Abstract
Paramecium bursaria (Ehrenberg 1831) is a ciliate species living in a symbiotic relationship with green algae. The aim of the study was to identify green algal symbionts of P. bursaria originating from distant geographical locations and to answer the question of whether the occurrence of endosymbiont taxa was correlated with a specific ciliate syngen (sexually separated sibling group). In a comparative analysis, we investigated 43 P. bursaria symbiont strains based on molecular features. Three DNA fragments were sequenced: two from the nuclear genomes—a fragment of the ITS1-5.8S rDNA-ITS2 region and a fragment of the gene encoding large subunit ribosomal RNA (28S rDNA), as well as a fragment of the plastid genome comprising the 3′rpl36-5′infA genes. The analysis of two ribosomal sequences showed the presence of 29 haplotypes (haplotype diversity Hd = 0.98736 for ITS1-5.8S rDNA-ITS2 and Hd = 0.908 for 28S rDNA) in the former two regions, and 36 haplotypes in the 3′rpl36-5′infA gene fragment (Hd = 0.984). The following symbiotic strains were identified: Chlorella vulgaris, Chlorella variabilis, Chlorella sorokiniana and Micractinium conductrix. We rejected the hypotheses concerning (i) the correlation between P. bursaria syngen and symbiotic species, and (ii) the relationship between symbiotic species and geographic distribution.
1. Introduction
The unicellular ciliate Paramecium bursaria (Peniculia, Oligohymenophorea) is a host of endosymbiotic algal species. The mutualistic symbiosis exhibited by P. bursaria suppresses the genetic change of the inhabitant and ensures a nutritionally stable environment. Doebeli and Knowlton [1] reported that the rate of nucleotide substitutions was lower in symbiotic algae than in free-living relatives and their corresponding inhabitants since their co-evolution from an ancient association. Paramecium spp. usually comprise several sexually separated sibling groups, termed “syngens”, which are morphologically indistinguishable. Currently, P. bursaria strains have been assigned to five syngens (R1 to R5), which may correspond to some syngens described by Bomford [2,3]. Each syngen in Bomford’s collection (which was lost) had specific geographical distributions. Based on some similarities between syngens from the “old” and “new” collections, it has been suggested that syngen R1 is widespread in Europe; syngen R2 is widespread in Europe, extending eastwards to Siberia and Australia; syngen R4 is fairly widespread in the USA; and syngen R3 is present in Russia, Japan, China and the USA; finally, syngen R5 is represented by only four strains from two locations in western Europe [4].
Symbiotic algae isolated from different Paramecium bursaria syngens are represented by Chlorella-like species belonging to two genetically distinct “European” and “American” populations [5]. Gaponova et al. [6] confirmed the existence of two groups of symbionts based on the analysis of rDNA PCR products of two different lengths, which corresponded to the southern (three introns) or northern (single intron) group. Phylogenetic analyses based on the 28S rDNA gene, ITS 1, 5.8S rDNA and ITS 2 sequences suggested the existence of five different endosymbionts: Chlorella vulgaris, Chlorella variabilis, Micractinium conductrix comb. nov., Choricystis minor (Choriocystis parasitica comb. nov.) and Coccomyxa simplex. Pröschold et al. [7] have confirmed the occurrence of two endosymbiont groups and found that Micractinium conductrix and Chlorella vulgaris belonged to the “European” population. Hoshina and Imamura [8] have found that Chlorella vulgaris is a symbiont of Paramecium bursaria strain. Chlorella variabilis represents the “American” population and has been found in Paramecium bursaria strains (CCAP211/84, 211/109 and 211/110) collected in the USA [7]. Algal symbionts of all P. bursaria strains of two different origins form one clade, but are split into two distinct lineages.
An evolutionary scenario for P. bursaria with respect to algal acquisition and subsequent switching assumes the coexistence of both species belonging to the “American” and “European” endosymbiont groups in one cell of ancestral P. bursaria. This sympatric relationship led to a continuous intron transmission. During evolution, the host “chose” one of the endosymbionts, and later “European” algae may have diverged into a lineage with a weakened host–algal partnership, in which accidental switching of the algae occurred twice [9,10].
Hoshina and Imamura [8] and Gaponova et al. [6] have shown that P. bursaria can contain different endosymbionts, depending on their origin. Nakahara et al. [11] identified an additional endosymbiont, Choricystis minor, in a strain from Florida (USA). Pröschold et al. [7] studied 17 strains of endosymbionts isolated from various hosts and different geographical locations. Phylogenetic analyses revealed that they were polyphyletic. The most studied ciliate, P. bursaria, harbors endosymbionts representing at least five different species: Coccomyxa sp., Choricystis minor, Micractinium conductrix, Chlorella vulgaris and Chlorella variabilis. C. vulgaris, C. variabilis and Micractinium conductrix are obligate endosymbionts of P. bursaria [7]. M. tetrahymenae forms a symbiotic association with Tetrahymena utriculariae only under anoxic or microaerobic conditions. Phylogenetic analyses using complex evolutionary models based on secondary structure have demonstrated that this endosymbiont represents a new species of Micractinium, which belongs to the so-called Chlorella clade (Trebouxiophyceae) [12].
In the present study, we investigated 43 strains of algal symbionts isolated from P. bursaria strains belonging to five syngens. The strains were collected in remote geographical locations. Twenty sequences of symbionts were available in GenBank (28S rDNA and ITS1-5.8S rDNA-ITS2 fragment). The strains of Coccomyxa chodatii, Stigeoclonium tenue, Stigeoclonium variabile, Parachlorella kessleri and Actinastrum hantzschii were used as outgroups. Three loci: a fragment of the ITS1-5.8S rDNA-ITS2 region and a fragment 28S rDNA, as well as chloroplast genes encoding ribosomal protein L36 (rpl36) and translation initiation factor IF-1 (infA) were applied to study phylogenetic relationships of symbiotic algae. The selected ribosomal primers were specific to symbiotic cells, which did not allow the simultaneous amplification of P. bursaria rDNA fragments. The 28S rDNA is characterized by higher variability than the 18S rDNA [8]. The ITS1-5.8S rDNA-ITS2 region is highly variable among the sequences of different species, while it is relatively conserved among the sequences of the same species of algae. Furthermore, this fragment is most commonly available in GenBank, which facilitates comparative analysis. The 3′rpl36-5′infA gene fragment has been selected due to the presence of an intergenic region, which is suspected to have more potential substitution sites than the gene-coding regions.
The main aim of the study was to determine the molecular phylogenetic relationships among green algal endosymbionts of P. bursaria in order to explore the history of the symbiosis events. We tried to answer whether endosymbiosis of a green algae in the host P. bursaria took place prior to the diversification of the host lineage into the various syngens or if endosymbionts are incorporated over and over again. In the latter case we assess whether endosymbionts are host-specific or if there is no relationship between host syngens and endosymbiont lineage.
2. Results
2.1. Syngen Identification
Identification of Paramecium bursaria syngens was performed by mating the studied strain with standard strains representing all mating types of each syngen. The number of symbiotic strains of algal species identified in each of the five P. bursaria syngens is presented in Table 1.

