Intercropping of Peanut–Tea Enhances Soil Enzymatic Activity and Soil Nutrient Status at Different Soil Profiles in Subtropical Southern China
Abstract
:1. Introduction
2. Results
2.1. Soil Organic Carbon
2.2. Fluctuations in Soil Total Nutrient Status
2.3. Variability in Soil Nutrient Availability
2.4. Soil Enzymatic Responses in Different Cropping Types
2.5. Correlation between Soil Nutrients and Soil Enzymatic Activity
2.6. Path Coefficients between Soil Nutrients and Soil Enzyme Activity
3. Discussion
4. Materials and Methods
4.1. Study Site
4.2. Experimental Design
4.3. Soil Sampling
4.4. Soil Chemical Analysis
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Factors Code | X1 | X2 | X3 | X4 | X5 | X6 | X7 | |
|---|---|---|---|---|---|---|---|---|
| Y1 | X1 | −0.534 * | −0.518 | −0.048 | 0.219 | −0.021 | −0.198 | −0.315 |
| X2 | 0.796 | 0.821 * | 0.066 | −0.320 | 0.099 | 0.304 | 0.550 | |
| X3 | 0.016 | 0.014 | 0.173 * | 0.014 | 0.022 | 0.112 | 0.054 | |
| X4 | 0.106 | 0.101 | −0.021 | −0.258 * | −0.080 | 0.041 | 0.065 | |
| X5 | 0.000 | −0.001 | −0.002 | −0.004 | −0.012 * | −0.001 | 0.000 | |
| X6 | −0.024 | −0.024 | −0.042 | 0.010 | −0.007 | −0.065 * | −0.027 | |
| X7 | −0.152 | −0.172 | −0.080 | 0.064 | 0.005 | −0.105 | −0.257 * | |
| Y2 | X1 | 0.689 * | 0.668 | 0.062 | −0.282 | 0.028 | 0.255 | 0.407 |
| X2 | 0.343 | 0.354 * | 0.028 | −0.138 | 0.042 | 0.131 | 0.237 | |
| X3 | −0.035 | −0.031 | −0.392 * | −0.031 | −0.051 | −0.255 | −0.122 | |
| X4 | −0.008 | −0.008 | 0.002 | 0.020* | 0.006 | −0.003 | −0.005 | |
| X5 | 0.009 | 0.027 | 0.029 | 0.070 | 0.225 * | 0.025 | −0.005 | |
| X6 | 0.194 | 0.194 | 0.341 | −0.084 | 0.058 | 0.524 * | 0.215 | |
| X7 | −0.115 | −0.131 | −0.060 | 0.049 | 0.004 | −0.080 | −0.195 * | |
| Y3 | X1 | −0.021 * | −0.020 | −0.002 | 0.009 | −0.001 | −0.008 | −0.012 |
| X2 | 0.430 | 0.443 * | 0.035 | −0.173 | 0.053 | 0.164 | 0.297 | |
| X3 | −0.035 | −0.032 | −0.394 * | −0.032 | −0.051 | −0.256 | −0.122 | |
| X4 | 0.064 | 0.061 | −0.013 | −0.157 * | −0.049 | 0.025 | 0.039 | |
| X5 | −0.005 | −0.014 | −0.015 | −0.036 | −0.115 * | −0.013 | 0.002 | |
| X6 | 0.086 | 0.086 | 0.151 | −0.037 | 0.026 | 0.233 * | 0.096 | |
| X7 | 0.162 | 0.184 | 0.085 | −0.069 | −0.006 | 0.113 | 0.275 * | |
| Y4 | X1 | 1.045 * | 1.014 | 0.094 | −0.428 | 0.042 | 0.387 | 0.617 |
| X2 | −0.124 | −0.128 * | −0.010 | 0.050 | −0.015 | −0.047 | −0.086 | |
| X3 | −0.004 | −0.003 | −0.042 * | −0.003 | −0.005 | −0.027 | −0.013 | |
| X4 | −0.033 | −0.031 | 0.006 | 0.080 * | 0.025 | −0.013 | −0.020 | |
| X5 | 0.005 | 0.016 | 0.017 | 0.042 | 0.134 * | 0.015 | −0.003 | |
| X6 | 0.026 | 0.026 | 0.045 | −0.011 | 0.008 | 0.069 * | 0.028 | |
| X7 | 0.028 | 0.032 | 0.015 | −0.012 | −0.001 | 0.020 | 0.048 * | |
| Y5 | X1 | 2.023 * | 1.962 | 0.182 | −0.829 | 0.081 | 0.749 | 1.194 |
| X2 | −2.007 | −0.069 * | −0.166 | 0.807 | −0.248 | −0.766 | −1.386 | |
| X3 | −0.016 | −0.014 | −0.181 * | −0.014 | −0.024 | −0.118 | −0.056 | |
| X4 | −0.027 | −0.025 | 0.005 | 0.065 * | 0.020 | −0.010 | −0.016 | |
| X5 | 0.003 | 0.008 | 0.008 | 0.020 | 0.065 * | 0.007 | −0.001 | |
| X6 | 0.090 | 0.090 | 0.159 | −0.039 | 0.027 | 0.244 * | 0.100 | |
| X7 | 0.100 | 0.113 | 0.052 | −0.042 | −0.003 | 0.069 | 0.169 * | |
| Factors Code | X1 | X2 | X3 | X4 | X5 | X6 | X7 | |
|---|---|---|---|---|---|---|---|---|
| Y1 | X1 | 0.104 * | 0.202 | −0.086 | 0.000 | 0.010 | −0.007 | 0.105 |
| X2 | 0.041 | 0.517 * | −0.050 | −0.003 | 0.004 | −0.033 | 0.039 | |
| X3 | 0.030 | 0.088 | −0.297 * | −0.015 | 0.007 | −0.068 | 0.174 | |
