A Revised Phylogeny of the Mentha spicata Clade Reveals Cryptic Species
Abstract
:1. Introduction
2. Results
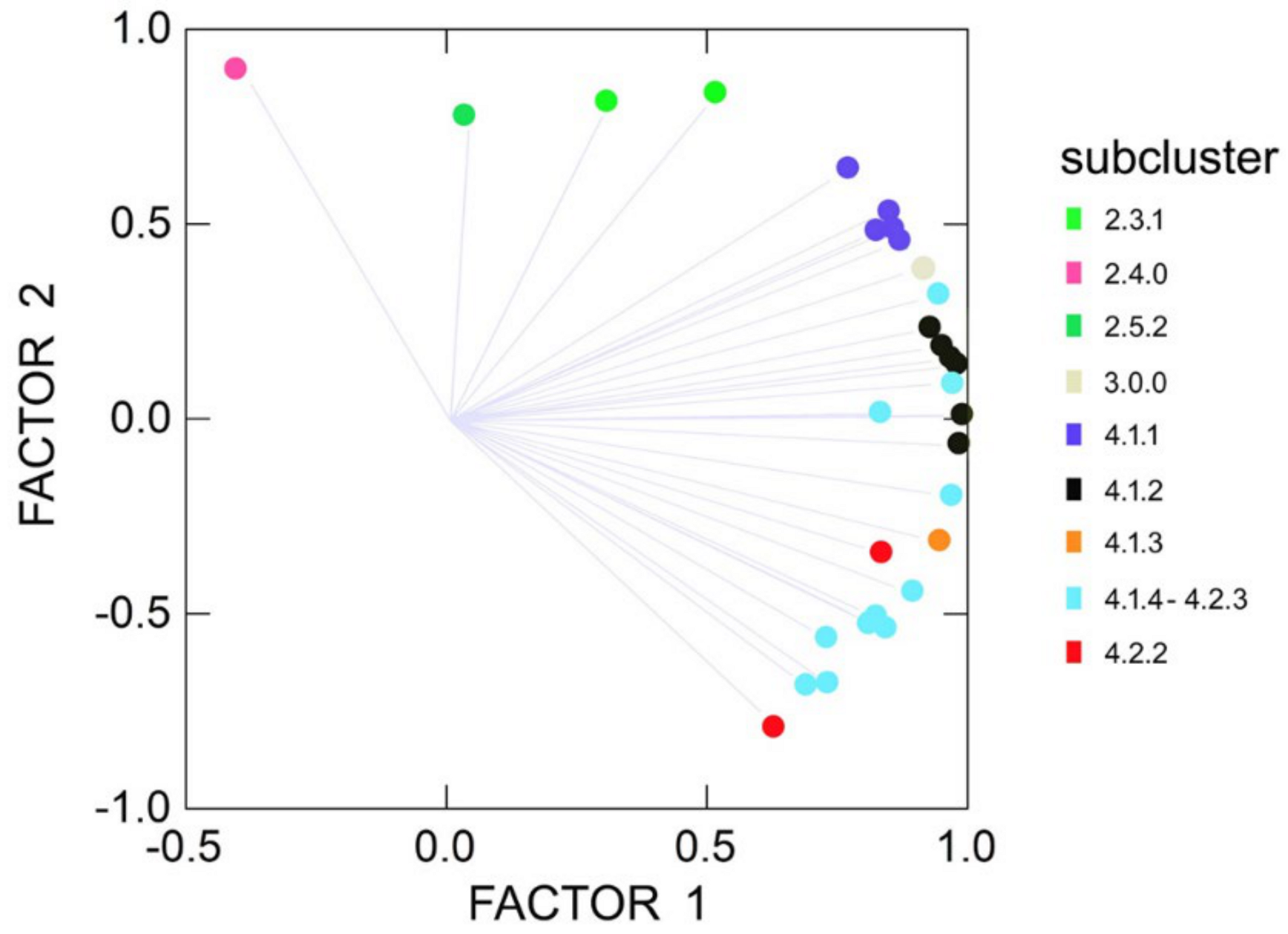
2.1. DNA Amplification, Phylogenetic Reconstruction and ITS Clusters
2.2. Start Codon Targeted Polymorphism Technique Integrated with ITS Sequences
2.3. Genetic Distances
2.4. Morphological Assessment of Genotypic Units
2.4.1. Variable Selection
2.4.2. Unconstrained Morphology
2.4.3. Constrained Morphology
3. Discussion
3.1. A Revised Phylogeny
3.2. Former Phylogenies in the Light of New Results
3.3. Perspectives in Mint Genetics
3.3.1. Nuclear DNA versus Chloroplast DNA
3.3.2. Intraindividual ITS Variability as A Mirror of Intraspecific Variation
3.4. Plasticity in Porous Genomes—Future Challenges in Mentha
3.5. The Hybrid Status of Mentha spicata Revisited
4. Materials and Methods
4.1. Sampling
4.2. DNA Extraction and Sequencing
4.3. Morphological Measurements
4.4. Data Analyses
4.4.1. Genetic Analyses
4.4.2. Morphological Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cutter, A.D.; Jovelin, R.; Dey, A. Molecular Hyperdiversity and Evolution in Very Large Populations. Mol. Ecol. 2013, 22, 2074–2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willmore, K.E.; Young, N.M.; Richtsmeier, J.T. Phenotypic Variability: Its Components, Measurement and Underlying Developmental Processes. Evol. Biol. 2007, 34, 99–120. [Google Scholar] [CrossRef]
- Alix, K.; Gérard, P.R.; Schwarzacher, T.; Heslop-Harrison, J.S. (Pat) Polyploidy and Interspecific Hybridization: Partners for Adaptation, Speciation and Evolution in Plants. Ann. Bot. 2017, 120, 183–194. [Google Scholar] [CrossRef]
- Lucchesi, J.C. Epigenetics, Nuclear Organization & Gene Function; Oxford University Press: Oxford, UK, 2019. [Google Scholar]
- Vereecken, N.J.; Cozzolino, S.; Schiestl, F.P. Hybrid Floral Scent Novelty Drives Pollinator Shift in Sexually Deceptive Orchids. BMC Evol. Biol. 2010, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rieseberg, L.H.; Wood, T.E.; Baack, E.J. The Nature of Plant Species. Nature 2006, 440, 524–527. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.; Raadik, T.A.; Burridge, C.P.; Georges, A. Global Biodiversity Assessment and Hyper-Cryptic Species Complexes: More Than One Species of Elephant in the Room? Syst. Biol. 2014, 63, 518–533. [Google Scholar] [CrossRef] [PubMed]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic Species as a Window on Diversity and Conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef]
- Jörger, K.M.; Schrödl, M. How to Describe a Cryptic Species? Practical Challenges of Molecular Taxonomy. Front. Zool. 2013, 10, 59. [Google Scholar] [CrossRef] [Green Version]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic Diversity and Conservation Units: Dealing with the Species-Population Continuum in the Age of Genomics. Front. Ecol. Evol. 2018, 6, 165. [Google Scholar] [CrossRef] [Green Version]
- Struck, T.H.; Feder, J.L.; Bendiksby, M.; Birkeland, S.; Cerca, J.; Gusarov, V.I.; Kistenich, S.; Larsson, K.H.; Liow, L.H.; Nowak, M.D.; et al. Finding Evolutionary Processes Hidden in Cryptic Species. Trends Ecol. Evol. 2018, 33, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Cerca, J. On the Origin of Cryptic Species: Insights from the Stygocapitella Species Complex. Ph.D. Thesis, University of Oslo, Natural History Museum Faculty of Mathematics and Natural Sciences, Oslo, Norway, 2019. [Google Scholar]
- Pentinsaari, M.; Vos, R.; Mutanen, M. Algorithmic Single-Locus Species Delimitation: Effects of Sampling Effort, Variation and Nonmonophyly in Four Methods and 1870 Species of Beetles. Mol. Ecol. Resour. 2017, 17, 393–404. [Google Scholar] [CrossRef]
- Luo, A.; Ling, C.; Ho, S.Y.W.; Zhu, C.-D. Comparison of Methods for Molecular Species Delimitation Across a Range of Speciation Scenarios. Syst. Biol. 2018, 67, 830–846. [Google Scholar] [CrossRef] [Green Version]
- Martoni, F.; Bulman, S.; Pitman, A.; Taylor, G.; Armstrong, K. DNA Barcoding Highlights Cryptic Diversity in the New Zealand Psylloidea (Hemiptera: Sternorrhyncha). Diversity 2018, 10, 50. [Google Scholar] [CrossRef] [Green Version]
- Rüegg, S.; Bräuchler, C.; Geist, J.; Heubl, G.; Melzer, A.; Raeder, U. Phenotypic Variation Disguises Genetic Differences among Najas Major and N. Marina, and Their Hybrids. Aquat. Bot. 2019, 153, 15–23. [Google Scholar] [CrossRef]
- Semenova, M.V.; Enina, O.L.; Shelepova, O.V. Intra- and Interspecific Variability of Mentha Arvensis L- and M. Canadensis L. Vavilovskii Zhurnal Genet. Sel. 2019, 23, 1067–1075. [Google Scholar] [CrossRef] [Green Version]
- Lexer, C.; Joseph, J.; van Loo, M.; Prenner, G.; Heinze, B.; Chase, M.W.; Kirkup, D. The Use of Digital Image-Based Morphometrics to Study the Phenotypic Mosaic in Taxa with Porous Genomes. TAXON 2009, 58, 5–20. [Google Scholar] [CrossRef]
- Pinho, C.; Hey, J. Divergence with Gene Flow: Models and Data. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 215–230. [Google Scholar] [CrossRef] [Green Version]
- Bräuchler, C.; Meimberg, H.; Heubl, G. Molecular Phylogeny of Menthinae (Lamiaceae, Nepetoideae, Mentheae)—Taxonomy, Biogeography and Conflicts. Mol. Phylogenet. Evol. 2010, 55, 501–523. [Google Scholar] [CrossRef]
- Zhao, F.; Chen, Y.-P.; Salmaki, Y.; Drew, B.T.; Wilson, T.C.; Scheen, A.-C.; Celep, F.; Bräuchler, C.; Bendiksby, M.; Wang, Q.; et al. An Updated Tribal Classification of Lamiaceae Based on Plastome Phylogenomics. BMC Biol. 2021, 19. [Google Scholar] [CrossRef]
- Briquet, J. Lieferung. Labiatae. IV. Teil, 3. Abteilung a, Bogen 18 bis 20. In Die natürlichen Pflanzenfamilien nebst ihren Gattungen und wichtigeren Arten insbesondere den Nutzpflanzen; Engler, A., Prantl, K., Eds.; Verlag von Wilhelm Engelmann: Leipzig, Germany, 1896. [Google Scholar]
- Tucker, A.O. Genetics and Breeding of the Genus Mentha: A Model for Other Polyploid Species with Secondary Constituents. J. Med. Act. Plants 2012, 1, 19–29. [Google Scholar] [CrossRef]
- Bräuchler, C. Delimitation and Revision of the Genus Thymbra (Lamiaceae). Phytotaxa 2018, 369, 15–27. [Google Scholar] [CrossRef]