Table 1.
The number of symbiotic strains of particular algal species identified in five syngens of Paramecium bursaria.
2.2. Geographical Distribution of Paramecium Bursaria Symbionts
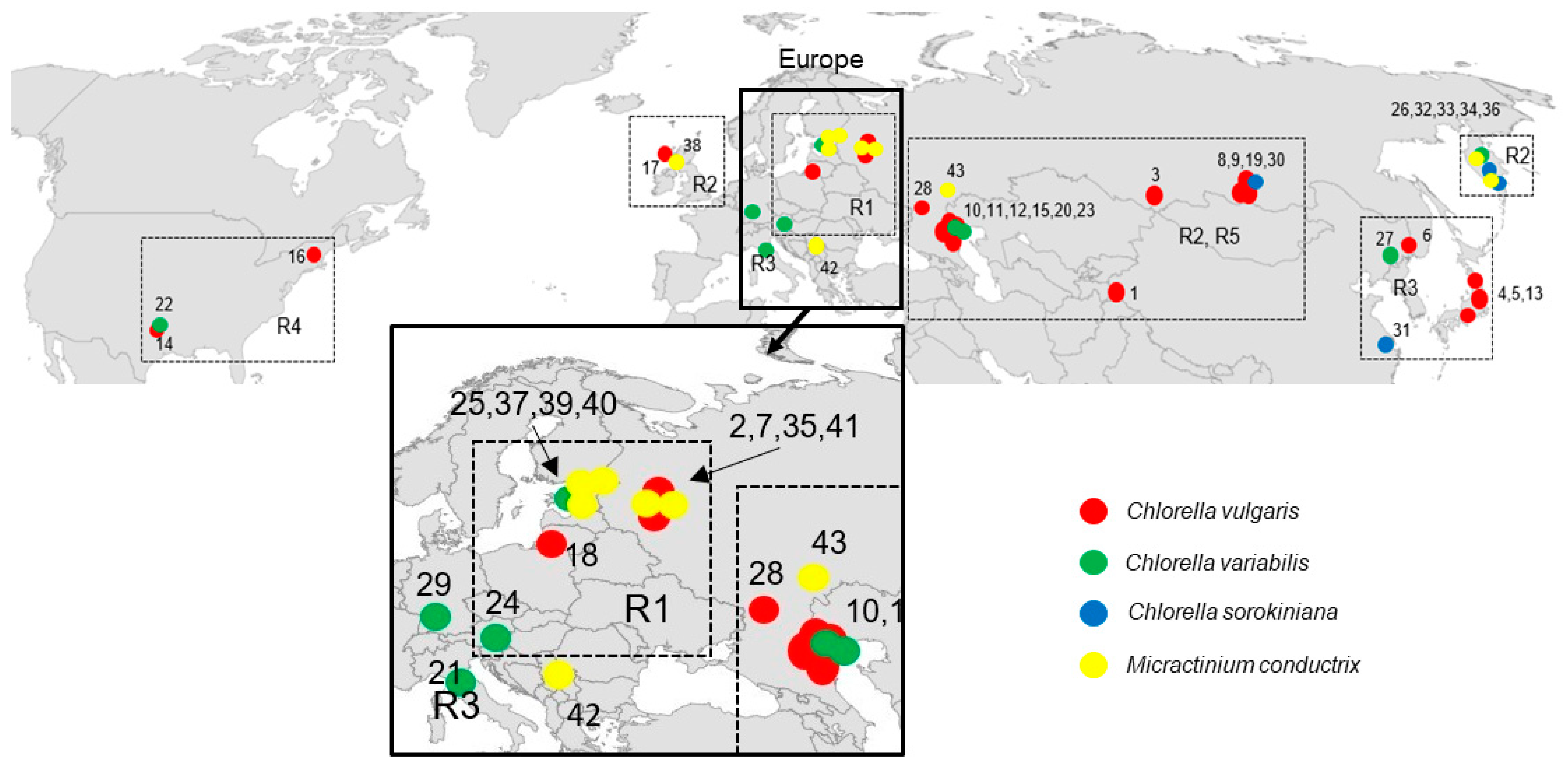
P. bursaria syngens and their geographical distribution are shown in Figure 1 and Table 2. Syngen R1 from central Asia (Tajikistan) harbored C. vulgaris strain but those from Europe (Wien) contained C. variabilis. Endosymbiotic Micractinium conductrix was isolated from the syngen originating from north-eastern Europe (St. Petersburg, Tver). Syngen R2 of P. bursaria was collected most frequently, and 10 endosymbionts from central Asia (Altai, Lake Baikal), eastern Europe (Astrakhan), eastern Europe (Tver, Yaroslavl, Kaliningrad), and Scotland (Europe) were assigned to C. vulgaris. Four strains from eastern Europe (Astrakhan), Far East (Kamchatka) and from Germany (Europe) belonged to C. variabilis. Two strains from Kamchatka and one from central Asia (Lake Baikal) were assigned to C. sorokiniana. Seven strains of Micractinium conductrix from Asia and Europe were found in this syngen. Green endosymbionts from syngen R3 sampled in Japan and Far East (Khabarovsk) belonged to the C. vulgaris clade, but C. variabilis (Khanka Nature Reserve) and C. sorokiniana strains were also found in China. One strain of C. variabilis was isolated in Europe (Italy). Strains isolated from syngen R4 of P. bursaria originating from the USA were assigned to C. vulgaris and C. variabilis. Endosymbionts isolated from syngen R5 originating from eastern Europe (Astrakhan) were assigned to C. vulgaris, while the strain isolated from the same P. bursaria syngen sampled in north-eastern Europe (St. Petersburg) was C. variabilis.
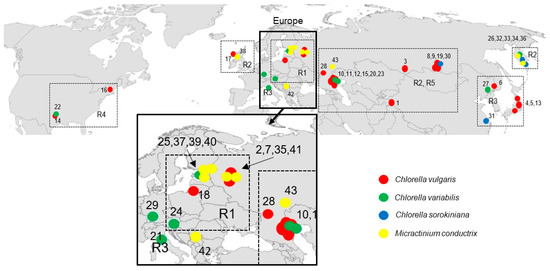
Figure 1.
Geographical distribution of Paramecium bursaria symbionts with numbers corresponding to those in Table 2.

Table 2.
Strains of symbiotic algae studied in the current survey.
2.3. Molecular Results
Results of the analysis of ITS1-5.8S-rDNA-ITS2, 28S rDNA and 3′rpl36-5′infA chloroplast gene fragments revealed similarity of the isolated strains to the species described as Chlorella vulgaris, Chlorella variabilis, Chlorella sorokiniana and Micractinium conductrix. Phylogenetic inference showed that these strains belonged to four distinct clades, thus the endosymbionts were polyphyletic.
2.3.1. Analysis of the ITS1-5.8S rDNA-ITS2 Fragment
Results of the analysis of the ITS1-5.8S rDNA-ITS2 fragments (543 bp) of 37 endosymbionts revealed the existence of 29 haplotypes in the studied dataset. The value of the interspecific haplotype diversity was Hd = 0.987 and the nucleotide diversity was π = 0.16040. Nucleotide frequencies were as follows: A = 20.5%, T = 22.6%, C = 30.1% and G = 26.8%.
