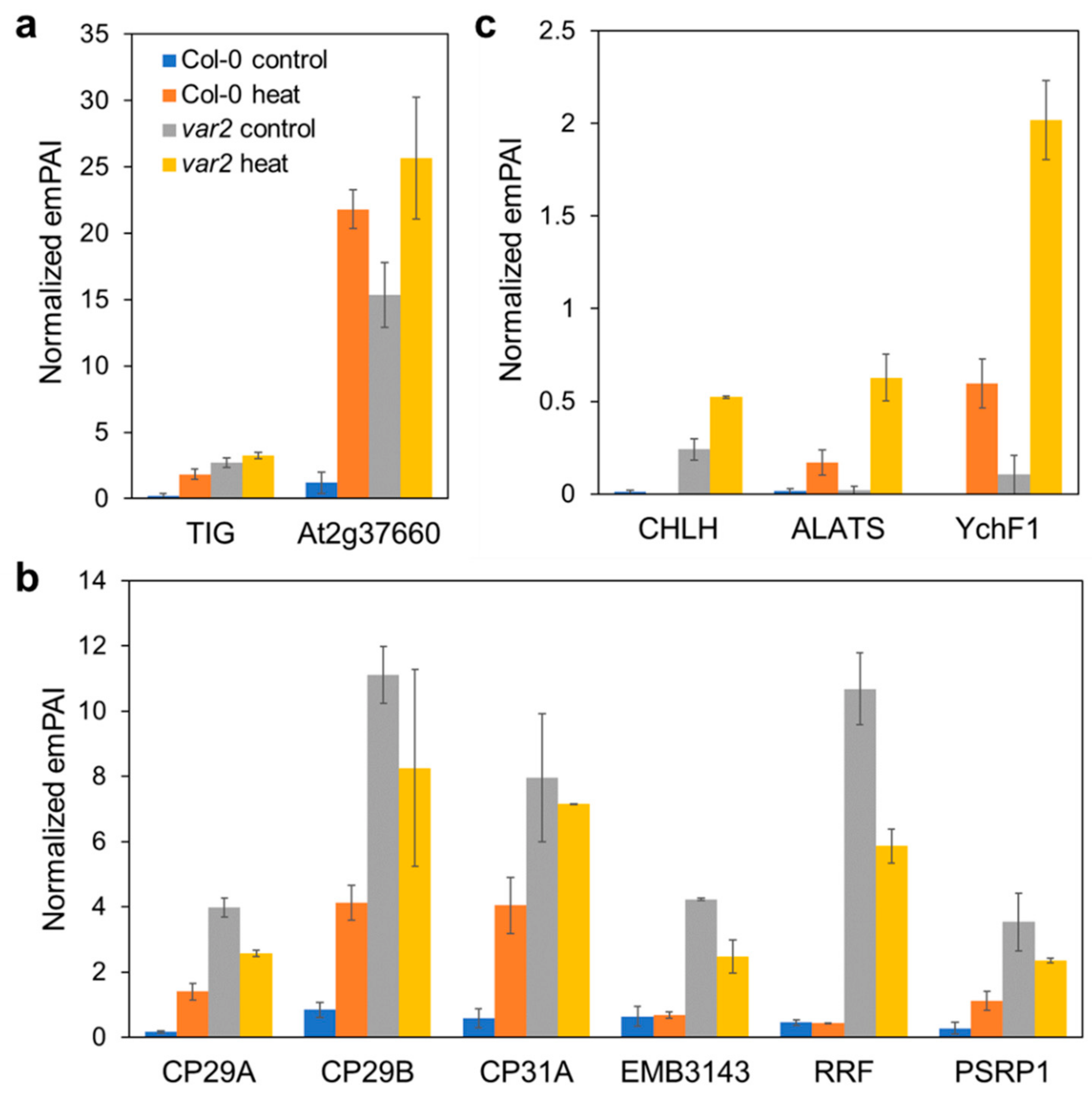
2.4. Altered Stromal Protein Distribution
2.5. Consequences on Extra-Plastid Proteostasis
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Condition and Stress Experiment
4.2. Thylakoid Isolation and Protein Extraction
4.3. SDS-PAGE and Immunoblotting
4.4. LC-MS/MS Analysis and Data Processing
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jarvis, P.; López-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787–802. [Google Scholar] [CrossRef]
- Pribil, M.; Labs, M.; Leister, D. Structure and dynamics of thylakoids in land plants. J. Exp. Bot. 2014, 65, 1955–1972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, Y.; Sakamoto, W. FtsH Protease in the Thylakoid Membrane: Physiological Functions and the Regulation of Protease Activity. Front. Plant Sci. 2018, 9, 855. [Google Scholar] [CrossRef] [Green Version]
- Van Wijk, K.J. Protein Maturation and Proteolysis in Plant Plastids, Mitochondria, and Peroxisomes. Annu. Rev. Plant Biol. 2015, 66, 75–111. [Google Scholar] [CrossRef]
- Nishimura, K.; Kato, Y.; Sakamoto, W. Essentials of Proteolytic Machineries in Chloroplasts. Mol. Plant 2017, 10, 4–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, W.; Zaltsman, A.; Adam, Z.; Takahashi, Y. Coordinated Regulation and Complex Formation of Yellow Variegated1 and Yellow Variegated2, Chloroplastic FtsH Metalloproteases Involved in the Repair Cycle of Photosystem II in Arabidopsis Thylakoid Membranes. Plant Cell 2003, 15, 2843–2855. [Google Scholar] [CrossRef] [Green Version]
- Kato, Y.; Miura, E.; Ido, K.; Ifuku, K.; Sakamoto, W. The Variegated Mutants Lacking Chloroplastic FtsHs Are Defective in D1 Degradation and Accumulate Reactive Oxygen Species. Plant Physiol. 2009, 151, 1790–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Ding, Y.; Zhu, C. Sensitivity and Responses of Chloroplasts to Heat Stress in Plants. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Allakhverdiev, S.I.; Kreslavski, V.D.; Klimov, V.V.; Los, D.A.; Carpentier, R.; Mohanty, P. Heat stress: An overview of molecular responses in photosynthesis. Photosynth. Res. 2008, 98, 541–550. [Google Scholar] [CrossRef]
- Takechi, K.; Sodmergen; Murata, M.; Motoyoshi, F.; Sakamoto, W. The yellow variegated (var2) locus encodes a homologue of FtsH, an ATP-dependent protease in arabidopsis. Plant Cell Physiol. 2000, 41, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially Modified Protein Abundance Index (emPAI) for Estimation of Absolute Protein Amount in Proteomics by the Number of Sequenced Peptides per Protein. Mol. Cell. Proteomics 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [Green Version]