- Chen, X.H.; Zhang, F.Y.; Yao, L. Chloroplast DNA Molecular Characterization and Leaf Volatiles Analysis of Mint (Mentha; Lamiaceae) Populations in China. Ind. Crops Prod. 2012, 37, 270–274. [Google Scholar] [CrossRef]
- Jabeen, A.; Guo, B.; Abbasi, B.H.; Shinwari, Z.K.; Mahmood, T. Phylogenetics of Selected Mentha Species on the Basis of Rps8, Rps11 and Rps14 Chloroplast Genes. J. Med. Plants Res. 2012, 6, 30–36. [Google Scholar] [CrossRef]
- Schanzer, I.A.; Semenova, M.V.; Shelepova, O.V.; Voronkova, T.V. Genetic Diversity and Natural Hybridization in Populations of Clonal Plants of Mentha Aquatica L. (Lamiaceae). Wulfenia 2012, 19, 131–139. [Google Scholar]
- Ahmed, S.M. Molecular Identification of Lavendula Dentata L., Mentha Longifolia (L.) Huds. and Mentha × Piperita L. by DNA Barcodes. Bangladesh J. Plant Taxon. 2018, 25, 149–157. [Google Scholar] [CrossRef]
- Ahmed, S.M.; Alamer, K.H. Discriminating Lamiaceae Species from Saudi Arabia Using Allozyme and Specific DNA Markers. Pak. J. Bot. 2018, 50, 969–975. [Google Scholar]
- Jedrzejczyk, I.; Rewers, M. Genome Size and ISSR Markers for Mentha L. (Lamiaceae) Genetic Diversity Assessment and Species Identification. Ind. Crops Prod. 2018, 120, 171–179. [Google Scholar] [CrossRef]
- Abasi, F.; Ahmad, I.; Khan, S.U.; Ahmad, K.S.; Ulfat, A.; Khurshid, R. Estimation of Genetic Diversity in Genus Mentha Collected From Azad Jammu And Kashmir, Pakistan. bioRxiv 2019. [Google Scholar] [CrossRef]
- Soilhi, Z.; Trindade, H.; Vicente, S.; Gouiaa, S.; Khoudi, H.; Mekki, M. Assessment of the Genetic Diversity and Relationships of a Collection of Mentha Spp. in Tunisia Using Morphological Traits and ISSR Markers. J. Hortic. Sci. Biotechnol. 2020, 95, 483–495. [Google Scholar] [CrossRef]
- Tucker, A.O.; Naczi, R.F.C. Mentha: An Overview of Its Classification and Relationships. In Mint. The Genus Mentha; Lawrence, B.M., Ed.; CRC Press: Boca Raton, FL, USA, 2006; pp. 1–40. [Google Scholar]
- Borissova, A.G. Genus 1301. Mentha L. In Flora of the U.S.S.R.; Shishkin, B.K., Landau, N., Eds.; Keterpress Enterprises: Jerusalem, Israel, 1977; pp. 427–450. ISBN 0-7065-1573-0. [Google Scholar]
- Jamzad, Z. Eremostachys Lanata and Mentha Mozaffarianii, Two New Labiatae from Iran. Iran. J. Bot. 1987, 3, 111–116. [Google Scholar]
- Jančić, R. Mentha Serbica R.Jančić, Sp. Nov- a New Species of the Genus Mentha L. (Lamiaceae). Glas. Prir. Muzeja U Beogr. Ser. B 1989, 43/44, 27–33. [Google Scholar]
- Conn, B.J.; Duval, D.J. Mentha Atrolilacina (Lamiaceae), a New Species from South Australia. Telopea 2010, 12, 521–524. [Google Scholar] [CrossRef]
- Gandoger, M. Menthae Novae, Imprimis Europaeae. Bull. Soc. Imp. Nat. Moscou 1883, 58, 14–102. [Google Scholar]
- Topitz, A. Ungarische Minzen. Ung. Bot. Bl. 1916, 15, 125–168. [Google Scholar]
- Duda, T.F.; Bolin, M.B.; Meyer, C.P.; Kohn, A.J. Hidden Diversity in a Hyperdiverse Gastropod Genus: Discovery of Previously Unidentified Members of a Conus Species Complex. Mol. Phylogenet. Evol. 2008, 49, 867–876. [Google Scholar] [CrossRef]
- Harley, R.M.; Brighton, C.A. Chromosome Numbers in the Genus Mentha L. Bot. J. Linn. Soc. 1977, 74, 71–96. [Google Scholar] [CrossRef]
- Schürhoff, P.N. Zytologische Und Genetische Untersuchungen an Mentha Und Ihre Bedeutung Für Die Pharmakognosie. Arch. Pharm. 1929, 267, 515–526. [Google Scholar] [CrossRef]
- Lietz, J. Beiträge Zur Zytologie Der Gattung Mentha; Friedrich-Wilhelms-Universität: Berlin, Germany, 1930. [Google Scholar]
- Ruttle, M.L. Cytological Embryological Studies Genus Mentha. Gartenbauwissenschaft 1931, 4, 428–468. [Google Scholar]
- Morton, J.K. The Chromosome Numbers of the British Menthae. Watsonia 1956, 3, 244–252. [Google Scholar]
- Harley, R.M. Taxonomic Studies in the Genus Mentha. Ph.D. Thesis, Oxford University, Oxford, UK, 1963. [Google Scholar]
- Harley, R.M. The Spicate Mints. Proc. Bot. Soc. Br. Isles 1967, 6, 369–372. [Google Scholar]
- Harley, R.M. Notes on the Genus Mentha (Labiatae). Bot. J. Linn. Soc. 1972, 65, 250–253. [Google Scholar]
- Harley, R.M. Mentha. In Flora Europaea, 3; Tutin, T.G., Heywood, V.H., Burges, N.A., Moore, D.M., Valentine, D.H., Walters, S.M., Webb, D.A., Eds.; Cambridge Universty Press: Cambridge, UK, 1972; pp. 183–186. [Google Scholar]
- Harley, R.M. Mentha. In Hybridization and the Flora of the British Isles; Stace, C.A., Ed.; Academic Press: London, UK, 1975; pp. 383–390. [Google Scholar]
- Harley, R.M. Mentha L. In Hybrid Flora of the British Isles; Stace, C.A., Preston, C.D., Pearman, D.A., Eds.; Botanical Society of Britain & Ireland: London, UK, 2016; pp. 255–262. [Google Scholar]
- Lebeau, J. Mentha L. (Menthe. Munt. Minze). In Nouvelle flore de la Belgique, du Grand-Duché de Luxembourg, du nord de la France et des Régions Voisines (Ptéridophytes et Spermatophytes); De Langhe, J.-E., Delvosalle, L., Duvigneaud, J., Lambinon, J., Vanden Berghen, C., Eds.; Édition du Patrimoine du Jardin Botanique National de Belgique: Meise, Belgium, 1973; pp. 411–414. [Google Scholar]
- Lebeau, J. Appellations Nouvelles de Mentha (Labiatae) Hybrides, et Proposition Du Rang Nouveau de Subhybride. Bull. Jard. Bot. Natl. Belg. Bull. Van Natl. Plantentuin Van Belg. 1974, 44, 249–257. [Google Scholar] [CrossRef]
- Lebeau, J. Nouvelles Mises Au Point Dans Le Genre Mentha. Nat. Mosana 1974, 27, 109–141. [Google Scholar]
- Gobert, V.; Moja, S.; Colson, M.; Taberlet, P. Hybridization in the Section Mentha (Lamiaceae) Inferred from AFLP Markers. Am. J. Bot. 2002, 89, 2017–2023. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.T.; Yu, X.; Liu, Y.; Liang, C.Y.; Li, W.L. Analysis of Genetic Variability and Relationships among Mentha L. Using the Limonene Synthase Gene, LS. Gene 2013, 524, 246–252. [Google Scholar] [CrossRef]
- Simon, C. An Evolving View of Phylogenetic Support. Syst. Biol. 2020, syaa068. [Google Scholar] [CrossRef]
- Khan, N.; Singh, S.; Dhawan, S.S. Development of Species Specific SCoT Markers and Analysis of Genetic Diversity among Mentha Genotypes. Int. J. Innov. Sci. Eng. Technol. 2017, 4, 145–156. [Google Scholar]
- Celenk, S.; Tarimcilar, G.; Bicakci, A.; Kaynak, G.; Malyer, H. A Palynological Study of the Genus Mentha L. (Lamiaceae). Bot. J. Linn. Soc. 2008, 157, 141–154. [Google Scholar] [CrossRef] [Green Version]