| X4 | 0.000 | 0.036 | −0.098 | −0.046 * | −0.001 | −0.018 | 0.132 | |
| X5 | 0.059 | 0.114 | −0.113 | 0.002 | 0.018 * | −0.072 | 0.102 | |
| X6 | 0.003 | 0.078 | −0.092 | −0.004 | 0.006 | −0.219 * | 0.049 | |
| X7 | 0.033 | 0.062 | −0.157 | −0.018 | 0.006 | −0.033 | 0.329 * | |
| Y2 | X1 | 0.469 * | −0.007 | −0.082 | 0.000 | 0.102 | −0.001 | 0.080 |
| X2 | 0.183 | −0.018 * | −0.048 | 0.015 | 0.039 | −0.006 | 0.030 | |
| X3 | 0.136 | −0.003 | −0.283 * | 0.069 | 0.068 | −0.011 | 0.133 | |
| X4 | 0.000 | −0.001 | −0.093 | 0.208 * | −0.009 | −0.003 | 0.100 | |
| X5 | 0.267 | −0.004 | −0.108 | −0.010 | 0.179 * | −0.012 | 0.078 | |
| X6 | 0.014 | −0.003 | −0.088 | 0.017 | 0.059 | −0.037 * | 0.038 | |
| X7 | 0.150 | −0.002 | −0.150 | 0.083 | 0.055 | −0.006 | 0.250 * | |
| Y3 | X1 | 0.707 * | −0.015 | −0.008 | 0.000 | 0.001 | −0.001 | 0.063 |
| X2 | 0.276 | −0.038 * | −0.005 | 0.011 | 0.000 | −0.003 | 0.024 | |
| X3 | 0.205 | −0.006 | −0.028 * | 0.053 | 0.000 | −0.005 | 0.104 | |
| X4 | 0.000 | −0.003 | −0.009 | 0.161 * | 0.000 | −0.001 | 0.078 | |
| X5 | 0.403 | −0.008 | −0.011 | −0.008 | 0.001 * | −0.006 | 0.061 | |
| X6 | 0.021 | −0.006 | −0.009 | 0.013 | 0.000 | −0.017 * | 0.029 | |
| X7 | 0.226 | −0.005 | −0.015 | 0.064 | 0.000 | −0.003 | 0.196 * | |
| Y4 | X1 | 0.219 * | 0.093 | 0.015 | 0.000 | 0.088 | −0.004 | 0.010 |
| X2 | 0.085 | 0.239 * | 0.009 | −0.010 | 0.034 | −0.020 | 0.004 | |
| X3 | 0.064 | 0.041 | 0.053 * | −0.046 | 0.059 | −0.041 | 0.016 | |
| X4 | 0.000 | 0.017 | 0.017 | −0.139 * | −0.008 | −0.010 | 0.012 | |
| X5 | 0.125 | 0.053 | 0.020 | 0.007 | 0.154 * | −0.043 | 0.010 | |
| X6 | 0.007 | 0.036 | 0.016 | −0.011 | 0.051 | −0.131 * | 0.005 | |
| X7 | 0.070 | 0.029 | 0.028 | −0.056 | 0.048 | −0.020 | 0.031 * | |
| Y5 | X1 | 0.243 * | 0.056 | 0.057 | 0.000 | 0.009 | −0.008 | 0.084 |
| X2 | 0.095 | 0.143 * | 0.033 | −0.003 | 0.003 | −0.041 | 0.032 | |
| X3 | 0.070 | 0.024 | 0.197 * | −0.012 | 0.006 | −0.085 | 0.140 | |
| X4 | 0.000 | 0.010 | 0.065 | −0.037 * | −0.001 | −0.022 | 0.106 | |
| X5 | 0.139 | 0.031 | 0.075 | 0.002 | 0.015 * | −0.090 | 0.082 | |
| X6 | 0.007 | 0.021 | 0.061 | −0.003 | 0.005 | −0.274 * | 0.040 | |
| X7 | 0.078 | 0.017 | 0.104 | −0.015 | 0.005 | −0.041 | 0.264 * | |
| Factors Code | X1 | X2 | X3 | X4 | X5 | X6 | X7 | |
|---|---|---|---|---|---|---|---|---|
| Y1 | X1 | 0.009 * | 0.083 | −0.107 | 0.000 | 0.001 | 0.016 | 0.079 |
| X2 | 0.004 | 0.213 * | −0.063 | −0.006 | 0.000 | 0.080 | 0.037 | |
| X3 | 0.003 | 0.036 | −0.368 * | −0.029 | 0.000 | 0.165 | 0.131 | |
| X4 | 0.000 | 0.015 | −0.121 | −0.089 * | 0.000 | 0.043 | 0.099 | |
| X5 | 0.005 | 0.047 | −0.140 | 0.004 | 0.001 * | 0.176 | 0.077 | |
| X6 | 0.000 | 0.032 | −0.114 | −0.007 | 0.000 | 0.532 * | 0.037 | |
| X7 | 0.003 | 0.026 | −0.195 | −0.036 | 0.000 | 0.080 | 0.248 * | |
| Y2 | X1 | 0.070 * | −0.016 | 0.262 | 0.000 | −0.317 | −0.014 | −0.162 |
| X2 | 0.027 | −0.041 * | 0.154 | 0.008 | −0.123 | −0.072 | −0.076 | |
| X3 | 0.020 | −0.007 | 0.905 * | 0.036 | −0.212 | −0.150 | −0.268 | |
| X4 | 0.000 | −0.003 | 0.299 | 0.110 * | 0.028 | −0.039 | −0.202 | |
| X5 | 0.040 | −0.009 | 0.344 | −0.006 | −0.557 * | −0.159 | −0.157 | |
| X6 | 0.002 | −0.006 | 0.281 | 0.009 | −0.184 | −0.483 * | −0.076 | |
| X7 | 0.022 | −0.005 | 0.480 | 0.044 | −0.173 | −0.072 | −0.506 * | |
| Y3 | X1 | 0.111 * | 0.108 | −0.041 | 0.000 | 0.123 | 0.011 | 0.023 |
| X2 | 0.043 | 0.278 * | −0.024 | 0.022 | 0.048 | 0.055 | 0.011 | |
| X3 | 0.032 | 0.047 | −0.142 * | 0.102 | 0.082 | 0.114 | 0.038 | |
| X4 | 0.000 | 0.019 | −0.047 | 0.309 * | −0.011 | 0.029 | 0.029 | |