- Ingelsson, B.; Vener, A.V. Phosphoproteomics of Arabidopsis chloroplasts reveals involvement of the STN7 kinase in phosphorylation of nucleoid protein pTAC16. FEBS Lett. 2012, 586, 1265–1271. [Google Scholar] [CrossRef] [Green Version]
- Zienkiewicz, M.; Ferenc, A.; Wasilewska, W.; Romanowska, E. High light stimulates Deg1-dependent cleavage of the minor LHCII antenna proteins CP26 and CP29 and the PsbS protein in Arabidopsis thaliana. Planta 2012, 235, 279–288. [Google Scholar] [CrossRef]
- Kavaliauskas, D.; Nissen, P.; Knudsen, C.R. The Busiest of All Ribosomal Assistants: Elongation Factor Tu. Biochemistry 2012, 51, 2642–2651. [Google Scholar] [CrossRef]
- Fu, J.; Momčilović, I.; Prasad, P.V.V. Roles of Protein Synthesis Elongation Factor EF-Tu in Heat Tolerance in Plants. J. Bot. 2012, 2012, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Cai, C.; Wang, Z.; Fan, B.; Zhu, C.; Chen, Z. Plastid Translation Elongation Factor Tu Is Prone to Heat-Induced Aggregation Despite Its Critical Role in Plant Heat Tolerance. Plant Physiol. 2018, 176, 3027–3045. [Google Scholar] [CrossRef] [Green Version]
- Kato, Y.; Sun, X.; Zhang, L.; Sakamoto, W. Cooperative D1 Degradation in the Photosystem II Repair Mediated by Chloroplastic Proteases in Arabidopsis. Plant Physiol. 2012, 159, 1428–1439. [Google Scholar] [CrossRef] [Green Version]
- Camp, P.J.; Randall, D.D. Purification and Characterization of the Pea Chloroplast Pyruvate Dehydrogenase Complex. Plant Physiol. 1985. [Google Scholar] [CrossRef] [Green Version]
- Mooney, B.P.; Miernyk, J.A.; Randall, D.D. Cloning and Characterization of the Dihydrolipoamide S -Acetyltransferase Subunit of the Plastid Pyruvate Dehydrogenase Complex (E2) from Arabidopsis. Plant Physiol. 1999, 120, 443–452. [Google Scholar] [CrossRef] [Green Version]
- Apel, K.; Hirt, H. REACTIVE OXYGEN SPECIES: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [Green Version]
- Dietz, K.J. Thiol-Based Peroxidases and Ascorbate Peroxidases: Why Plants Rely on Multiple Peroxidase Systems in the Photosynthesizing Chloroplast? Mol. Cells 2016, 39, 20–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zielińska-Dawidziak, M.; Siger, A. Effect of elevated accumulation of iron in ferritin on the antioxidants content in soybean sprouts. Eur. Food Res. Technol. 2012, 234, 1005–1012. [Google Scholar] [CrossRef] [Green Version]
- Kmiec, B.; Teixeira, P.F.; Berntsson, R.P.A.; Murcha, M.W.; Branca, R.M.M.; Radomiljac, J.D.; Regberg, J.; Sèensson, L.M.; Bakali, A.; Langel, Ü.; et al. Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts. Proc. Natl. Acad. Sci. USA 2013. [Google Scholar] [CrossRef] [Green Version]
- Kmiec, B.; Branca, R.M.M.; Berkowitz, O.; Li, L.; Wang, Y.; Murcha, M.W.; Whelan, J.; Lehtiö, J.; Glaser, E.; Teixeira, P.F. Accumulation of endogenous peptides triggers a pathogen stress response in Arabidopsis thaliana. Plant J. 2018, 96, 705–715. [Google Scholar] [CrossRef] [Green Version]