- Panjeshahin, Z.; Sharifi-Sirchi, G.-R.; Samsampour, D. Genetic and Morphological Diversity of Wild Mint “Mentha Longifolia (L.) Hudson Subsp. Noeana (Briq.) Briq.” in South and Southeastern Iran. J. Med. Plants Prod. 2018, 1, 105–115. [Google Scholar] [CrossRef]
- Ahmad, I.; Khan, S.U.; Khan, A.; Amjad, M.S.; Abbasi, F. Reassessment of Mentha Species from Kunhar River Catchment Using Morphological and Molecular Markers. ANADOLU 2018, 28, 2018. [Google Scholar]
- El-Kashoury, E.S.A.; El-Askary, H.I.; Kandil, Z.A.; Salem, M.A. Botanical and Genetic Characterization of Mentha Suaveolens Ehrh. Cultivated in Egypt. Pharmacogn. J. 2013, 5, 228–237. [Google Scholar] [CrossRef]
- Bokić, B.S. Morfološka i Fitohemijska Karakterizacija Predstavnika Sekcija Pulegium (Mill.) Lam. & DC. 1805 i Mentha (Mentha L., Lamiaceae) Sa Balkanskog Poluostrva i Južnog Dela Panonske Nizije. Ph.D. Thesis, Universitei U Novom Sadu, Prirodno-matematički Fakultet Departman za Biologiju I Ekologiju, Novi Sad, Serbia, 2021. [Google Scholar]
- Youssef, A.I.; Deeb, G.H.; Bitar, G. A Study of Genetic Variations of Genetic Types of Mentha Aquatica Distributed in Coastal Region Using PCR-RAPD Technique. Tishreen Univ. J. Res. Sci. Stud. Biol. Sci. Ser. 2011, 33, 183–198. [Google Scholar]
- Rodrigues, L.; Póvoa, O.; van den Berg, C.; Figueiredo, A.C.; Moldão, M.; Monteiro, A. Genetic Diversity in Mentha Cervina Based on Morphological Traits, Essential Oils Profile and ISSRs Markers. Biochem. Syst. Ecol. 2013, 51, 50–59. [Google Scholar] [CrossRef]
- Gobert, V.; Moja, S.; Taberlet, P.; Wink, M. Heterogeneity of Three Molecular Data Partition Phylogenies of Mints Related to M. x Piperita (Mentha; Lamiaceae). Plant Biol. 2006, 8, 470–485. [Google Scholar] [CrossRef] [PubMed]
- Kalfagianni, A. DNA Barcoding of Native Plants; Use of «DNA Barcodes» ITS, MatK and TrnH-PsbA to Identify Native Plants of Genus Mentha in Chios (Greece) and Erythrae Peninsula (Turkey). Postgraduate studies Program Conservation of Biodiversity and Sustainable Exploitation of Native Plants (BNP). Ph.D. Thesis, Aristotle University of Thessaloniki School of Biology, Thessaloniki, Greece, 2012. [Google Scholar]
- Tucker, A.O.; Harley, R.M.; Fairbrothers, D.E. The Linnaean Types of Mentha (Lamiaceae). TAXON 1980, 29, 233–255. [Google Scholar] [CrossRef]
- Qin, Y.; Li, M.; Cao, Y.; Gao, Y.; Zhang, W. Molecular Thresholds of ITS2 and Their Implications for Molecular Evolution and Species Identification in Seed Plants. Sci. Rep. 2017, 7, 17316. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for Primary Species Delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Hedge, I.C. A Global Survey of the Biogeography of the Labiatae. In Advances in Labiatae Science; Harley, R.M., Reynolds, T., Eds.; Royal Botanic Gardens, Kew: Kew, UK, 1992; pp. 7–17. [Google Scholar]
- Schemske, D.W.; Bradshaw, H.D. Pollinator Preference and the Evolution of Floral Traits in Monkeyflowers (Mimulus). Proc. Natl. Acad. Sci. USA 1999, 96, 11910–11915. [Google Scholar] [CrossRef] [Green Version]
- Cozzolino, S.; Scopece, G. Specificity in Pollination and Consequences for Postmating Reproductive Isolation in Deceptive Mediterranean Orchids. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2008, 363, 3037–3046. [Google Scholar] [CrossRef] [Green Version]
- Lowry, D.B.; Modliszewski, J.L.; Wright, K.M.; Wu, C.A.; Willis, J.H. The Strength and Genetic Basis of Reproductive Isolating Barriers in Flowering Plants. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2008, 363, 3009–3021. [Google Scholar] [CrossRef] [Green Version]
- Farré-Armengol, G.; Filella, I.; Llusia, J.; Peñuelas, J. Relationships among Floral VOC Emissions, Floral Rewards and Visits of Pollinators in Five Plant Species of a Mediterranean Shrubland. Plant Ecol. Evol. 2015, 148, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Rezende, L.; Suzigan, J.; Amorim, F.W.; Moraes, A.P. Can Plant Hybridization and Polyploidy Lead to Pollinator Shift? Acta Bot. Bras. 2020, 34, 229–242. [Google Scholar] [CrossRef]
- Warren, J. Is Wind-Mediated Passive Leaf Movement an Effective Form of Herbivore Defence? Plant Ecol. Evol. 2015, 148, 52–56. [Google Scholar] [CrossRef]
- Schwarzbach, A.E.; Donovan, L.A.; Rieseberg, L.H. Transgressive Character Expression in a Hybrid Sunflower Species. Am. J. Bot. 2001, 88, 270–277. [Google Scholar] [CrossRef]
- Rosenthal, D.M.; Schwarzbach, A.E.; Donovan, L.A.; Raymond, O.; Rieseberg, L.H. Phenotypic Differentiation between Three Ancient Hybrid Taxa and Their Parental Species. Int. J. Plant Sci. 2002, 163, 387–398. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, B.M. A Study of the Monoterpene Interrelationships in the Genus Mentha with Special Reference to the Origin of Pulegone and Menthofuran. Ph.D. Thesis, Faculty of Science and Engineering, University of Groningen, Groningen, The Netherlands, 1978. [Google Scholar]
- Kokkini, S. Chemical Races Within the Genus Mentha L. In Essential Oils and Waxes. Modern Methods of Plant Analysis; Linskens, H.F., Jackson, J.F., Eds.; Springer: Berlin/Heidelberg, Germany, 1991; Volume 12, pp. 63–78. [Google Scholar]
- Voirin, B.; Bayet, C.; Faure, O.; Jullien, F. Free Flavonoid Aglycones as Markers of Parentage in Mentha Aquatica, M. Citrata, M. Spicata and M. x Piperita. Phytochemistry 1999, 50, 1189–1193. [Google Scholar] [CrossRef]
- Shasany, A.K.; Shukla, A.K.; Gupta, S.; Rajkumar, S.; Khanuja, S.P.S. AFLP Analysis for Genetic Relationships among Mentha Species. Plant Genet. Resour. Newsl. IPGRIFAO 2005, 144, 14–19. [Google Scholar]
- Chengyuan, L.; Wei-lin, L.; Yong, Z.; BoHeng, X.; Xiao-yue, W. Study on Morphological Diversity of Mentha Haplocalyx Briq. Med. Plant 2011, 2, 1–3. [Google Scholar]
- Barzin, G.; Mazooji, A.; Salimpour, F. Chemotaxonomy of Four Varieties of Mentha Longifolia L. Using Essential Oil Composition Markers. J. Biodivers. Environ. Sci. JBES 2014, 5, 172–176. [Google Scholar]
- Hoque, A.; Rumman, R.; Akter, K.T.; Choudhury, T.I.; Faisal, W. Morphological Characterization and Evaluation of Twenty Two Mint Germplasm. Res. Rev. J. Crop Sci. Technol. 2014, 4, 4–12. [Google Scholar] [CrossRef]