| X5 | 0.063 | 0.061 | −0.054 | −0.015 | 0.216 * | 0.121 | 0.022 | |
| X6 | 0.003 | 0.042 | −0.044 | 0.025 | 0.071 | 0.368 * | 0.011 | |
| X7 | 0.036 | 0.033 | −0.075 | 0.124 | 0.067 | 0.055 | 0.072 * | |
| Y4 | X1 | 0.043 * | 0.174 | −0.173 | 0.000 | 0.084 | 0.027 | −0.005 |
| X2 | 0.017 | 0.445 * | −0.102 | 0.017 | 0.033 | 0.133 | −0.002 | |
| X3 | 0.012 | 0.076 | −0.598 * | 0.082 | 0.056 | 0.274 | −0.008 | |
| X4 | 0.000 | 0.031 | −0.197 | 0.249 * | −0.007 | 0.071 | −0.006 | |
| X5 | 0.025 | 0.098 | −0.227 | −0.012 | 0.148 * | 0.292 | −0.005 | |
| X6 | 0.001 | 0.067 | −0.185 | 0.020 | 0.049 | 0.885 * | −0.002 | |
| X7 | 0.014 | 0.053 | −0.317 | 0.100 | 0.046 | 0.133 | −0.015 * | |
| Y5 | X1 | −0.200 * | 0.170 | 0.026 | 0.000 | −0.113 | 0.005 | 0.088 |
| X2 | −0.078 | 0.436 * | 0.015 | −0.009 | −0.044 | 0.026 | 0.041 | |
| X3 | −0.058 | 0.074 | 0.088 * | −0.044 | −0.076 | 0.054 | 0.145 | |
| X4 | 0.000 | 0.031 | 0.029 | −0.132 * | 0.010 | 0.014 | 0.110 | |
| X5 | −0.114 | 0.096 | 0.033 | 0.007 | −0.199 * | 0.057 | 0.085 | |
| X6 | −0.006 | 0.065 | 0.027 | −0.011 | −0.066 | 0.174 * | 0.041 | |
| X7 | −0.064 | 0.052 | 0.047 | −0.053 | −0.062 | 0.026 | 0.274 * | |
References
- Brooker, R.W.; Bennett, A.E.; Cong, W.; Daniell, T.J.; George, T.S.; Hallett, P.D.; Hawes, C.; Iannetta, P.P.M.; Jones, H.G.; Karley, A.J. Improving intercropping: A synthesis of research in agronomy, plant physiology and ecology. New Phytol. 2015, 206, 107–117. [Google Scholar] [CrossRef]
- Ananthi, T.; Amanullah, M.M.; Al-Tawaha, A.R.M.S. A review on maize-legume intercropping for enhancing the productivity and soil fertility for sustainable agriculture in India. Adv. Environ. Biol. 2017, 11, 49–64. [Google Scholar]
- Farooq, T.H.; Shakoor, A.; Wu, X.; Li, Y.; Rashid, M.H.U.; Zhang, X.; Gilani, M.M.; Kumar, U.; Chen, X.; Yan, W. Perspectives of plantation forests in the sustainable forest development of China. iForest 2021, 14, 166. [Google Scholar] [CrossRef]
- Farooq, T.H.; Yan, W.; Rashid, M.H.U.; Tigabu, M.; Gilani, M.M.; Zou, X.H.; Wu, P.F. Chinese fir (Cunninghamia Lanceolata) a green gold of China with continues decline in its productivity over the successive rotations: A review. Appl. Ecol. Environ. Res. 2019, 17, 11055–11067. [Google Scholar] [CrossRef]
- Farooq, T.H.; Wu, W.; Tigabu, M.; Ma, X.; He, Z.; Rashid, M.H.; Gilani, M.M.; Wu, P.F. Growth, biomass production and root development of Chinese fir in relation to initial planting density. Appl. Ecol. Environ. Res. 2019, 17, 3553–3566. [Google Scholar] [CrossRef]
- Farooq, T.H.; Ma, X.; Rashid, M.H.U.; Wu, W.; Xu, J.; Tarin, M.W.K.; He, Z.; Wu, P. Impact of stand density on soil quality in Chinese Fir (Cunninghamia Lanceolata) monoculture. Appl. Ecol. Environ. Res. 2019, 17, 3553–3566. [Google Scholar] [CrossRef]
- Powlson, D.S.; Gregory, P.J.; Whalley, W.R.; Quinton, J.N.; Hopkins, D.W.; Whitmore, A.P.; Hirsch, P.R.; Goulding, K.W.T. Soil management in relation to sustainable agriculture and ecosystem services. Food Policy 2011, 36, S72–S87. [Google Scholar] [CrossRef]
- Ghosh, P.K.; Bandyopadhyay, K.K.; Wanjari, R.H.; Manna, M.C.; Misra, A.K.; Mohanty, M.; Rao, A.S. Legume effect for enhancing productivity and nutrient use-efficiency in major cropping systems–an Indian perspective: A review. J. Sustain. Agric. 2007, 30, 59–86. [Google Scholar] [CrossRef]
- Rowe, H.; Withers, P.J.A.; Baas, P.; Chan, N.I.; Doody, D.; Holiman, J.; Jacobs, B.; Li, H.; MacDonald, G.K.; McDowell, R. Integrating legacy soil phosphorus into sustainable nutrient management strategies for future food, bioenergy and water security. Nutr. Cycl. Agro Ecosyst. 2016, 104, 393–412. [Google Scholar] [CrossRef]