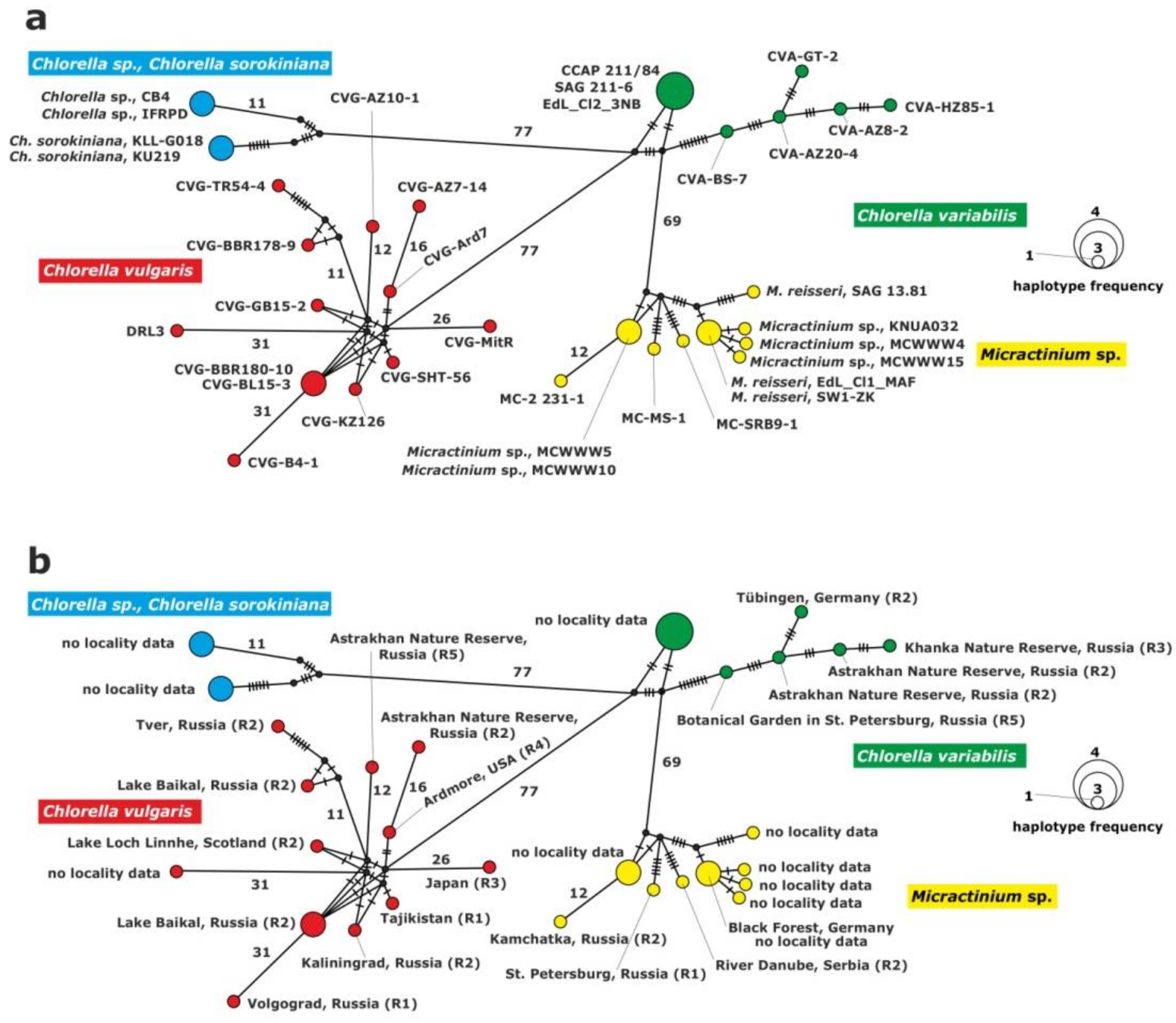
The haplotype network of the ITS1-5.8S rDNA-ITS2 fragment was constructed for the inference and visualization of genetic relationships between green endosymbionts of P. bursaria (Figure 2). Four haplogroups were identified for the rDNA fragment in the studied strains, i.e., C. vulgaris, C. variabilis, C. sorokiniana and M. conductrix. The clade of C. vulgaris was composed of 12 haplotypes; one of them comprised two strains isolated from P. bursaria syngen R2: CVG-BBR-180-10 and CVG-BL15-3 sampled from the Baikal Lake (central Asia). The clade of C. variabilis included six haplotypes. Three strains: CCAP 211/84, SAG 211-6 and Edl_Cl2_3NB from GenBank formed a common haplotype. The remaining strains represented single haplotypes.
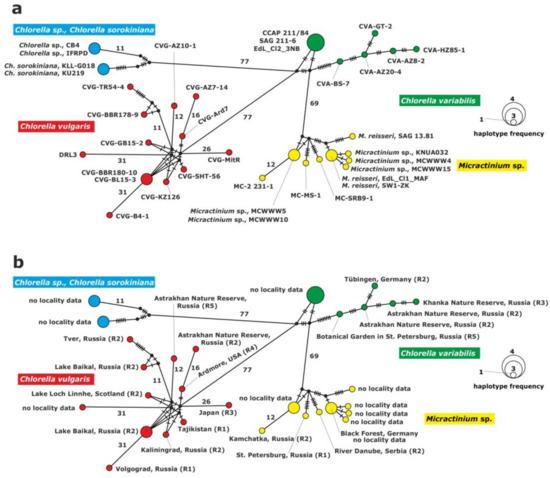
Figure 2.
Haplotype network constructed for 37 symbiotic alga strains based on the comparison of the ITS1-5.8S rDNA-ITS2 sequences, (a) with strain abbreviations, (b) geographical origin of P. bursaria strains and syngens. The size of the dots is proportional to haplotype frequency. Median vectors that represent hypothetical intermediates or unsampled haplotypes are shown as black dots. Hatch marks on individual branches represent nucleotide substitutions between individual haplotypes (corresponding number was assigned for more than 10). Haplotypes marked as “no locality data” were acquired from GenBank.
The clade of C. sorokiniana was composed of two unique haplotypes. The first one consisted of two Chlorella sp. strains, CB4 and IFRPD, and the second one of Chlorella sorokiniana KLL-G018 and KU219 from GenBank.
The following clade, Micractinium, included nine haplotypes and seven of them represented unique haplotypes; two of them were composed of two strains: Micractinium sp., MCWWW5 and MCWWW10 from GenBank, and the second haplotype: Micractinium reisseri EDL_Cl1_MAF from GenBank and SW1-ZK1 from Germany. There were 88 to 112 differences between C. variabilis and C. sorokiniana, 81 to 128 between C. vulgaris and C. variabilis, 72 to 100 between C. variabilis and Micractinium, 149 to 192 between Micractinium and C. vulgaris and 168 to 204 differences between C. vulgaris and C. sorokiniana. Intraspecific variation among haplotypes was the result of several substitutions (Table 2, Figure 2).