- Hanafy, D.M.; Prenzler, P.D.; Hill, R.A.; Burrows, G.E. Leaf Micromorphology of 19 Mentha Taxa. Aust. J. Bot. 2019, 67, 463. [Google Scholar] [CrossRef]
- Venkatesha, K.T.; Singh, V.R.; Padalia, R.; Verma, R.; Upadyay, R.; Kumar, R.; Chauhan, A. Genetic Variability, D2 Analysis and Characters Association among Quantitative and Qualitative Traits of Spearmint (Mentha Spicata L.). Trends Phytochem. Res. 2019, 3, 101–108. [Google Scholar]
- Bokić, B.S.; Rat, M.M.; Kladar, N.V.; Anačkov, G.T.; Božin, B.N. Chemical Diversity of Volatile Compounds of Mints from Southern Part of Pannonian Plain and Balkan Peninsula—New Data. Chem. Biodivers. 2020, 17. [Google Scholar] [CrossRef] [PubMed]
- Mamadalieva, N.Z.; Hussain, H.; Xiao, J. Recent Advances in Genus Mentha: Phytochemistry, Antimicrobial Effects, and Food Applications. Food Front. 2020, 1–24. [Google Scholar] [CrossRef]
- Mack, C. Morphologische, Mikroskopische Und Analytisch-Biochemische Untersuchungen Zur Differenzierung Verschiedener Arten Und Sorten Der Gattung Mentha/Claudia Mack; Institut für Pflanzenwissenschaften, Karl-Franzens-Universität: Graz, Austria, 2010. [Google Scholar]
- Srivastava, A.; Shasany, A.K.; Kumar, S.; Khanuja, S.P.S.; Bahl, J.R.; Sharma, S. Plant Genetic Resources Newsletter—Genetic Diversity Assessment of Mentha Spicata L. Germplasm through RAPD Analysis. Plant Genet. Resour. Newsl. 2002, 130, 1–5. [Google Scholar]
- Bunsawatt, J.; Elliott, N.E.; Hertweck, K.L.; Sproles, E.; Alice, L.A. Phylogenetics of Mentha (Lamiaceae): Evidence from Chloroplast DNA Sequences. Syst. Bot. 2004, 29, 959–964. [Google Scholar] [CrossRef]
- Vining, K.J.; Zhang, Q.; Tucker, A.O.; Smith, C.; Davis, T.M. Mentha Longifolia (L.) L.: A Model Species for Mint Genetic Research. HortScience 2005, 40, 1225–1229. [Google Scholar] [CrossRef]
- Momeni, S.; Shiran, B.; Razmjoo, K. Genetic Variation in Iranian Mints on the Bases of RAPD Analysis. Pak. J. Biol. Sci. 2006, 9, 1898–1904. [Google Scholar] [CrossRef]
- Al-Rawashdeh, I.M. Molecular Taxonomy Among Mentha Spicata, Mentha Longifolia and Ziziphora Tenuior Populations Using the RAPD Technique. Jordan J. Biol. Sci. 2011, 4, 63–70. [Google Scholar]
- Theodoridis, S.; Stefanaki, A.; Tezcan, M.; Aki, C.; Kokkini, S.; Vlachonasios, K.E. DNA Barcoding in Native Plants of the Labiatae (Lamiaceae) Family from Chios Island (Greece) and the Adjacent Çeşme-Karaburun Peninsula (Turkey). Mol. Ecol. Resour. 2012, 12, 620–633. [Google Scholar] [CrossRef]
- Kumar, B.; Kumar, U.; Yadav, H.K. Identification of EST–SSRs and Molecular Diversity Analysis in Mentha Piperita. Crop J. 2015, 3, 335–342. [Google Scholar] [CrossRef] [Green Version]
- Capuzzo, A.; Maffei, M.E. Molecular Fingerprinting of Peppermint (Mentha Piperita) and Some Mentha Hybrids by Sequencing and RFLP Analysis of the 5S RRNA Non-Transcribed Spacer (NTS) Region. Plant Biosyst. 2016, 150, 236–243. [Google Scholar] [CrossRef]
- Sabboura, D.; Yacoub, R.; Lawand, S. Assessment of Genetic Relationships among Mint Species. Int. J. ChemTech Res. 2016, 9, 462–468. [Google Scholar]
- Biswas, K.; Rohira, H.; Biswas, R. Molecular Studies of In-Vitro Propagated Three Mentha Species on “KFA+”Media. Int. J. Appl. 2017, 12, 211–217. [Google Scholar]
- Vining, K.J.; Johnson, S.R.; Ahkami, A.; Lange, I.; Parrish, A.N.; Trapp, S.C.; Croteau, R.B.; Straub, S.C.K.; Pandelova, I.; Lange, B.M. Draft Genome Sequence of Mentha Longifolia and Development of Resources for Mint Cultivar Improvement. Mol. Plant 2017, 10, 323–339. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, L.; Hua, Y.; Zhao, M.; Li, S.; Sun, H.; Lv, Y.; Wang, Y. The Complete Chloroplast Genome of Mentha Spicata, an Endangered Species Native to South Europe. Mitochondrial DNA Part B Resour. 2017, 2, 907–909. [Google Scholar] [CrossRef] [Green Version]
- Vining, K.J.; Pandelova, I.; Hummer, K.E.; Bassil, N.V.; Contreras, R.; Neill, K.; Chen, H.; Parrish, A.N.; Lange, B.M. Genetic Diversity Survey of Mentha Aquatica L. and Mentha Suaveolens Ehrh., Mint Crop Ancestors. Genet. Resour. Crop Evol. 2019, 66, 825–845. [Google Scholar] [CrossRef]
- Moshrefi-Araghi, A.; Nemati, H.; Azizi, M.; Moshtaghi, N.; Shoor, M. Study of Genetic Diversity of Some Genotypes of Iranian Wild Mint (Mentha Longifolia L.) Using ISSR Marker and Its Correlation with Dry Yield and Essential Oil Content. Agric. Biotechnol. J. 2020, 12, 117–140. [Google Scholar] [CrossRef]
- Briquet, J. Fragmenta Monographiae Labiaterum—Fascicule Ier. Révision Systématique Des Groupes Spécifiques et Subspécifiques Dans Le Sous-Genre Menthastrum Du Genre Mentha. Bull. Trav. Société Bot. Genève 1889, 5, 20–122. [Google Scholar]
- Bunsawatt, J. Mentha (Lamiaceae) Phylogenetic Analysis Using Chloroplast TRNL-TRNF and Nuclear Ribosomal DNA ITS Sequences. Master’s Thesis, Western Kentucky University, Bowling Green, KY, USA, 2002. [Google Scholar]
- Capuzzo, A.; Maffei, M.E. Molecular Fingerprinting of Some Mentha Species by Sequencing and RFLP Analysis of the 5S-RRNA Non-Transcribed Spacer Region. Plant Biosyst. 2014, 148, 683–690. [Google Scholar] [CrossRef] [Green Version]
- Tucker, A.; Chambers, H. Mentha Canadensis L. (Lamiaceae): A Relict Amphidiploid from the Lower Tertiary. Taxon 2002, 51, 703. [Google Scholar] [CrossRef]
- Thakur, V.V.; Tiwari, S.; Tripathi, N.; Sapre, S. DNA Barcoding and Phylogenetic Analyses of Mentha Species Using RbcL Sequences. Ann. Phytomed. 2016, 5, 59–62. [Google Scholar]
- Choupani, A.; Shojaeiyan, A.; Maleki, M. Genetic Relationships of Iranian Endemic Mint Species, Mentha Mozaffariani Jamzad and Some Other Mint Species Revealed by ISSR Markers. Biotechnologia 2019, 100, 19–28. [Google Scholar] [CrossRef]
- Yaghini, H.; Sabzalian, M.R.; Rahimmalek, M.; Garavand, T.; Maleki, A.; Mirlohi, A. Seed Set in Inter Specific Crosses of Male Sterile Mentha Spicata with Mentha Longifolia. Euphytica 2020, 216. [Google Scholar] [CrossRef]
- Rahimmalek, M. Study of Genetic Relationships of Some Mint Species Using R-ISSR Markers. Argicult. Biotechnol. 2011, 10, 11–17. [Google Scholar]
- Zinodini, A.; Farshad Far, M.; Safari, H.; Moradi, F.; Shirvani, H. Study of Genetic Relationships of Some Mint Species Using ISSR Markers. Crop Biotechnol. 2014, 3, 11–21. [Google Scholar]