- Wagner, U.; Edwards, R.; Dixon, D.P.; Mauch, F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 2002, 49, 515–532. [Google Scholar] [CrossRef]
- Dogra, V.; Duan, J.; Lee, K.P.; Kim, C. Impaired PSII proteostasis triggers a UPR-like response in the var2 mutant of Arabidopsis. J. Exp. Bot. 2019, 70, 3075–3088. [Google Scholar] [CrossRef]
- Lu, S.C. Glutathione synthesis. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3143–3153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira Dos Santos, C.; Rey, P. Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci. 2006, 11, 329–334. [Google Scholar] [CrossRef]
- Nikkanen, L.; Rintamäki, E. Thioredoxin-dependent regulatory networks in chloroplasts under fluctuating light conditions. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikkanen, L.; Rintamäki, E. Chloroplast thioredoxin systems dynamically regulate photosynthesis in plants. Biochem. J. 2019, 476, 1159–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balmer, Y.; Koller, A.; del Val, G.; Manieri, W.; Schurmann, P.; Buchanan, B.B. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. USA 2003, 100, 370–375. [Google Scholar] [CrossRef] [Green Version]
- Okegawa, Y.; Motohashi, K. Chloroplastic thioredoxin m functions as a major regulator of Calvin cycle enzymes during photosynthesis in vivo. Plant J. 2015, 84, 900–913. [Google Scholar] [CrossRef]
- Da, Q.; Wang, P.; Wang, M.; Sun, T.; Jin, H.; Liu, B.; Wang, J.; Grimm, B.; Wang, H.-B. Thioredoxin and NADPH-Dependent Thioredoxin Reductase C Regulation of Tetrapyrrole Biosynthesis. Plant Physiol. 2017, 175, 652–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheibe, R. Malate valves to balance cellular energy supply. Physiol. Plant. 2004, 120, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Thormählen, I.; Zupok, A.; Rescher, J.; Leger, J.; Weissenberger, S.; Groysman, J.; Orwat, A.; Chatel-Innocenti, G.; Issakidis-Bourguet, E.; Armbruster, U.; et al. Thioredoxins Play a Crucial Role in Dynamic Acclimation of Photosynthesis in Fluctuating Light. Mol. Plant 2017, 10, 168–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz-Barrio, R.; Fernández-San Millán, A.; Carballeda, J.; Corral-Martínez, P.; Seguí-Simarro, J.M.; Farran, I. Chaperone-like properties of tobacco plastid thioredoxins f and m. J. Exp. Bot. 2012, 63, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Rohr, M.; Ries, F.; Herkt, C.; Gotsmann, V.L.; Westrich, L.D.; Gries, K.; Trösch, R.; Christmann, J.; Chaux-Jukic, F.; Jung, M.; et al. The Role of Plastidic Trigger Factor Serving Protein Biogenesis in Green Algae and Land Plants. Plant Physiol. 2019, 179, 1093–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kupsch, C.; Ruwe, H.; Gusewski, S.; Tillich, M.; Small, I.; Schmitz-Linneweber, C. Arabidopsis Chloroplast RNA Binding Proteins CP31A and CP29A Associate with Large Transcript Pools and Confer Cold Stress Tolerance by Influencing Multiple Chloroplast RNA Processing Steps. Plant Cell 2012, 24, 4266–4280. [Google Scholar] [CrossRef] [Green Version]