2.3.2. Analysis of the 28S rDNA Fragment
Results of the analysis of 28S rDNA fragments (555 bp) of 43 symbionts isolated from different P. bursaria strains showed the presence of 29 haplotypes. The value of the interspecific haplotype diversity was Hd = 0.908 and the nucleotide diversity was π = 0.03165. Nucleotide frequencies were as follows: A = 26.7%, T = 18.7%, C = 23.8% and G = 30.8%.
The haplotype network of the 28S rDNA fragment grouped the strains into four clades: C. vulgaris, C. variabilis, C. sorokiniana and Micractinium. The clade of C. variabilis was composed of 10 unique haplotypes with 2 to 9 substitutions between them (Figure 3).
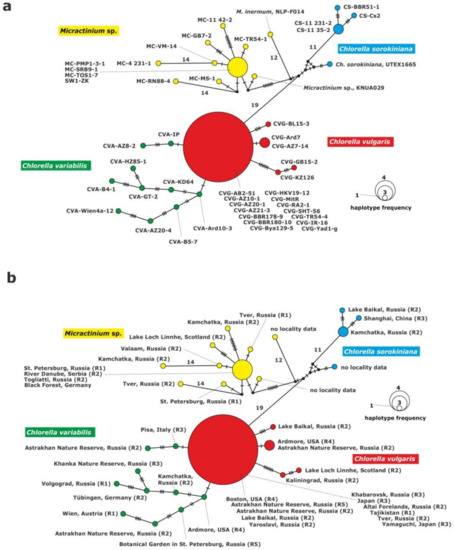
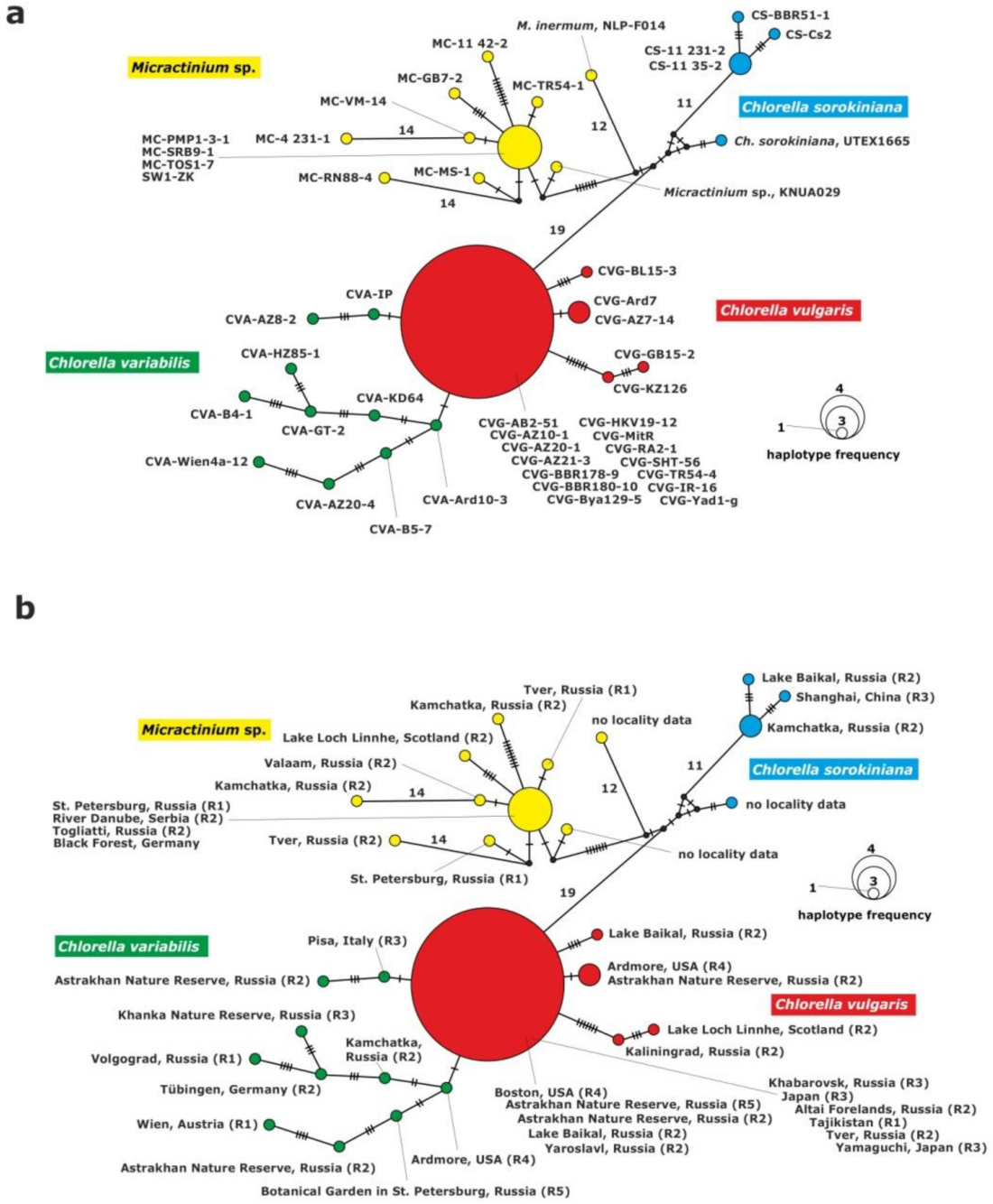
Figure 3.
Haplotype network constructed for 43 symbiotic algae of P. bursaria strains based on sequence comparison of the 28S rDNA gene fragment, (a) with strain abbreviations, (b) geographical origin of P. bursaria strains and syngens. The size of the dots is proportional to haplotype frequency. Median vectors that represent hypothetical intermediates or un-sampled haplotypes are shown as black dots. Hatch marks on individual branches represent nucleotide substitutions between individual haplotypes (corresponding number was assigned for more than 10). Haplotypes marked as “no locality data” were acquired from GenBank.
The C. vulgaris clade consisted of five unique haplotypes. One of them included 14 strains: CVG-Bya129-5 (Yaroslavl) and CVG-TR54-4 (Tver) from eastern Europe, CVG-SHT56 (Tajikistan) from central Asia, CVG-RA2-1 (Altai) and CVG-BBR178-9, CVG-BBR180-10 (Baikal Lake) from central Asia, CVG-AZ10-1, CVG-AZ20-1, CVG-AZ21-3, (Astrakhan) from eastern Europe, CVG-HKV19-12 (Khabarovsk) from the Far East, CVG-JR-16, CVG-MitR and CVG-Yad1-g from Japan, CVG-AB2-51 (Boston) from USA. The second haplotype was composed of two strains: CVG-AZ7-14 (Astakhan) from eastern Europe and CVG-Ard7 (Ardmore) from USAj. The other haplotypes were represented by the following single strains: CVG-BL15-3, CVG-KZ-126 and CVG-GB15-2. The C. variabilis clade was composed of 10 single strains.