- Ibrahim, H.M.M. Assessment of Genetic Diversity and Relationships of Five Mentha Species Using RAPD Marker. Curr. Sci. Int. 2017, 6, 271–277. [Google Scholar]
- Smolik, M.; Jadczak, D.; Rzepka-Plevneš, D.; Sękowska, A. Morphological and Genetic Variability of Chosen Mentha Species. Herba Pol. J. 2007, 53, 90–97. [Google Scholar]
- De Matos Rodrigues, L.S. Phytochemical and Genetic Diversity in Mentha Species: Assessment, Valorization and Conservation. Ph.D. Thesis, Universidade Tecnica de Lisboa, Lisbon, Portugal, 2012. [Google Scholar]
- Li, B.; Cantino, P.D.; Olmstead, R.G.; Bramley, G.L.C.; Xiang, C.-L.; Ma, Z.-H.; Tan, Y.-H.; Zhang, D.-X. A Large-Scale Chloroplast Phylogeny of the Lamiaceae Sheds New Light on Its Subfamilial Classification. Sci. Rep. 2016, 6, 34343. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Qi, Z.-C.; Liu, L.-X.; Ohi-Toma, T.; Lee, J.; Hsieh, T.-H.; Fu, C.-X.; Cameron, K.M.; Qiu, Y.-X. Molecular Phylogenetics and Biogeography of the Mint Tribe Elsholtzieae (Nepetoideae, Lamiaceae), with an Emphasis on Its Diversification in East Asia. Sci. Rep. 2017, 7, 2057. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Li, M.; Liao, B.; Shi, X.; Xu, Y. DNA Barcoding Analysis and Phylogenetic Relation of Mangroves in Guangdong Province, China. Forests 2019, 10, 56. [Google Scholar] [CrossRef] [Green Version]
- De Mattia, F.; Bruni, I.; Galimberti, A.; Cattaneo, F.; Casiraghi, M.; Labra, M. A Comparative Study of Different DNA Barcoding Markers for the Identification of Some Members of Lamiacaea. Food Res. Int. 2011, 44, 693–702. [Google Scholar] [CrossRef]
- Vilas, R.; Criscione, C.D.; Blouin, M.S. A Comparison between Mitochondrial DNA and the Ribosomal Internal Transcribed Regions in Prospecting for Cryptic Species of Platyhelminth Parasites. Parasitology 2005, 131, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Drew, B.T.; Sytsma, K.J. Phylogenetics, Biogeography, and Staminal Evolution in the Tribe Mentheae (Lamiaceae). Am. J. Bot. 2012, 99, 933–953. [Google Scholar] [CrossRef] [Green Version]
- Draisma, S.G.A.; Eurlings, M.C.M.; Lim, P.-E. High Intra-Individual Sequence Variation in the Nuclear RDNA LSU-5S Intergenic Spacer in the Sargassaceae (Fucales, Phaeophyceae). J. Appl. Phycol. 2012, 24, 1373–1379. [Google Scholar] [CrossRef]
- Babineau, M.; Gagnon, E.; Bruneau, A. Phylogenetic Utility of 19 Low Copy Nuclear Genes in Closely Related Genera and Species of Caesalpinioid Legumes. South Afr. J. Bot. 2013, 89, 94–105. [Google Scholar] [CrossRef]
- Chen, J.; Zeng, Y.-F.; Liao, W.-J.; Yan, P.-C.; Zhang, J.-G. A Novel Set of Single-Copy Nuclear Gene Markers in White Oak and Implications for Species Delimitation. Tree Genet. Genomes 2017, 13, 50. [Google Scholar] [CrossRef]
- Yasuda, N.; Taquet, C.; Nagai, S.; Fortes, M.; Fan, T.-Y.; Harii, S.; Yoshida, T.; Sito, Y.; Nadaoka, K. Genetic Diversity, Paraphyly and Incomplete Lineage Sorting of MtDNA, ITS2 and Microsatellite Flanking Region in Closely Related Heliopora Species (Octocorallia). Mol. Phylogenet. Evol. 2015, 93, 161–171. [Google Scholar] [CrossRef]
- Waugh, J. DNA Barcoding in Animal Species: Progress, Potential and Pitfalls. BioEssays News Rev. Mol. Cell. Dev. Biol. 2007, 29, 188–197. [Google Scholar] [CrossRef]
- Chase, M.W.; Salamin, N.; Wilkinson, M.; Dunwell, J.M.; Kesanakurthi, R.P.; Haidar, N.; Savolainen, V. Land Plants and DNA Barcodes: Short-Term and Long-Term Goals. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2005, 360, 1889–1895. [Google Scholar] [CrossRef] [Green Version]
- Saini, A.; Reddy, S.K.; Jawali, N. Intra-Individual and Intra-Species Heterogeneity in Nuclear RDNA ITS Region of Vigna Species from Subgenus Ceratotropis. Genet. Res. 2008, 90, 299–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiragaki, K.; Yokoi, S.; Tezuka, T. Phylogenetic Analysis and Molecular Diversity of Capsicum Based on RDNA-ITS Region. Horticulturae 2020, 6, 87. [Google Scholar] [CrossRef]
- Bower, J.E.; Cooper, R.D.; Beebe, N.W. Internal Repetition and Intraindividual Variation in the RDNA ITS1 of the Anopheles Punctulatus Group (Diptera: Culicidae): Multiple Units and Rates of Turnover. J. Mol. Evol. 2009, 68, 66–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFadden, C.S.; Sánchez, J.A.; France, S.C. Molecular Phylogenetic Insights into the Evolution of Octocorallia: A Review. Integr. Comp. Biol. 2010, 50, 389–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuan, Y.; Wu, Y.; Li, P.; Liu, R.; Luo, Y.; Yuan, J.; Xiang, Z.; He, N. Molecular Phylogeny of Mulberries Reconstructed from ITS and Two CpDNA Sequences. PeerJ 2019, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, B.; Zeng, X.-M.; Gao, X.-F.; Jin, D.-P.; Zhang, L.-B. ITS Non-Concerted Evolution and Rampant Hybridization in the Legume Genus Lespedeza (Fabaceae). Sci. Rep. 2017, 7, 40057. [Google Scholar] [CrossRef] [Green Version]
- Wan, D.; Sun, Y.; Zhang, X.; Bai, X.; Wang, J.; Wang, A.; Milne, R. Multiple ITS Copies Reveal Extensive Hybridization within Rheum (Polygonaceae), a Genus That Has Undergone Rapid Radiation. PLoS ONE 2014, 9, e89769. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, M.C. Incomplete Concerted Evolution in the Non-Hybrid Diploid Clematis Fremontii S. Watson (Ranunculaceae). Master’s Thesis, University of Tennessee, Chattanooga, TN, USA, 2009. [Google Scholar]
- King, M.G.; Roalson, E.H. Exploring Evolutionary Dynamics of NrDNA in Carex Subgenus Vignea (Cyperaceae). Syst. Bot. 2008, 33, 514–524. [Google Scholar] [CrossRef]
- Meimberg, H.; Abele, T.; Bräuchler, C.; McKay, J.K.; Pérez de Paz, P.L.; Heubl, G. Molecular Evidence for Adaptive Radiation of Micromeria Benth. (Lamiaceae) on the Canary Islands as Inferred from Chloroplast and Nuclear DNA Sequences and ISSR Fingerprint Data. Mol. Phylogenet. Evol. 2006, 41, 566–578. [Google Scholar] [CrossRef]
- Wang, X.; Chen, X.; Yang, P.; Wang, L.; Han, J. Barcoding the Dendrobium (Orchidaceae) Species and Analysis of the Intragenomic Variation Based on the Internal Transcribed Spacer 2. BioMed Res. Int. 2017, 2017, 2734960. [Google Scholar] [CrossRef] [Green Version]