- Tillich, M.; Hardel, S.L.; Kupsch, C.; Armbruster, U.; Delannoy, E.; Gualberto, J.M.; Lehwark, P.; Leister, D.; Small, I.D.; Schmitz-Linneweber, C. Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. USA 2009, 106, 6002–6007. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.R.; Dönhöfer, A.; Barat, C.; Marquez, V.; Datta, P.P.; Fucini, P.; Wilson, D.N.; Agrawal, R.K. PSRP1 Is Not a Ribosomal Protein, but a Ribosome-binding Factor That Is Recycled by the Ribosome-recycling Factor (RRF) and Elongation Factor G (EF-G). J. Biol. Chem. 2010, 285, 4006–4014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallace, E.W.J.; Kear-Scott, J.L.; Pilipenko, E.V.; Schwartz, M.H.; Laskowski, P.R.; Rojek, A.E.; Katanski, C.D.; Riback, J.A.; Dion, M.F.; Franks, A.M.; et al. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell 2015, 162, 1286–1298. [Google Scholar] [CrossRef] [Green Version]
- Cherkasov, V.; Hofmann, S.; Druffel-Augustin, S.; Mogk, A.; Tyedmers, J.; Stoecklin, G.; Bukau, B. Coordination of Translational Control and Protein Homeostasis during Severe Heat Stress. Curr. Biol. 2013, 23, 2452–2462. [Google Scholar] [CrossRef] [Green Version]
- Burke, K.A.; Yates, E.A.; Legleiter, J. Biophysical Insights into How Surfaces, Including Lipid Membranes, Modulate Protein Aggregation Related to Neurodegeneration. Front. Neurol. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Staehelin, L.A. Chloroplast structure: From chlorophyll granules to supra-molecular architecture of thylakoid membranes. Photosynth. Res. 2003, 76, 185–196. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, N.S.; Haynes, C.M. Folding the Mitochondrial UPR into the Integrated Stress Response. Trends Cell Biol. 2020, 30, 428–439. [Google Scholar] [CrossRef]
- Ramundo, S.; Casero, D.; Mühlhaus, T.; Hemme, D.; Sommer, F.; Crèvecoeur, M.; Rahire, M.; Schroda, M.; Rusch, J.; Goodenough, U.; et al. Conditional Depletion of the Chlamydomonas Chloroplast ClpP Protease Activates Nuclear Genes Involved in Autophagy and Plastid Protein Quality Control. Plant Cell 2014, 26, 2201–2222. [Google Scholar] [CrossRef] [Green Version]