The Micractinium clade was composed of 10 haplotypes. One of them included four strains from Europe: MC-PMP1-3-1, (St. Petersburg, north-eastern Europe), MC-SRB9-1 (Serbia, southern Europe), MC-TOS1-7 (Togliatii, south-eastern Europe) and SW1-ZK (Germany, western Europe). The other nine corresponded to single strains: MC-4 231-1, MC-VM-14, MC-RN88-4, MC-MS-1, MC-GB7-2, MC-11 42-2, MC-TR54-1, NLP-F014 and KNUA029.
The last clade consisted of C. sorokiniana representatives, and included four haplotypes. One haplotype was formed by two strains from the Far East origin: CS-11 231-2 and CS-11 35-2 (Kamchatka) and the other two represented single strains: CS-BBR51-1 and CS-Cs2.
2.3.3. Analysis of the rpl36-infA Genes Fragment
Results of the rpl36-infA gene fragment (267 bp) analysis in symbionts isolated from 43 P. bursaria strains showed the presence of 36 haplotypes. The value of the interspecific haplotype diversity was Hd = 0.984, and the nucleotide diversity was π = 0.07886. Nucleotide frequencies were as follows: A = 29.6%, T = 36.0%, C = 18.5% and G = 15.9%.
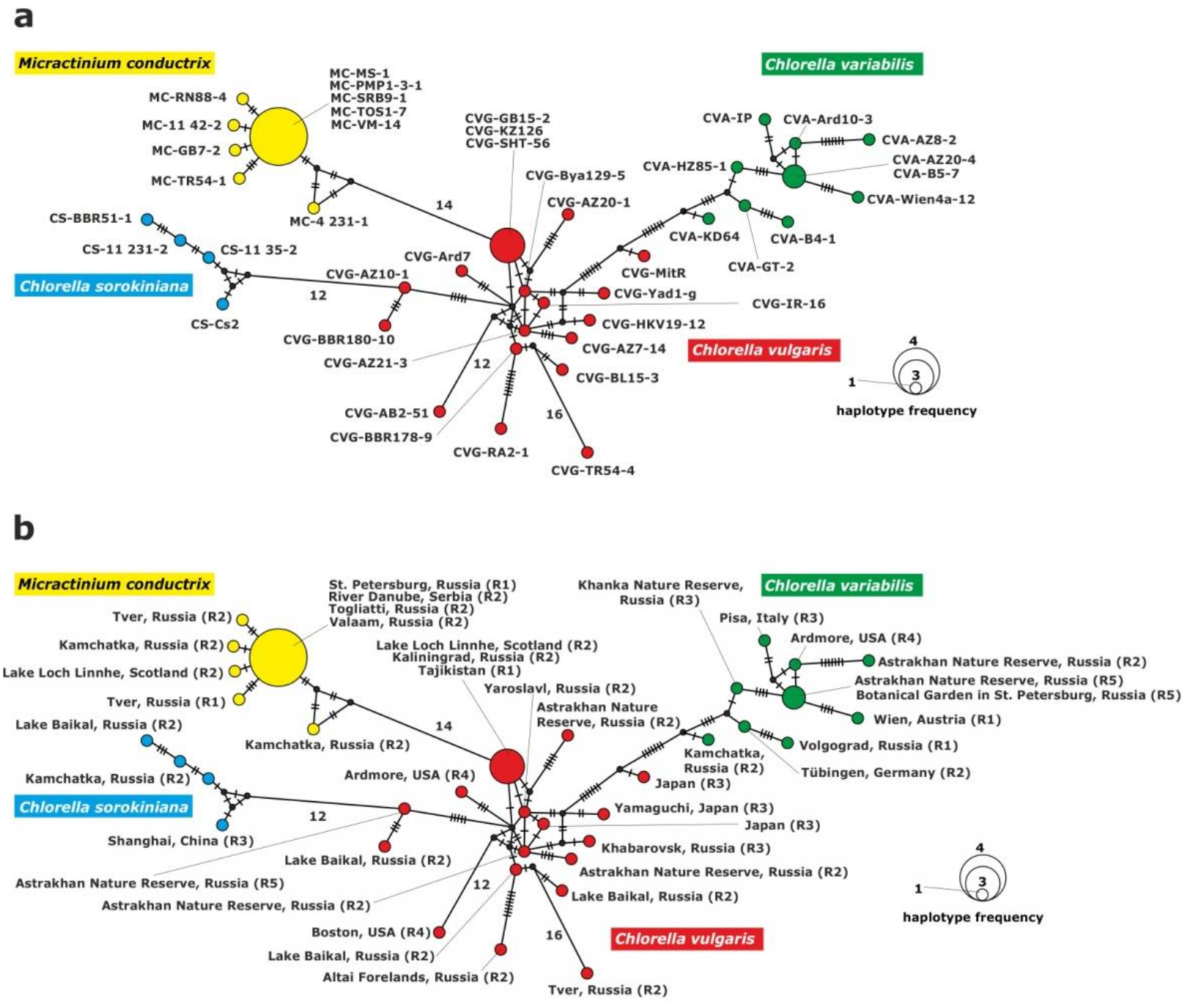
The haplotype network of chloroplast gene fragments grouped the strains into four clades: C. vulgaris, C. variabilis, C. sorokiniana and M. conductrix (Figure 4). The C. vulgaris clade included 17 haplotypes; one haplotype was represented by three strains. Two strains from Europe: CVG-GB15-2 (Scotland), CVG-KZ-126 (Kaliningrad) isolated from P. bursaria syngen R2, and one strain from central Asia: CVG-SHT-56 (Tajikistan) from syngen R1. The remaining haplotypes consisted of single strains.
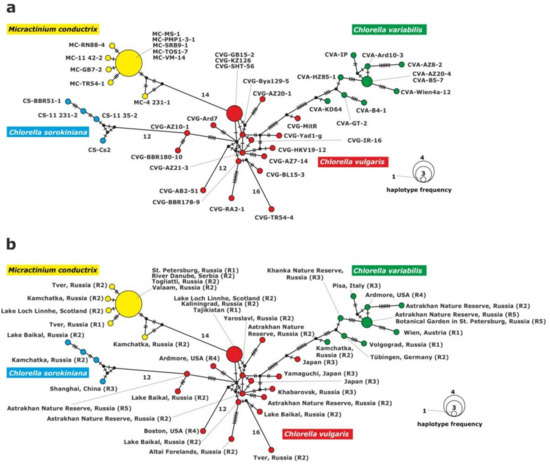
Figure 4.
Haplotype network constructed for 43 symbiotic algae strains based on the comparison of the 3′rpl36-5′infA genes sequences, (a) with strain abbreviations, (b) geographical origin of P. bursaria strains and syngens. The size of the dots is proportional to haplotype frequency. Median vectors that represent hypothetical intermediates or unsampled haplotypes are shown as black dots. Hatch marks on individual branches represent nucleotide substitutions between individual haplotypes (corresponding number was assigned for more than 10).