- Nieto Feliner, G.; Rosselló, J.A. Better the Devil You Know? Guidelines for Insightful Utilization of NrDNA ITS in Species-Level Evolutionary Studies in Plants. Mol. Phylogenet. Evol. 2007, 44, 911–919. [Google Scholar] [CrossRef] [Green Version]
- Estensmo, E.L.F.; Maurice, S.; Morgado, L.; Martin-Sanchez, P.M.; Skrede, I.; Kauserud, H. The Influence of Intraspecific Sequence Variation during DNA Metabarcoding: A Case Study of Eleven Fungal Species. Mol. Ecol. Resour. 2020. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B. Nuclear Ribosomal Internal Transcribed Spacer (ITS) Region as a Universal DNA Barcode Marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [Green Version]
- Rojas, A.; Dvir, E.; Farkas, R.; Sarma, K.; Borthakur, S.; Jabbar, A.; Markovics, A.; Otranto, D.; Baneth, G. Phylogenetic Analysis of Spirocerca Lupi and Spirocerca Vulpis Reveal High Genetic Diversity and Intra-Individual Variation. Parasit. Vectors 2018, 11, 639. [Google Scholar] [CrossRef] [Green Version]
- Alquezar, D.E.; Hemmerter, S.; Cooper, R.D.; Beebe, N.W. Incomplete Concerted Evolution and Reproductive Isolation at the RDNA Locus Uncovers Nine Cryptic Species within Anopheles Longirostris from Papua New Guinea. BMC Evol. Biol. 2010, 10, 392. [Google Scholar] [CrossRef] [Green Version]
- Fujisawa, T.; Barraclough, T.G. Delimiting Species Using Single-Locus Data and the Generalized Mixed Yule Coalescent Approach: A Revised Method and Evaluation on Simulated Data Sets. Syst. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef] [Green Version]
- Flot, J.-F. CHAMPURU 1.0: A Computer Software for Unraveling Mixtures of Two DNA Sequences of Unequal Lengths. Mol. Ecol. Notes 2007, 7, 974–977. [Google Scholar] [CrossRef]
- Popp, M. Disentangling the Reticulate History of Polyploids in Silene (Caryophyllaceae). Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology n°924. Ph.D. Thesis, Uppsala University, Uppsala, Sweden, 2004. [Google Scholar]
- Wu, C.I. The Genic View of the Process of Speciation. J. Evol. Biol. 2001, 14, 851–865. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. Ancient WGD Events as Drivers of Key Innovations in Angiosperms. Curr. Opin. Plant Biol. 2016, 30, 159–165. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wang, Y.; Lei, F.; Liu, Y.; Wang, H.; Chen, J. Incomplete Lineage Sorting and Introgression in the Diversification of Chinese Spot-Billed Ducks and Mallards. Curr. Zool. 2019, 65, 589–597. [Google Scholar] [CrossRef] [Green Version]
- Šarić-Kundalić, B.; Fialová, S.; Dobeš, C.; Ölzant, S.; Tekel’ová, D.; Grančai, D.; Reznicek, G.; Saukel, J. Multivariate Numerical Taxonomy of Mentha Species, Hybrids, Varieties and Cultivars. Sci. Pharm. 2009, 77, 851–876. [Google Scholar] [CrossRef]
- Mogosan, C.; Vostinaru, O.; Oprean, R.; Heghes, C.; Filip, L.; Balica, G.; Moldovan, R.I. A Comparative Analysis of the Chemical Composition, Anti-Inflammatory, and Antinociceptive Effects of the Essential Oils from Three Species of Mentha Cultivated in Romania. Molecules 2017, 22, 263. [Google Scholar] [CrossRef] [PubMed]
- Harder, L.D.; Strelin, M.M.; Clocher, I.C.; Kulbaba, M.W.; Aizen, M.A. The Dynamic Mosaic Phenotypes of Flowering Plants. New Phytol. 2019, 224, 1021–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacco, T.; Peracino, V.; Maffei, M.E. Phenotypic Plasticity in Mentha Viridis Lavanduliodora. J. Essent. Oil Res. 1992, 4, 491–496. [Google Scholar] [CrossRef]
- Maffei, M.E.; Scannerini, S. UV-B Effect on Photomorphogenesis and Essential Oil Composition in Peppermint (Mentha Piperita L.). J. Essent. Oil Res. 2000, 12, 523–529. [Google Scholar] [CrossRef]
- Mishra, A.; Jain, P.; Lal, R.K.; Dhawan, S.S. Trichomes and Yield Traits in Mentha Arvensis: Genotype Performance and Stability Evaluation. J. Herbs Spices Med. Plants 2018, 24, 1–14. [Google Scholar] [CrossRef]
- Singh, V.R.; Lal, R.K. Genotype × Environment Interaction, Genetic Variability and Inheritance Pattern in Breeding Lines Including Varieties/Cultivars of Menthol Mint (Mentha Arvensis L.). J. Med. Aromat. Plant Sci. 2020, 42, 145–156. [Google Scholar]
- Bertea, C.M.; Mucciarelli, M. Anatomy, physiology, biosynthesis, molecular biology, tissue culture and biotechnology of mint essential oil production Biosynthesis, Molecular Biology, Tissue Culture, and Biotechnology of Mint Essential Oil Production. In Mint. The Genus Mentha; Lawrence, B.M., Ed.; CRC Press: Boca Raton, FL, USA, 2007; pp. 41–85. ISBN 4213.6.20069. [Google Scholar]
- Stayton, C.T. Are Our Phylomorphospace Plots so Terribly Tangled? An Investigation of Disorder in Data Simulated under Adaptive and Nonadaptive Models. Curr. Zool. 2020, 66, 565–574. [Google Scholar] [CrossRef]
- Phillips, J.D.; Gillis, D.J.; Hanner, R.H. Incomplete Estimates of Genetic Diversity within Species: Implications for DNA Barcoding. Ecol. Evol. 2019, 9, 2996–3010. [Google Scholar] [CrossRef]
- Formia, A.; Broderick, A.C.; Glen, F.; Godley, B.J.; Hays, G.C.; Bruford, M.W. Genetic Composition of the Ascension Island Green Turtle Rookery Based on Mitochondrial DNA: Implications for Sampling and Diversity. Endanger. Species Res. 2007, 3, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Egeland, T.; Salas, A. Estimating Haplotype Frequency and Coverage of Databases. PLoS ONE 2008, 3, e3988. [Google Scholar] [CrossRef] [Green Version]
- Luo, A.; Lan, H.; Ling, C.; Zhang, A.; Shi, L.; Ho, S.Y.W.; Zhu, C. A Simulation Study of Sample Size for DNA Barcoding. Ecol. Evol. 2015, 5, 5869–5879. [Google Scholar] [CrossRef] [Green Version]
- Jullien, F. Mint. In Biotechnology in Agriculture and Forestry 59—Transgenic Crops IV; Nagata, T., Lörz, H., Widholm, J.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 435–466. [Google Scholar]
- Vining, K.J.; Hummer, K.E.; Bassil, N.V.; Lange, B.M.; Khoury, C.K.; Carver, D. Crop Wild Relatives as Germplasm Resource for Cultivar Improvement in Mint (Mentha L.). Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Elliott, N.E. Testing Hypotheses of Hybridization in Mentha Spicata and M. Canadensis Using Molecular Data. Available online: https://digitalcommons.murraystate.edu/postersatthecapitol/2005/WKU/5/ (accessed on 19 April 2021).