- Gebru, H. A review on the comparative advantages of intercropping to mono-cropping system. J. Biol. Agric. Healthc. 2015, 5, 1–13. [Google Scholar]
- Powell, J.M.; Williams, T.O. Livestock, Nutrient Cycling and Sustainable Agriculture in the West African Sahel; International Institute for Environment and Development: London, UK, 1993. [Google Scholar]
- Nyawade, S.O.; Gachene, C.K.K.; Karanja, N.N.; Gitari, H.I.; Schulte-Geldermann, E.; Parker, M.L. Controlling soil erosion in smallholder potato farming systems using legume intercrops. Geoderma Reg. 2019, 17, 00225. [Google Scholar] [CrossRef]
- Seran, T.H.; Brintha, I. Review on maize based intercropping. J. Agron. 2010, 9, 135–145. [Google Scholar] [CrossRef] [Green Version]
- Dong, B.; Wu, B.; Hong, W.; Li, X.; Li, Z.; Xue, L.; Huang, Y. Transcriptome analysis of the tea oil camellia (Camellia oleifera) reveals candidate drought stress genes. PLoS ONE 2017, 12, 0181835. [Google Scholar] [CrossRef] [PubMed]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Peanut (Arachis hypogaea L.): A prospective legume crop to offer multiple health benefits under changing climate. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1325–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Wu, L.; Jun, C.; Khan, M.A.; Luo, X.; Lin, W. Biochemical and microbial properties of rhizospheres under maize/peanut intercropping. J. Integr. Agric. 2016, 15, 101–110. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Ding, N.; Shi, J.; Wu, M.; Liao, H.; Xu, J. Profiling of microbial PLFAs: Implications for interspecific interactions due to intercropping which increase phosphorus uptake in phosphorus limited acidic soils. Soil Biol. Biochem. 2013, 57, 625–634. [Google Scholar] [CrossRef]
- Burns, R.G.; DeForest, J.L.; Marxsen, J.; Sinsabaugh, R.L.; Stromberger, M.E.; Wallenstein, M.D.; Weintraub, M.N.; Zoppini, A. Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol. Biochem. 2013, 58, 216–234. [Google Scholar] [CrossRef]
- Das, S.K.; Varma, A. Role of enzymes in maintaining soil health. In Soil Enzymology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 25–42. [Google Scholar]
- Neher, D.A. Role of nematodes in soil health and their use as indicators. J. Nematol. 2001, 33, 161. [Google Scholar]
- Farooq, T.H.; Tigabu, M.; Ma, X.; Zou, X.; Liu, A.; Odén, P.C.; Wu, P. Nutrient uptake, allocation and biochemical changes in two Chinese fir cuttings under heterogeneous phosphorus supply. IForest 2018, 11, 411–417. [Google Scholar] [CrossRef] [Green Version]
- Farooq, T.H.; Yan, W.; Chen, X.; Shakoor, A.; Rashid, M.H.U.; Gilani, M.M.; He, Z.; Wu, P. Dynamics of canopy development of Cunninghamia lanceolata mid-age plantation in relation to foliar nitrogen and soil quality influenced by stand density. Glob. Ecol. Conserv. 2020, 24, 01209. [Google Scholar] [CrossRef]
- Wu, P.; Wang, G.; Farooq, T.H.; Li, Q.; Zou, X.; Ma, X. Low phosphorus and competition affect Chinese fir cutting growth and root organic acid content: Does neighboring root activity aggravate P nutrient deficiency? J. Soils Sediments 2017, 17, 2775–2785. [Google Scholar] [CrossRef]
- Loveland, P.; Webb, J. Is there a critical level of organic matter in the agricultural soils of temperate regions: A review. Soil Tillage Res. 2003, 70, 1–18. [Google Scholar] [CrossRef]