The clade of C. variabilis consisted of nine haplotypes and eight of them included single strains. Strain CVA-B5-7 (St. Petersburg, north-eastern Europe) from syngen R5 and strain CVA-AZ20-4 (Astrakhan, eastern Europe) from syngen R2 belonged to the ninth haplotype.
The C. sorokiniana clade was composed of four unique haplotypes corresponding to single strains.
The M. conductrix clade included six haplotypes, five of them represented single strains and one haplotype was composed of the five following strains: MC-PMP1-3-1 and MC-MS-1 (St. Petersburg, north-eastern Europe), isolated from syngen R1, MC-SRB9-1 (Serbia, southern Europe), MC-TOS1-7 (Togliatii, south-eastern Europe), and MC-VM-14 (Valaam, northern Europe) isolated from syngen R2.
3. Discussion
Paramecium bursaria is an archetypical outbreeder, which presumably means that its effective population size is large. P. bursaria is divided into five syngens which are characterized by a specific geographical distribution. Nyberg [22] concluded that P. bursaria syngens, as extreme outbreeders, should be globally distributed, but Bomford [2] and Greczek-Stachura et al. [4] postulated that most sibling species were restricted to certain geographical regions, and thus adapted to specific conditions. Based on the comparison of syngens from Bomford’s collection and new syngen annotations, it is known that syngens R3 and R4 have been found in the United States [23], and syngen R3 has been reported later in China [24]. According to the study by Hoshina et al. [25], P. bursaria strains from Japan were also classified as syngen R3. Two syngens, R1 and R2, are only of Eurasian origin, and have been recorded at various locations from Great Britain to central Siberia; in addition, two strains of syngen 2 have been found in one locality in Australia. Syngen R3 strains have been isolated in far-eastern Russia and south-eastern Siberia (but never western Siberia), China, Japan, and the USA. Recently, this syngen has been reported in Europe, namely in Austria and in Italy (although the strain from Pisa was collected in a botanical garden, where it could have been brought along with some tropical plants). Syngen 4 strains are restricted to the USA. Strains belonging to syngen 5 have been found in the Volga delta, known for its great migration routes of waterfowl that are suspected transmitters of paramecia [4,26]. The current investigation of different syngens of P. bursaria collected in Europe, Asia and North America confirmed the previous knowledge about their biogeography. P. bursaria syngen R1 has been found in central Asia and north-eastern Europe. Strains of syngen R2 have been found in Asia and Europe. Syngen R3 was sampled in Japan, Far East and China. Strains of syngen R4 originate from the USA and syngen R5 strains are derived from eastern Europe and north-eastern Europe (Figure 1, Table 2).
The existence of syngens is the result of the process of speciation. The key question regarding evolution is: what are the driving forces behind initial speciation of Paramecium bursaria? Geographic isolation is often the main speciation factor, but its significance in protists is uncertain as there is still disagreement over their distribution—whether it is cosmopolitan or endemic.
If P. bursaria syngens are hosting the same species of endosymbiotic algae, they can be sympatric or other speciation mechanisms may play a leading role. Therefore, in our opinion, identification of species of endosymbiotic algae can explain a possible process of co-evolution. In the present study, we have identified four species of endosymbiotic algae, i.e., C. vulgaris, C. variabilis, C. sorokiniana and M. conductrix. Spanner et al. [27], based on ITS-2 sequencing, identified Chlorella variabilis and Micractinium conductrix in Paramecium bursaria cells. The two above endosymbionts have been identified in strains belonging to syngens R1 and R2 of P. bursaria, which originated from Europe. Moreover, we have found C. vulgaris and C. variabilis in all five syngens of P. bursaria, M. conductrix was present in syngen R1 and R2, and C. sorokiniana in syngen R2 and R3 (Table 1). Gaponova et al. [6] have also found M. conductrix in P. bursaria isolates collected in North Karelia (Russia). Overall, it seems that M. conductrix occurs only in Europe, whereas C. variabilis is distributed worldwide. Hoshina et al. [5,10] established the geographical distribution of Micractinium sp. in the regions of England, Germany, Austria and northern Karelia, which was consistent with the results obtained by Luo et al. [17,28]. Strains belonging to the American group derived from USA, Japan, China and southern Australia carried symbiotic algae classified as Chlorella vulgaris and Chlorella variabilis [7]. Hoshina and Imamura [9] identified the strains from Kaliningrad as C. vulgaris, similar to our findings i.e., the strain isolated from syngen R2. Pröschold et al. [7] have suggested that C. variabilis is characteristic of the American but not the European group; however, according to our results, the strains from St. Petersburg and Valaam as well as strains from central Europe (Pisa, River Danube in Serbia) have been assigned to C. variabilis and M. conductrix.
Our findings suggests that there is no correlation between P. bursaria syngen and the species of symbiont, as was previously argued by Weis [29]. Similarly, Reisser et al. [30] stated that P. bursaria strains of American or European origin formed a stable symbiosis with symbionts of both groups. Then, Meier and Wiessner [31] demonstrated that P. bursaria could eliminate symbionts and subsequently be reinfected by new symbionts. Summerer et al. [32] mixed two aposymbiotic P. bursaria strains with symbiotic and free-living Chlorella strains. Symbioses were formed with endosymbiotic Chlorella, with the exception of those from H. viridis and free-living algae. Similarly, in the current survey we demonstrated that there is no strong relationship between species of symbionts and the geographical distribution of their host, P. bursaria. This may be explained by the ancestral aposymbiotic ciliate P. bursaria possibly having acquired different species of green algae and later diverging into a lineage with a host-algal partnership where accidental algal change may have occurred. Summerer et al. [33] analyzed nuclear 18S rDNA, the ITS1 region and chloroplast 16S rDNA from algal symbionts of P. bursaria strains originating from two lakes in Austria. These strains formed a clade with two distinct lineages, suggesting the existence of a biogeographic pattern. Genetic differences between symbiotic algae are 10 times higher than between free-living algae. This suggests that multiple symbiotic origins are more likely than the divergence of one symbiotic species to different symbiotic algae existing currently [25]. The endosymbiotic lifestyle has evolved many times in green algae, as evidenced by the presence of numerous haplotypes of endosymbiotic algae in the haplotype network based on the nuclear ITS1-5.8S rDNA-ITS2 fragment, 28S rDNA fragment and 3′rpl36-5′infA gene sequences. Endosymbionts of the Chlorellaceae species, which also serve as specific hosts for large dsDNA viruses known as chloroviruses, do not cluster together, providing strong evidence for independent transitions to endosymbiosis [34].
Therefore, we suppose that the speciation of P. bursaria syngens was an earlier evolutionarily event than the establishment of symbiosis, as evidenced by the diversity of symbionts and their lack of specificity.
4. Materials and Methods
4.1. Strain Cultivation and Strain Crosses
Paramecium bursaria strains were cultivated on a lettuce medium according to Sonneborn [35], fed Klebsiella pneumoniae (SMC) and stored at 18 °C (12L/12D). We investigated 43 symbiotic strains isolated from P. bursaria cells derived from different geographical locations. We also analyzed 20 sequences of symbiotic algae available in GenBank and strains of Coccomyxa chodatii, Stigeoclonium tenue, Stigeoclonium variabile, Parachlorella kessleri and Actinastrum hantzschii as outgroups (Table 2).