- Bohne, A.-V.; Schwarz, C.; Schottkowski, M.; Lidschreiber, M.; Piotrowski, M.; Zerges, W.; Nickelsen, J. Reciprocal Regulation of Protein Synthesis and Carbon Metabolism for Thylakoid Membrane Biogenesis. PLoS Biol. 2013, 11, e1001482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chodasiewicz, M.; Sokolowska, E.M.; Nelson-Dittrich, A.C.; Masiuk, A.; Beltran, J.C.M.; Nelson, A.D.L.; Skirycz, A. Identification and Characterization of the Heat-Induced Plastidial Stress Granules Reveal New Insight Into Arabidopsis Stress Response. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, M.; Boronat, S.; Marte, L.; Vega, M.; Pérez, P.; Ayté, J.; Hidalgo, E. Chaperone-Facilitated Aggregation of Thermo-Sensitive Proteins Shields Them from Degradation during Heat Stress. Cell Rep. 2020, 30, 2430–2443.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grousl, T.; Ivanov, P.; Malcova, I.; Pompach, P.; Frydlova, I.; Slaba, R.; Senohrabkova, L.; Novakova, L.; Hasek, J. Heat Shock-Induced Accumulation of Translation Elongation and Termination Factors Precedes Assembly of Stress Granules in S. cerevisiae. PLoS ONE 2013, 8, e57083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kedersha, N.; Ivanov, P.; Anderson, P. Stress granules and cell signaling: More than just a passing phase? Trends Biochem. Sci. 2013, 38, 494–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Zheng, L.; Jia, J.; Guo, J.; Zheng, M.; Zhao, J.; Shao, J.; Liu, X.; An, L.; Yu, F.; et al. Chloroplast Translation Elongation Factor EF-Tu/SVR11 Is Involved in var2-Mediated Leaf Variegation and Leaf Development in Arabidopsis. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Dogra, V.; Li, M.; Singh, S.; Li, M.; Kim, C. Oxidative post-translational modification of EXECUTER1 is required for singlet oxygen sensing in plastids. Nat. Commun. 2019, 10, 2834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Zhao, J.; Jia, M.; Xu, N.; Liang, S.; Shao, J.; Qi, Y.; Liu, X.; An, L.; Yu, F. Balance between Cytosolic and Chloroplast Translation Affects Leaf Variegation. Plant Physiol. 2018, 176, 804–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, L.; Zhou, C.; Jin, E.; Kucharavy, A.; Zhang, Y.; Wen, Z.; Florens, L.; Li, R. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 2017, 543, 443–446. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishimura, K.; Nakagawa, R.; Hachisuga, C.; Nakajima Munekage, Y. Deciphering the Proteotoxic Stress Responses Triggered by the Perturbed Thylakoid Proteostasis in Arabidopsis. Plants 2021, 10, 519. https://doi.org/10.3390/plants10030519
Nishimura K, Nakagawa R, Hachisuga C, Nakajima Munekage Y. Deciphering the Proteotoxic Stress Responses Triggered by the Perturbed Thylakoid Proteostasis in Arabidopsis. Plants. 2021; 10(3):519. https://doi.org/10.3390/plants10030519
Chicago/Turabian StyleNishimura, Kenji, Reiko Nakagawa, Chisato Hachisuga, and Yuri Nakajima Munekage. 2021. "Deciphering the Proteotoxic Stress Responses Triggered by the Perturbed Thylakoid Proteostasis in Arabidopsis" Plants 10, no. 3: 519. https://doi.org/10.3390/plants10030519