- Jiang, F.; Drohan, P.J.; Cibin, R.; Preisendanz, H.E.; White, C.M.; Veith, T.L. Reallocating crop rotation patterns improves water quality and maintains crop yield. Agric. Syst. 2021, 187, 103015. [Google Scholar] [CrossRef]
- Li, W.; Zheng, Z.; Li, T.; Zhang, X.; Wang, Y.; Yu, H.; He, S.; Liu, T. Effect of tea plantation age on the distribution of soil organic carbon fractions within water-stable aggregates in the hilly region of Western Sichuan, China. Catena 2015, 133, 198–205. [Google Scholar] [CrossRef]
- Lu, W.; Shen, X.; Chen, Y. Effects of intercropping peanut on soil nutrient status and microbial activity within young Camellia oleifera plantation. Commun. Soil Sci. Plant Anal. 2019, 50, 1232–1238. [Google Scholar] [CrossRef]
- Hu, C.H.; Wang, P.Q.; Zhang, P.P.; Nie, X.M.; Li, B.B.; Tai, L.; Liu, W.T.; Li, W.Q.; Chen, K.M. NADPH oxidases: The vital performers and center hubs during plant growth and signaling. Cells 2020, 9, 437. [Google Scholar] [CrossRef] [Green Version]
- Cameron, K.C.; Di, H.J.; Moir, J.L. Nitrogen losses from the soil/plant system: A review. Ann. Appl. Biol. 2013, 162, 145–173. [Google Scholar] [CrossRef]
- Dubey, R.S.; Pessarakli, M. Physiological mechanisms of nitrogen absorption and assimilation in plants under stressful conditions. Handb. Plant Crop Physiol. 1995, 605–625. [Google Scholar]
- Jensen, E.S. Grain yield, symbiotic N2 fixation and interspecific competition for inorganic N in pea-barley intercrops. Plant Soil 1996, 182, 25–38. [Google Scholar] [CrossRef]
- Jensen, E.S.; Carlsson, G.; Hauggaard-Nielsen, H. Intercropping of grain legumes and cereals improves the use of soil N resources and reduces the requirement for synthetic fertilizer N: A global-scale analysis. Agron. Sustain. Dev. 2020, 40, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.C.; Li, F.M.; Shan, L. Switchgrass and milkvetch intercropping under 2: 1 row-replacement in semiarid region, northwest China: Aboveground biomass and water use efficiency. Eur. J. Agron. 2008, 28, 485–492. [Google Scholar] [CrossRef]
- Ikramul Haq, M.; Maqbool, M.M.; Ali, A.; Farooq, S.; Khan, S.; Saddiq, M.S.; Khan, K.A.; Ali, S.; Ifnan Khan, M.; Hussain, A.; et al. Optimizing planting geometry for Barley-Egyptian clover intercropping system in semi-arid sub-tropical climate. PLoS ONE 2020, 15, 0233171. [Google Scholar] [CrossRef] [PubMed]
- Giller, K.E. Nitrogen Fixation in Tropical Cropping Systems; Cabi: Oxfordshire, UK, 2001. [Google Scholar]
- Singh, D.K.; Kumar, S. Nitrate reductase, arginine deaminase, urease and dehydrogenase activities in natural soil (ridges with forest) and in cotton soil after acetamiprid treatments. Chemosphere 2008, 71, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Xue, L.; Feng, Y.; Liu, Y.; Song, Z.; Mandal, S.; Yang, L.; Sun, Q.; Xing, B. Hydrochar reduced NH3 volatilization from rice paddy soil: Microbial-aging rather than water-washing is recommended before application. J. Clean. Prod. 2020, 268, 122233. [Google Scholar] [CrossRef]
- Li, L.; Li, S.-M.; Sun, J.H.; Zhou, L.L.; Bao, X.G.; Zhang, H.-G.; Zhang, F.-S. Diversity enhances agricultural productivity via rhizosphere phosphorus facilitation on phosphorus-deficient soils. Proc. Natl. Acad. Sci. USA 2007, 104, 11192–11196. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Li, C.; Mi, G.; Li, L.; Yuan, L.; Jiang, R.; Zhang, F. Maximizing root/rhizosphere efficiency to improve crop productivity and nutrient use efficiency in intensive agriculture of China. J. Exp. Bot. 2013, 64, 1181–1192. [Google Scholar] [CrossRef]