Identification of P. bursaria syngens was performed by mating reaction of a studied strain with standard strains representing all mating types of each syngen. The studied strains were assigned to a certain syngen based on the occurrence of strong clumping at the beginning of the mating reaction, the presence of mating couples and survival of F1 progeny.
4.2. Molecular Methods
Symbiotic DNA was extracted using the GeneJET Plant Genomic DNA Purification Kit (ThermoScientific) according to the protocol. Dense P. bursaria culture (1.5 mL) was harvested from a liquid culture by centrifugation. Then, the pellet was sonicated on ice for 10 s at 40 W. Subsequently, the standard extraction protocol was followed. The ITS1-5.8S rDNA-ITS2 fragment was amplified using the following primers pairs: ITS1 [32]/ITS2R (primer designed for the present study, Table 3) and ITS1F/ITS2R (primers designed for the present study, Table 3) according to the protocol with the following parameters: initial denaturation at 95 °C for 5 min followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 54 °C for 2 min, extension at 72 °C for 3 min and a final extension at 72 °C for 5 min.

Table 3.
Primers used in the present study.
The fragment of a 28S rDNA was amplified by polymerase chain reaction (PCR) using the HLR0F/HLR4R primer pair [8,37] (Table 3), according to the protocol described by Hoshina et al. [38]. The fragment of 3′rpl36-5′infA genes was amplified using the UCP2F and UCP2R primer set (Table 3), according to Provan et al. [36]. After amplification, PCR products were separated by electrophoresis in 1% agarose gel for 1 h at 95 V and then gel-purified using NucleoSpin Extract II (Macherey-Nagel, Düren, Germany). Sequencing reaction was performed in both directions using the BigDye Terminator v3.1 kit (Applied Biosystems, Foster City, USA). Sequencing products were precipitated using Ex Terminator (A&A Biotechnology, Gdynia, Poland).
4.3. Data Analyzes
Sequences were examined and corrected using Chromas Lite (Technylesium), and aligned using BioEdit [39]. The analysis of haplotype diversity (Hd) and nucleotide diversity (π) was carried out using DnaSP v5.10.01 [39]. The analysis of nucleotide frequencies and identification of the best nucleotide substitution models for maximum likelihood tree reconstruction (T92 + G for three loci) were conducted using Mega v5.1. Haplotype networks were constructed using the Median Joining method implemented in the Network 4.6.1.3 software [40,41].
5. Conclusions
The ITS1-5.8S rDNA-ITS2 fragment is the most appropriate molecular marker to identify and resolve evolutionary relationship between symbionts of Paramecium bursaria. We assigned symbiotic algae of P. bursaria to four species: Chlorella vulgaris, Chlorella variabilis, Chlorella sorokiniana and Micractinium conductrix. The division of P. bursaria endosymbionts into the American and European groups and the correlation between P. bursaria syngen and a symbiotic species has not been confirmed. No strong relationships have been found between symbiotic species and geographical distribution of their host P. bursaria.
Molecular markers: ITS1-5.8S rDNA-ITS2, 28S rDNA fragments and 3′rpl36-5′infA gene fragments are useful molecular tools for distinguishing closely related taxa of P. bursaria symbionts. The ITS1-5.8S rDNA-ITS2 fragment is the most appropriate due to its high interspecific and low intraspecific variability. Additionally, the application of two independent genome fragments (nuclear and chloroplast) increases the reliability of the results.
Author Contributions
Conceptualization, M.G.-S. and M.R.; methodology, M.G.-S. and S.T.; software, M.G.-S., P.Z.L. and S.T.; validation, M.G.-S., P.Z.L. and S.T.; formal analysis, P.Z.L. and M.G.-S.; investigation, M.G.-S., P.Z.L.; resources, M.G.-S., P.Z.L. and S.T.; data curation, P.Z.L.; writing—original draft preparation, M.G.-S., P.Z.L. and K.M.; writing—review and editing, M.G.-S., P.Z.L., S.T., M.R., K.M.; visualization, M.G.-S., P.Z.L. and K.M.; supervision, M.G.-S.; project administration, M.G.-S. and K.M.; funding acquisition, M.G.-S. All authors have read and agreed to the published version of the manuscript
Funding
This research was funded by Pedagogical University of Krakow.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Doebeli, M.; Knowlton, N. The evolution of interspecific mutualisms. Proc. Natl. Acad. Sci. USA 1998, 95, 8676–8680. [Google Scholar] [CrossRef] [PubMed]
- Bomford, B. The syngens of Paramecium bursaria: New mating types and intersyngenic mating reactions. J. Protozool. 1966, 13, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Witcherman, R. The Biology of Paramecium, 2nd ed.; Plenum Press: New York, NY, USA, 1985. [Google Scholar]
- Greczek-Stachura, M.; Potekhin, A.; Przyboś, E.; Rautian, M.; Skoblo, I.; Tarcz, S. Identification of Paramecium bursaria syngens through molecular markers—Comparative analysis of three loci in the nuclear and mitochondrial DNA. Protist 2012, 163, 671–685. [Google Scholar] [CrossRef]
- Hoshina, R.; Iwataki, M.; Imamura, N. Chlorella variabilis and Micractinium reisseri sp. nov. (Chlorellaceae, Trebouxiophyceae): Redescription of the endosymbiotic green algae of Paramecium bursaria (Peniculia, Oligohymenophorea) in the 120th year. Phycol. Res. 2010, 58, 188–201. [Google Scholar] [CrossRef]
- Gapanova, I.N.; Andronov, E.E.; Migunova, A.V.; Vorobyev, K.P.; Chizhevskaja, E.P.; Kvitko, K.V. Genomic dactyloscopy of Chlorella sp., symbionts of Paramecium bursaria. Protistology 2007, 4, 311–317. [Google Scholar]
- Pröschold, T.; Darienko, T.; Silva, P.C.; Reisser, W.; Krienitz, L. The systematics of Zoochlorella revisited employing an integrative approach. Environ. Microbiol. 2011, 13, 350–364. [Google Scholar] [CrossRef] [PubMed]
- Hoshina, R.; Imamura, N. Multiple origins of the symbioses in Paramecium bursaria. Protist 2008, 159, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Hoshina, R.; Imamura, N. Origins of algal symbionts of Paramecium bursaria. In Endosymbionts in Paramecium. Microbiology Monographs; Fujishima, M., Ed.; Springer GmbH: Berlin/Heidelberg, Germany, 2009; Volume 12, pp. 1–29. [Google Scholar] [CrossRef]
- Hoshina, R.; Imamura, N. Phyogenetically close group I introns with difefrent positions among Paramecium bursaria pfotobionts imply a primitive stage of intron diversification. Mol. Biol. Evol. 2009, 26, 1309–1319. [Google Scholar] [CrossRef]