- Bräuchler, C.; Meimberg, H.; Heubl, G. Molecular Phylogeny of the Genera Digitalis L. and Isoplexis (Lindley) Loudon (Veronicaceae) Based on ITS- and TrnL-F Sequences. Plant Syst. Evol. 2004, 248, 111–128. [Google Scholar] [CrossRef]
- Stephens, M.; Smith, N.J.; Donnelly, P. A New Statistical Method for Haplotype Reconstruction from Population Data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef] [Green Version]
- Stephens, M.; Scheet, P. Accounting for Decay of Linkage Disequilibrium in Haplotype Inference and Missing-Data Imputation. Am. J. Hum. Genet. 2005, 76, 449–462. [Google Scholar] [CrossRef] [Green Version]
- Flot, J.-F.; Tillier, A.; Samadi, S.; Tillier, S. Phase Determination from Direct Sequencing of Length-Variable DNA Regions. Mol. Ecol. Notes 2006, 6, 627–630. [Google Scholar] [CrossRef]
- Khan, N.; Dhawan, S.S. Role of Molecular Markers in Assessing Genetic Diversity in Mentha: A Review. Sci. J. Genet. Gene Ther. 2016, 2, 022–026. [Google Scholar] [CrossRef] [Green Version]
- Salama, A.M.; Osman, E.A.; El-Tantawy, A.A. Taxonomical Studies on Four Mentha Species Grown in Egypt through Morpho-Anatomical Characters and SCOT Genetic Markers. Plant Arch. 2019, 19, 2273–2286. [Google Scholar]
- Bylesjö, M.; Segura, V.; Soolanayakanahally, R.Y.; Rae, A.M.; Trygg, J.; Gustafsson, P.; Jansson, S.; Street, N.R. LAMINA: A Tool for Rapid Quantification of Leaf Size and Shape Parameters. BMC Plant Biol. 2008, 8. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Kuma, K.I.; Toh, H.; Miyata, T. MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10. Virus Evol. 2018, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spöri, Y.; Flot, J. HaplowebMaker and CoMa: Two Web Tools to Delimit Species Using Haplowebs and Conspecificity Matrices. Methods Ecol. Evol. 2020, 11, 1434–1438. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Flot, J.-F.; Couloux, A.; Tillier, S. Haplowebs as a Graphical Tool for Delimiting Species: A Revival of Doyle’s “Field for Recombination” Approach and Its Application to the Coral Genus Pocillopora in Clipperton. BMC Evol. Biol. 2010, 10, 372. [Google Scholar] [CrossRef]
- Debortoli, N.; Li, X.; Eyres, I.; Fontaneto, D.; Hespeels, B.; Tang, C.Q.; Flot, J.-F.; Van Doninck, K. Genetic Exchange among Bdelloid Rotifers Is More Likely Due to Horizontal Gene Transfer Than to Meiotic Sex. Curr. Biol. 2016, 26, 723–732. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S. Applied Multivariate Techniques; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1996. [Google Scholar]
- Ripley, B.; Venables, B.; Bates, D.M.; Hornik, K.; Gebhardt, A.; Firth, D. Package ‘MASS’. Support Functions and Datasets for Venables and Ripley’s MASS. Available online: https://cran.r-project.org/web/packages/MASS/MASS.pdf/ (accessed on 19 April 2021).
- Witten, D. Package ‘penalizedLDA’. Package Penalized Classification Using Fishers’s Linear Discriminant. Available online: https://cran.r-project.org/web/packages/penalizedLDA/penalizedLDA.pdf (accessed on 19 April 2021).
- Friedman, J.; Hastie, T.; Tibshirani, R.; Narasimhan, B.; Tay, K.; Simon, N.; Qian, J. Package ‘glmnet’. Lasso and Elastic-Net Regularized Generalized Linear Models. Available online: https://cran.r-project.org/web/packages/glmnet/glmnet.pdf (accessed on 19 April 2021).
- Cadima, J.; Orestes Cerdeira, J.; Duarte Silva, P.; Minhoto, M. Package ‘subselect’. Selecting Variable Subsets. Available online: https://cran.r-project.org/web/packages/subselect/subselect.pdf (accessed on 19 April 2021).