- Wang, X.; Gao, Y.; Zhang, H.; Shao, Z.; Sun, B.; Gao, Q. Enhancement of rhizosphere citric acid and decrease of NO3−/NH4+ ratio by root interactions facilitate N fixation and transfer. Plant Soil 2020, 447, 169–182. [Google Scholar] [CrossRef]
- Rasool, B.; Ramzani, P.M.A.; Zubair, M.; Khan, M.A.; Lewińska, K.; Turan, V.; Karczewska, A.; Khan, S.A.; Farhad, M.; Tauqeer, H.M.; et al. Impacts of oxalic acid-activated phosphate rock and root-induced changes on lead bioavailability in rhizosphere and distribution in mung bean plant. Environ. Pollut. 2021, 116903. [Google Scholar] [CrossRef]
- Maurya, P.R.; Lal, R. Effects of different mulch materials on soil properties and on the root growth and yield of maize (Zea mays) and cowpea (Vigna unguiculata). F. Crop. Res. 1981, 4, 33–45. [Google Scholar] [CrossRef]
- Dick, R.P. Soil enzyme activities as indicators of soil quality. Defin. Soil Qual. Sustain. Environ. 1994, 35, 107–124. [Google Scholar]
- Sherene, T. Role of soil enzymes in nutrient transformation: A review. Bio. Bull. 2017, 3, 109–131. [Google Scholar]
- Han, S.; Delgado-Baquerizo, M.; Luo, X.; Liu, Y.; Van Nostrand, J.D.; Chen, W.; Zhou, J.; Huang, Q. Soil aggregate size-dependent relationships between microbial functional diversity and multifunctionality. Soil Biol. Biochem. 2021, 154, 108143. [Google Scholar] [CrossRef]
- Roohi, M.; Arif, M.S.; Yasmeen, T.; Riaz, M.; Rizwan, M.; Shahzad, S.M.; Ali, S.; Bragazza, L. Effects of cropping system and fertilization regime on soil phosphorous are mediated by rhizosphere-microbial processes in a semi-arid agroecosystem. J. Environ. Manag. 2020, 271, 111033. [Google Scholar] [CrossRef] [PubMed]
- Jan, M.T.; Roberts, P.; Tonheim, S.K.; Jones, D.L. Protein breakdown represents a major bottleneck in nitrogen cycling in grassland soils. Soil Biol. Biochem. 2009, 41, 2272–2282. [Google Scholar] [CrossRef]
- Kwiatkowski, C.A.; Harasim, E.; Feledyn-Szewczyk, B.; Antonkiewicz, J. Enzymatic activity of loess soil in organic and conventional farming systems. Agriculture 2020, 10, 135. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Rossi, F.; Deng, S.; Liu, Y.; Wang, G.; Adessi, A.; De Philippis, R. Macromolecular and chemical features of the excreted extracellular polysaccharides in induced biological soil crusts of different ages. Soil Biol. Biochem. 2014, 78, 1–9. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, J.; Zheng, X.; Zhang, J.; Zhou, P.; Song, X.; Song, W.; Wang, S. Wheat straw and biochar effect on soil carbon fractions, enzyme activities, and nutrients in a tobacco field. Can. J. Soil Sci. 2021, 101, 1–12. [Google Scholar] [CrossRef]
- Rashid, M.H.U.; Tigabu, M.; Chen, H.; Farooq, T.H.; Ma, X.; Wu, P. Calcium-mediated adaptive responses to low phosphorus stress in Chinese fir. Trees 2020, 34, 825–834. [Google Scholar] [CrossRef]
- Deng, S.P.; Tabatabai, M.A. Effect of tillage and residue management on enzyme activities in soils: III. Phosphatases and arylsulfatase. Biol. Fertil. Soils 1997, 24, 141–146. [Google Scholar] [CrossRef]
- Zhang, W.; Gruszewski, H.A.; Chevone, B.I.; Nessler, C.L. An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol. 2008, 146, 431–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uren, N.C. Types, amounts, and possible functions of compounds released into the rhizosphere by soil-grown plants. Rhizosph. Biochem. Org. Subst. Soil Plant Interface 2007, 2, 1–21. [Google Scholar]
- Wen, F.; VanEtten, H.D.; Tsaprailis, G.; Hawes, M.C. Extracellular proteins in pea root tip and border cell exudates. Plant Physiol. 2007, 143, 773–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mobley, H.L.T.; Hu, L.-T.; Foxall, P.A. Helicobacter pylori urease: Properties and role in pathogenesis. Scand. J. Gastroenterol. 1991, 26, 39–46. [Google Scholar] [CrossRef]