- Nakahara, M.; Handa, S.; Watanabe, S.; Deguchi, H. Choricystis minor as a new symbiont of simultaneous two-species association with Paramecium bursaria and implications for its phylogeny. Symbiosis 2004, 36, 127–151. [Google Scholar]
- Pröschold, T.; Pitsch, G.; Darienko, T. Micractinium tetrahymenae (Trebouxiophyceae, Chlorophyta), a new endosymbiont isolated from ciliates. Diversity 2020, 12, 200. [Google Scholar] [CrossRef]
- Zagata, P.; Greczek-Stachura, M.; Tarcz, S.; Rautian, M. The evolutionary relationships between endosymbiotic green algae of Paramecium bursaria syngens originating from different geographical locations. Folia Biol. 2016, 64, 47–54. [Google Scholar] [CrossRef]
- Rosenberg, J.N.; Kobayashi, N.; Barnes, A.; Noel, E.A.; Btenbaugh, M.J.; Oyler, G.A. Comparative analyses of three Chlorella species in response to light and sugar reveal distinctive lipid accumulation patterns in the microalga C. sorokiniana. PLoS ONE 2014, 9, e92460. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.W.; Jo, S.W.; Cho, H.W.; Nam, S.W.; Shin, W.; Park, K.M.; Lee, K.I.; Yoon, H.S. Phylogeny, morphology, and physiology of Micractinium strains isolated from shallow ephemeral freshwater in Antarctica. Phycol. Res. 2015, 63, 212–218. [Google Scholar] [CrossRef]
- Park, K.C.; Whitney, C.G.; Kozera, C.; O’Leary, S.J.; McGinn, P.J. Seasonal isolation of microalgae from municipal wastewater for remediation and biofuel applications. J. App. Microbiol. 2015, 119, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Pröschold, T.; Bock, C.; Krienitz, L. Generic concept in Chlorella-related coccoid green algae (Chlorophyta, Trebouxiophyceae). Plant Biol. 2010, 12, 545–553. [Google Scholar] [CrossRef]
- Kaplan-Levy, R.N.; Alster-Gloukhovski, A.; Benyamini, Y.; Zohary, T. Lake Kinneret phytoplankton: Integrating classical and molecular taxonomy. Hydrobiologia 2016, 764, 283–302. [Google Scholar] [CrossRef]
- Darienko, T.; Gustavs, L.; Eggert, A.; Wolf, W.; Pröschold, T. Evaluating the species boundaries of green microalgae (Coccomyxa, Trebouxiophyceae, Chlorophyta) using integrative taxonomy and DNA barcoding with further implications for the species identification in environmental samples. PLoS ONE 2015, 10, e0127838. [Google Scholar] [CrossRef]
- Caisova, L.; Marin, B.; Melkonian, M. A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist 2013, 164, 482–496. [Google Scholar] [CrossRef]
- Ustinova, I.; Krienitz, L.; Huss, V.A.R. Closteriopsis acicularis (G.M. Smith) Belcher et Swale is a fusiform alga closely related to Chlorella kessleri Fott et Novakova (Chlorophyta, Trebouxiophyceae). Eur. J. Phycol. 2001, 36, 341–351. [Google Scholar] [CrossRef]
- Nyberg, D. The species concept and breeding systems. In Paramecium; Görtz, H.D., Ed.; Springer: Berlin/Heidelberg, Germany, 1988. [Google Scholar] [CrossRef]
- Jennings, H.S. Sex reaction types and their interrelations in Paramecium bursaria: I. Proc. Natl. Acad. Sci. USA 1938, 24, 112–177. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-T. Varieties and mating types in Paramecium bursaria. II. Variety and mating types found in China. J. Exp. Zool. 1956, 132, 266–268. [Google Scholar] [CrossRef]
- Hoshina, R.; Hayashi, S.; Imamura, N. Intraspecific genetic divergnce of Paramecium bursaria and re-construction of paramecian phylogentic tree. Acta Protozool. 2006, 45, 377–386. [Google Scholar]
- Zagata, P.; Greczek-Stachura, M.; Tarcz, S.; Rautian, M. Molecular identification of Paramecium bursaria syngens and studies on geographic distribution using mitochondrial cytochrome C oxidase subunit I (COI). Folia Biol. 2015, 63, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Spanner, C.; Darienko, T.; Biehler, T.; Sonntag, B.; Pröschold, T. Endosymbiotic green algae in Paramecium bursaria: A new isolation method and a simple diagnostic PCR approach for the identification. Diversity 2020, 12, 240. [Google Scholar] [CrossRef]
- Luo, W.; Pflugmacher, S.; Pröschold, T.; Walz, N.; Krienitz, L. Genotype versus phenotype variability in Chlorella and Micractinium (Chlorophyta, Trebouxiophyceae). Protist 2006, 157, 315–333. [Google Scholar] [CrossRef]
- Weis, D.S. Correlation of infectivity and concanavalin a agglutinability of algae exsymbiotic from Paramecium bursaria. J. Protozool. 1978, 25, 366–370. [Google Scholar] [CrossRef]
- Reisser, W.; Vietze, S.; Widowski, M. Taxonomic studies on endocytobiotic chlorophycean algae isolated from different American and European strains of Paramecium bursaria. Symbiosis 1988, 6, 253–270. [Google Scholar]
- Meier, R.; Wiessner, W. Infection of algae-free Paramecium bursaria with symbiotic Chlorella sp. isolated from green paramecia II: A time study. J. Cell Sci. 1989, 93, 571–579. [Google Scholar] [CrossRef]
- Summerer, M.; Sonntag, B.; Sommaruga, R. An experimental test of the symbiosis specificity between the ciliate Paramecium bursaria and strains of the unicellular green alga Chlorella. Environ. Microbiol. 2007, 9, 2117–2122. [Google Scholar] [CrossRef]
- Summerer, M.; Sonntag, B.; Sommaruga, R. Ciliate-symbiont specificity of freshwater endosymbiotic Chlorella (Trebouxiophyceae, Chlorophyta). J. Phycol. 2008, 44, 77–84. [Google Scholar] [CrossRef]
- Fan, W.; Guo, W.; Van Etten, J.L.; Mower, J.P. Multiple origins of endosymbionts in Chlorellaceae with no reductive effects on the plastid or mitochondrial genomes. Sci. Rep. 2017, 7, 10101. [Google Scholar] [CrossRef]
- Sonneborn, T.M. Methods in Paramecium research. In Methods in Cell Biology; Prescott, E.D.M., Ed.; Academic Press: New York, NY, USA, 1970; Volume 3, pp. 241–339. [Google Scholar] [CrossRef]
- Provan, J.; Murphy, S.; Maggs, C.A. Universal plastid primers for Chlorophyta and Rodophyta. Eur. J. Phycol. 2004, 39, 43–50. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.M., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Hoshina, A.R.; Kamako, S.I.; Imamura, N. Phylogenetic position of endosymbiotic green algae in Paramecium bursaria Ehrenberg from Japan. Plant Biol. 2004, 6, 447–453. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Expertise in Software for Genetics aand Engineering. Available online: http://www.fluxus-engineering.com/ (accessed on 2 October 2020).
- Bandelt, H.J.; Forster, P.; Röthl, A. Median-Joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).