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Springer Series in Statistics; Springer: New York, NY, USA, 2002. [Google Scholar]
- Heylen, O. Principal Component Analysis of Nested Binary Data. In Characteristics and Application to Ecology; University Center for Statistics, KU Leuven: Leuven, Belgium, 2007. [Google Scholar]
- Witten, D.M.; Tibshirani, R. Penalized Classification Using Fisher’s Linear Discriminant. J. R. Stat. Soc. Ser. B Stat. Methodol. 2011, 73, 753–772. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H.; Chang, W.; Henry, L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D. Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics—Ggplot2-Package; RStudio: Boston, MA, USA, 2020. [Google Scholar]
- Reynolds, A. Cubemaker; Altius Institute: Seattle, WA, USA, 2015. [Google Scholar]






| Accession Number | Plant Labels (Only 1–2 Boxes Means Informal Label, 3 Boxes Means ‘Officially’ Labeled) | Label Assignment to One of the Parental Species (Criterium Least ‘Pattern Disturbing’ Parent) | Perceived Taxa | Morphotype (Ward’s Group Ranking) | Cluster ITS | Subcluster ITS + HAPL + SCOT | Specimen Unit | ||
|---|---|---|---|---|---|---|---|---|---|
| 286 | M. spicata (subsp. glabrata) or M. × cordifolia | 1 | 4.2 | 4.2.2/4.2.1 | 4.2.2 × 4.2.1 | ||||
| 455 | 1 | 2.5 | 2.5.2 | 2.5.2 | |||||
| 562 (V) | 1 | 2.5 | 2.5.2 | 2.5.2 | |||||
| 627 | 1 | 1.0 | - | 1.0.0 | |||||
| 217 | 1 | 2.5 | 2.5.1 | 2.5.1 | |||||
| 555 (V) | 1 | 4.2 | 4.2.1 | 4.2.1 | |||||
| 499 | 1 | 2.1 | - | 2.1.0 | |||||
| 222 | 1 | 4.1/4.2 | 4.1.1/4.2.1 | 4.1.1 × 4.2.1 | |||||
| 409 | 1 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 570 | M. longifolia incl. M. asiatica, M. longifolia subsp. hymalaiensis, M. longifolia subsp. capensis | 1 | 2.3 | 2.3.2 | 2.3.2 | ||||
| 53 | 2 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 564 (V) | 2 | 2.2 | - | 2.2.0 | |||||
| 567 (V) | 2 | 2.2 | - | 2.2.0 | |||||
| 571 | 2 | 2.3 | 2.3.1 | 2.3.1 | |||||
| 595 | M. longifolia | 2 | 3.0/4.2 | 3.0/4.2.1 | 3.0.0 × 4.2.1 | ||||
| 71 | 2 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 212 | 2 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 130 | 2 | 3.0 | - | 3.0.0 | |||||
| 191 | 2 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 308 | 2 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 312 | 2 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 456 | 2 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 11 | 3 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 77 | M. longifolia or M. spicata subsp. tomentosa/subsp. condensata/M. microphylla | 3 | 2.5 | 2.5.1 | 2.5.1 | ||||
| 250 | 3 | 2.5 | 2.5.2 | 2.5.2 | |||||
| 283 | 3 | 2.5 | 2.5.1 | 2.5.1 | |||||
| 319 | 3 | 2.4 | - | 2.4.0 | |||||
| 311 | 3 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 248 | 3 | 2.5 | 2.5.1 | 2.5.1 | |||||
| 306 | 3 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 450 | 3 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | |||||
| 313 | M. longifolia (probably) or exceptionally M. spicata (?) | 4 | 1.0 | - | 1.0.0 | ||||
| 591 | 4 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 632 | 4 | 1.0 | - | 1.0.0 | |||||
| 274 | M. longifolia or hybrid | 4 | 4.1 | 4.1.1 | 4.1.1 | ||||
| 553 | 4 | 2.5 | 2.5.2 | 2.5.2 | |||||
| 512 | 4 | 2.5 | 2.5.1 | 2.5.1 | |||||
| 545 | 4 | 4.1 | 4.1.2 | 4.1.2 | |||||
| 9 | 4 | 2.3 | 2.3.1 | 2.3.1 | |||||
| 91 | 4 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | |||||
| 203 | 4 | 4.2 | 4.2.2 | 4.2.2 | |||||
| 395 | 5 | 4.1/4.2 | 4.1.1/4.2.1 | 4.1.1 × 4.2.1 | |||||
| 547 | 5 | 2.5 | 2.5.1 | 2.5.1 | |||||
| 565 (V) | 5 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 508 | M. spicata subsp. spicata or M. × villosa | 5 | 2.5 | 2.5.1 | 2.5.1 | ||||
| 391 | 5 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | |||||
| 473 | 5 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | |||||
| 486 | 5 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | |||||
| 530 | 5 | 4.1 | 4.1.3 | 4.1.3 | |||||
| 515 | 5 | 4.2 | 4.2.1 | 4.2.1 | |||||
| 383 | 5 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | |||||
| 526 | 5 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | |||||
| 264 | M. suaveolens (subsp. suaveolens) or its hybrids including M. × villosa (subsp. alopecuroides) and M. × rotundifolia | 6 | 4.1 | 4.1.2 | 4.1.2 | ||||
| 102 | 6 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | |||||
| 144 | 6 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | |||||
| 40 | 6 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 23 | 6 | 4.1 | 4.1.2 | 4.1.2 | |||||
| 185 | 6 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | |||||
| 196 | 6 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | |||||
| 13 | 6 | 4.1 | 4.1.2 | 4.1.2 | |||||
| 170 | 6 | 4.1 | 4.1.2 | 4.1.2 | |||||
| 115 | M. suaveolens subsp. insularis or M. timija | 7 | 4.1/4.2 | 4.1.1/4.2.4 | 4.1.1 × 4.2.4 | ||||
| 87 | 7 | 4.3/3.0 | - | 4.3.0 × 3.0.0 | |||||
| 314 | 7 | 2.5 | 2.5.2 | 2.5.2 | |||||
| 63 | 7 | 4.1 | 4.1.1 | 4.1.1 | |||||
| 200 | M. suaveolens (subsp. suaveolens) or M. × rotundifolia | 7 | 4.1/4.2 | 4.1.4/4.2.3 | 4.1.4 × 4.2.3 | ||||
| 190 | 7 | 4.2 | 4.2.2 | 4.2.2 | |||||
| 76 | M. spicata (subsp. spicata) | 7 | 2.5 | 2.5.1 | 2.5.1 | ||||
| 19 | 7 | 4.1 | 4.1.2 | 4.1.2 | |||||
| 10 | 7 | 4.1 | 4.1.3 | 4.1.3 | |||||
| Specimen Unit | 1.0.0 | 2.1.0 | 2.2.0 | 2.3.1 | 2.3.2 | 2.4.0 | 2.5.1 | 2.5.2 | 3.0.0 | 4.1.1 | 4.1.4 × 4.2.3 | 4.2.1 × | 4.2.2 | 4.2.4 × | 4.3.0 × |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.0.0 | 0.0 | ||||||||||||||
| 2.1.0 | 14.9 | 0.0 | |||||||||||||
| 2.2.0 | 18.8 | 7.3 | 0.0 | ||||||||||||
| 2.3.1 | 12.9 | 3.4 | 2.9 | 0.0 | |||||||||||
| 2.3.2 | 12.4 | 4.1 | 3.0 | 0.6 | 0.0 | ||||||||||
| 2.4.0 | 12.7 | 4.7 | 3.2 | 1.2 | 1.5 | 0.0 | |||||||||
| 2.5.1 | 13.5 | 4.7 | 4.1 | 1.2 | 1.5 | 1.3 | 0.0 | ||||||||
| 2.5.2 | 14.3 | 5.0 | 4.4 | 1.5 | 1.8 | 1.6 | 1.0 | 0.0 | |||||||
| 3.0.0 | 13.3 | 4.7 | 3.4 | 1.2 | 1.5 | 1.5 | 1.5 | 1.8 | 0.0 | ||||||
| 4.1.1 | 12.8 | 4.7 | 3.0 | 1.2 | 1.5 | 1.5 | 1.5 | 1.8 | 0. | 0.0 | |||||
| 4.1.4 × 4.2.3 | 13.3 | 5.3 | 4.1 | 1.8 | 2.1 | 2.1 | 1.9 | 2.4 | 0.6 | 0.7 | 0.0 | ||||
| 4.2.1 × | 13.1 | 5.0 | 3.9 | 1.5 | 1.8 | 1.8 | 1.8 | 2.1 | 0.6 | 1.0 | 0.1 | 0.0 | |||
| 4.2.2 | 18.3 | 6.6 | 4.6 | 2.5 | 3.0 | 2.3 | 3.0 | 3.4 | 1.1 | 1.3 | 0.7 | 0.9 | 0.0 | ||
| 4.2.4 × | 13.5 | 5.2 | 3.9 | 1.6 | 1.9 | 1.9 | 1.9 | 2.2 | 0.4 | 0.3 | 0.4 | 0.6 | 1.3 | 0.0 | |
| 4.3.0 × | 13.5 | 5.5 | 4.4 | 1.9 | 2.2 | 2.2 | 2.2 | 2.5 | 0.7 | 0.9 | 1.0 | 1.2 | 2.0 | 0.9 | 0.0 |
| interspecific | |||||||||||||||
| (in between) | |||||||||||||||
| sister species | |||||||||||||||
| (in between) | |||||||||||||||
| intraspecific | |||||||||||||||
| (no taxonomic consideration) | |||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heylen, O.C.G.; Debortoli, N.; Marescaux, J.; Olofsson, J.K. A Revised Phylogeny of the Mentha spicata Clade Reveals Cryptic Species. Plants 2021, 10, 819. https://doi.org/10.3390/plants10040819
Heylen OCG, Debortoli N, Marescaux J, Olofsson JK. A Revised Phylogeny of the Mentha spicata Clade Reveals Cryptic Species. Plants. 2021; 10(4):819. https://doi.org/10.3390/plants10040819
Chicago/Turabian StyleHeylen, Olivier C. G., Nicolas Debortoli, Jonathan Marescaux, and Jill K. Olofsson. 2021. "A Revised Phylogeny of the Mentha spicata Clade Reveals Cryptic Species" Plants 10, no. 4: 819. https://doi.org/10.3390/plants10040819
APA StyleHeylen, O. C. G., Debortoli, N., Marescaux, J., & Olofsson, J. K. (2021). A Revised Phylogeny of the Mentha spicata Clade Reveals Cryptic Species. Plants, 10(4), 819. https://doi.org/10.3390/plants10040819