- Ansari, M.W.; Trivedi, D.K.; Sahoo, R.K.; Gill, S.S.; Tuteja, N. A critical review on fungi mediated plant responses with special emphasis to Piriformospora indica on improved production and protection of crops. Plant Physiol. Biochem. 2013, 70, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.H.; Wang, Q.; Ruan, X.; Pan, C.D.; Jiang, D.A. Phenolics and plant allelopathy. Molecules 2010, 15, 8933–8952. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Cao, F.; Wang, G. Soil microbiological properties and enzyme activity in Ginkgo–tea agroforestry compared with monoculture. Agrofor. Syst. 2013, 87, 1201–1210. [Google Scholar] [CrossRef]
- Udawatta, R.P.; Kremer, R.J.; Adamson, B.W.; Anderson, S.H. Variations in soil aggregate stability and enzyme activities in a temperate agroforestry practice. Appl. Soil Ecol. 2008, 39, 153–160. [Google Scholar] [CrossRef]
- Shakoor, A.; Shakoor, S.; Rehman, A.; Ashraf, F.; Abdullah, M.; Shahzad, S.M.; Farooq, T.H.; Ashraf, M.; Manzoor, M.A.; Altaf, M.; et al. Effect of animal manure, crop type, climate zone, and soil attributes on greenhouse gas emissions from agricultural soils—A global meta-analysis. J. Clean. Prod. 2020, 124019. [Google Scholar] [CrossRef]
- Shakoor, A.; Shahzad, S.M.; Chatterjee, N.; Arif, M.S.; Farooq, T.H.; Altaf, M.M.; Tufail, M.A.; Dar, A.A.; Mehmood, T. Nitrous oxide emission from agricultural soils: Application of animal manure or biochar? A global meta-analysis. J. Environ. Manag. 2021, 285, 112170. [Google Scholar] [CrossRef]
- Shakoor, A.; Shahbaz, M.; Farooq, T.H.; Sahar, N.E.; Shahzad, S.M.; Altaf, M.M.; Ashraf, M. A global meta-analysis of greenhouse gases emission and crop yield under no-tillage as compared to conventional tillage. Sci. Total Environ. 2021, 750, 142299. [Google Scholar] [CrossRef]
- Hu, W.; Jiao, Z.; Wu, F.; Liu, Y.; Dong, M.; Ma, X.; Fan, T.; An, L.; Feng, H. Long-term effects of fertilizer on soil enzymatic activity of wheat field soil in Loess Plateau, China. Ecotoxicology 2014, 23, 2069–2080. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, T.; Li, F.; Lemcoff, J.H.; Cohen, S. Fertilization regulates soil enzymatic activity and fertility dynamics in a cucumber field. Sci. Hortic. 2008, 116, 21–26. [Google Scholar] [CrossRef]
- Zhao, S.; Chen, X.; Deng, S.; Dong, X.; Song, A.; Yao, J.; Fang, W.; Chen, F. The effects of fungicide, soil fumigant, bio-organic fertilizer and their combined application on chrysanthemum Fusarium wilt controlling, soil enzyme activities and microbial properties. Molecules 2016, 21, 526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, N.; Yu, M.; Cui, M.; Luo, Z.; Feng, Y.; Cao, S.; Sun, Y.; Li, Y. Effects of different ectomycorrhizal fungal inoculates on the growth of Pinus tabulaeformis seedlings under greenhouse conditions. Forests 2016, 7, 316. [Google Scholar] [CrossRef] [Green Version]
- Ryan, J.; Estefan, G.; Rashid, A. Soil and Plant Analysis Laboratory Manual; ICARDA: Beirut, Lebanon, 2001; ISBN 9291271187. [Google Scholar]
- Bray, R.H.; Kurtz, L.T. Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 1945, 59, 39–46. [Google Scholar] [CrossRef]
- Schollenberger, C.J.; Simon, R.H. Determination of exchange capacity and exchangeable bases in soil—ammonium acetate method. Soil Sci. 1945, 59, 13–24. [Google Scholar] [CrossRef]
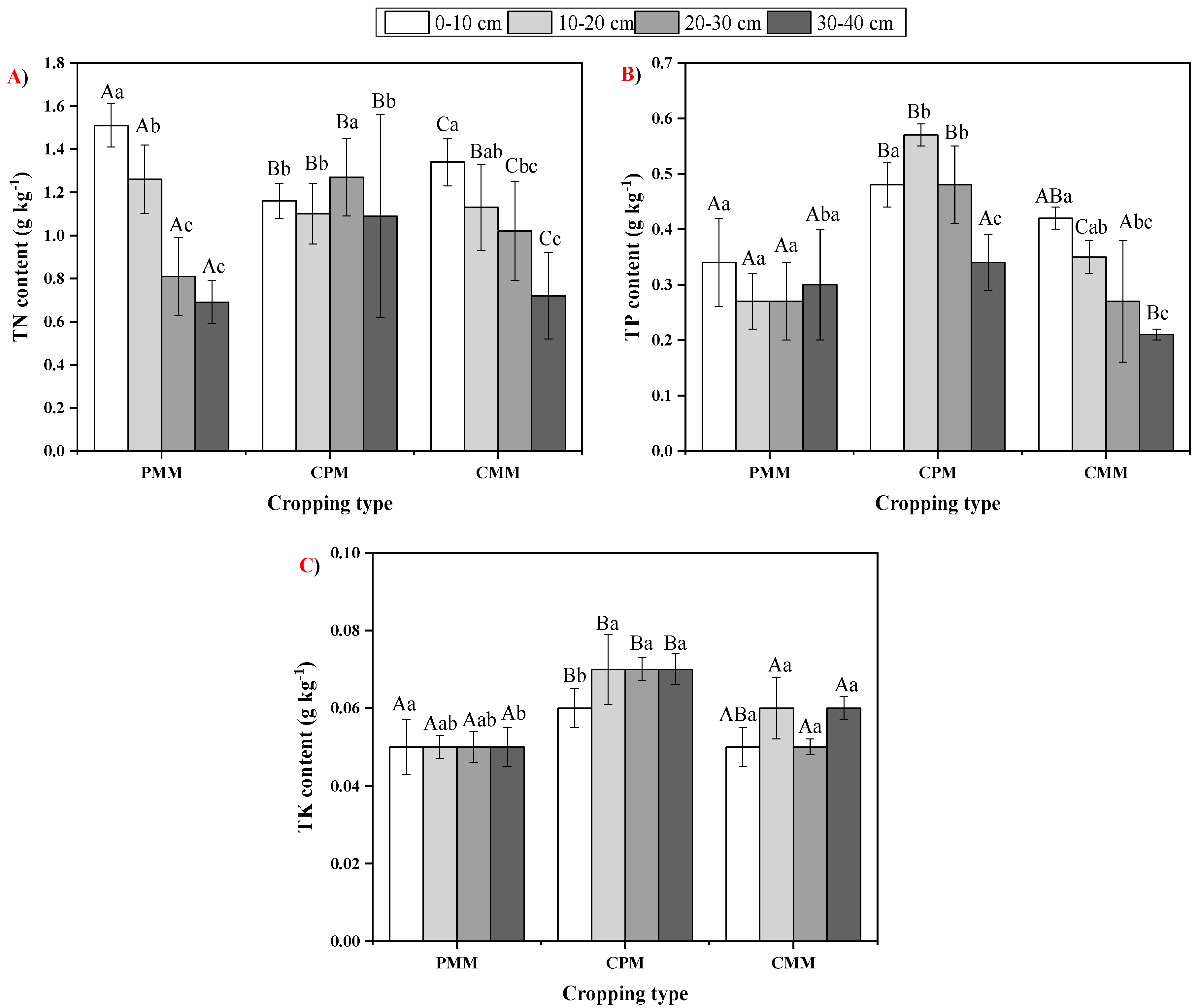
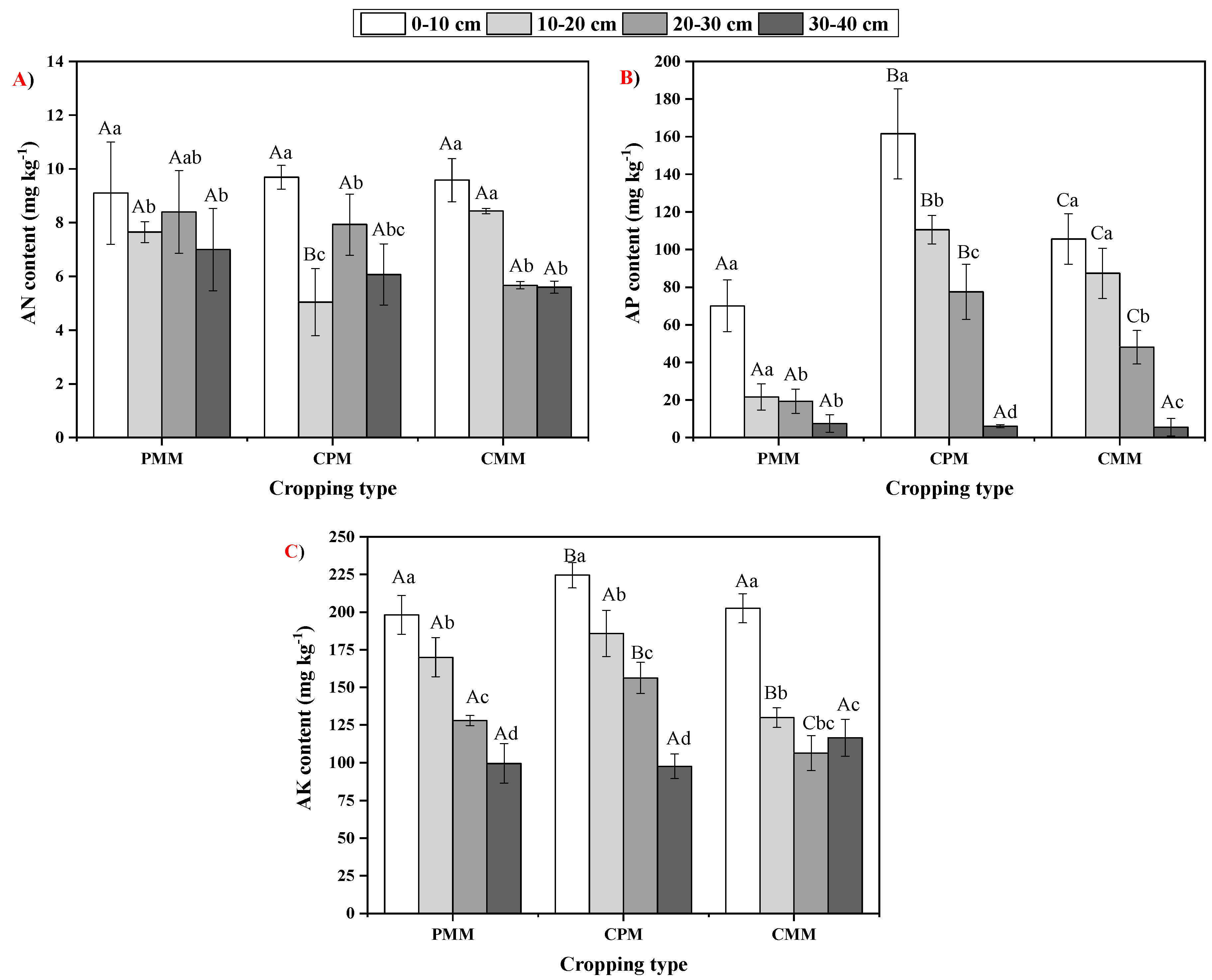

| Cropping Model | 0–10 cm | 10–20 cm | 20–30 cm | 30–40 cm |
|---|---|---|---|---|
| PMM | 19.93 ± 1.5 Aa | 16.61 ± 1.39 Ab | 8.62 ± 2.11 Ac | 6.56 ± 1.79 Ac |
| CPM | 16.77 ± 0.77 Ba | 12.56 ± 4.48 Ba | 14.37 ± 6.17 Bab | 7.68 ± 2.22 Bb |
| CMM | 13.9 ± 1.65 Ca | 12.15 ± 1.93 Ba | 9.89 ± 3.04 Ca | 5.61 ± 3.11 Cb |
| Cropping Model | Soil Enzyme | SOC | TN | TP | TK | AN | AP | AK |
|---|---|---|---|---|---|---|---|---|
| PMM | Sucrase | 0.21 | 0.22 | 0.04 | −0.27 | 0.01 | 0.08 | 0.07 |
| Protease | 0.45 | 0.47 | −0.05 | −0.14 | 0.29 | 0.37 | 0.17 | |
| Urease | 0.68 * | 0.71 * | −0.15 | −0.5 * | −0.14 | 0.25 | 0.58 * | |
| Acid phosphatase | 0.94 * | 0.92 ** | 0.13 | −0.29 | 0.19 | 0.41 | 0.57 * | |
| Catalase | 0.17 | 0.05 | 0.06 | −0.02 | −0.11 | 0.2 | −0.03 | |
| CPM | Sucrase | 0.33 | 0.51 * | −0.08 | 0 | 0.11 | −0.18 | 0.22 |
| Protease | 0.56 * | 0.21 | 0.11 | 0.2 | 0.39 | 0.32 | 0.38 | |
| Urease | 0.75 * | 0.26 | 0.32 | 0.23 | 0.43 * | 0.03 | 0.46 * | |
| Acid phosphatase | 0.42 ** | 0.34 | 0.15 | −0.11 | 0.33 | −0.03 | 0.13 | |
| Catalase | 0.44 * | 0.26 | 0.34 | 0.12 | 0.25 | −0.14 | 0.41 * | |
| CMM | Sucrase | 0.35 | 0.41 | 0.38 | −0.05 | 0.03 | 0.41 * | 0.31 |
| Protease | 0.05 | −0.01 | 0.17 | −0.07 | −0.55 * | 0.19 | −0.31 | |
| Urease | 0.41 * | 0.52 * | 0.52 * | 0.23 | 0.23 | 0.48 * | 0.49 * | |
| Acid phosphatase | 0.51 ** | 0.62 * | 0.58 * | 0.11 | 0.12 | 0.62 * | 0.44 ** | |
| Catalase | 0.43 * | 0.52 * | 0.54 * | 0.01 | −0.08 | 0.54 * | 0.42 * |
| Cropping Model | Soil Depth | Cropping Model × Soil Depth | ||||
|---|---|---|---|---|---|---|
| Variable | F | P | F | P | F | P |
| TN | 122.6 | ** | 1233.6 | * | 365.5 | ** |
| TP | 870.2 | * | 1364.4 | ** | 102.1 | *** |
| TK | 683.3 | ** | 65.3 | 0.274 | 89.2 | ** |
| AN | 42.9 | ** | 78.3 | ** | 51.2 | ** |
| AP | 632.4 | ** | 896.7 | ** | 3546.5 | ** |
| AK | 536.2 | * | 336.3 | *** | 1456.5 | ** |
| Protease | 256.3 | 0.066 | 256.5 | ** | 785.4 | 0.064 |
| Sucrase | 158.9 | ** | 447.3 | ** | 1235.9 | * |
| A. phosphatase | 1244.3 | * | 380.3 | 0.066 | 3225.0 | ** |
| Urease | 1563.2 | ** | 125.6 | ** | 2132.5 | ** |
| Catalase | 853.2 | ** | 0.072 | ** | 125.2 | ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farooq, T.H.; Kumar, U.; Mo, J.; Shakoor, A.; Wang, J.; Rashid, M.H.U.; Tufail, M.A.; Chen, X.; Yan, W. Intercropping of Peanut–Tea Enhances Soil Enzymatic Activity and Soil Nutrient Status at Different Soil Profiles in Subtropical Southern China. Plants 2021, 10, 881. https://doi.org/10.3390/plants10050881
Farooq TH, Kumar U, Mo J, Shakoor A, Wang J, Rashid MHU, Tufail MA, Chen X, Yan W. Intercropping of Peanut–Tea Enhances Soil Enzymatic Activity and Soil Nutrient Status at Different Soil Profiles in Subtropical Southern China. Plants. 2021; 10(5):881. https://doi.org/10.3390/plants10050881
Chicago/Turabian StyleFarooq, Taimoor Hassan, Uttam Kumar, Jing Mo, Awais Shakoor, Jun Wang, Muhammad Haroon U. Rashid, Muhammad Aammar Tufail, Xiaoyong Chen, and Wende Yan. 2021. "Intercropping of Peanut–Tea Enhances Soil Enzymatic Activity and Soil Nutrient Status at Different Soil Profiles in Subtropical Southern China" Plants 10, no. 5: 881. https://doi.org/10.3390/plants10050881
APA StyleFarooq, T. H., Kumar, U., Mo, J., Shakoor, A., Wang, J., Rashid, M. H. U., Tufail, M. A., Chen, X., & Yan, W. (2021). Intercropping of Peanut–Tea Enhances Soil Enzymatic Activity and Soil Nutrient Status at Different Soil Profiles in Subtropical Southern China. Plants, 10(5), 881. https://doi.org/10.3390/plants10050881








