Genome-Wide and Comprehensive Analysis of the Multiple Stress-Related CAF1 (CCR4-Associated Factor 1) Family and Its Expression in Poplar
Abstract
:1. Introduction
2. Results
2.1. Identification of the CAF1 Proteins in P. trichocarpa
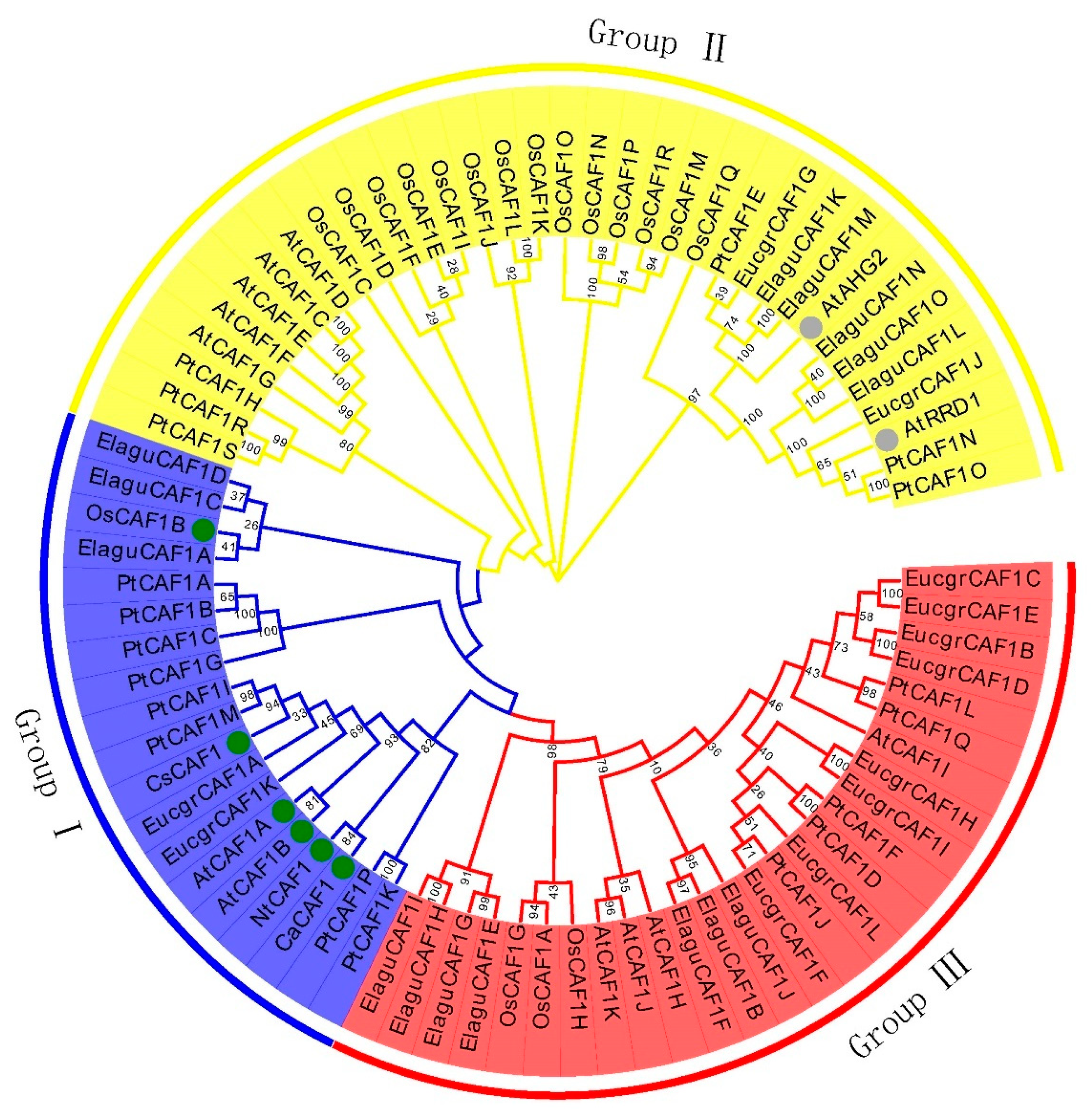
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
2.3. Gene Structure and Motif Composition of PtCAF1 Gene Family
2.4. Protein Structure Prediction of CAF1
2.5. Promoter cis-Element Analysis
2.6. Chromosomal Distribution and Synteny Analysis of PtCAF1 Genes and Ka/Ks Calculation
2.7. Expression Patterns of PtCAF1 Genes in Different Plant Tissues
2.8. Expression Profiling of PtCAF1 in Response to Different Treatments
3. Discussion
4. Materials and Methods
4.1. Identification and Characterization of the CAF1 Family in P. trichocarpa
4.2. Sequence Analysis
4.3. Multiple Sequence Alignment and Phylogenetic Analysis of CAF1 Proteins
4.4. Prediction of Protein Spatial Structure
4.5. Chromosomal Locations, Gene Duplication of PtCAF1 Genes, and Calculation of Ka/Ks
4.6. Transcriptional Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reverdatto, S.V.; Dutko, J.A.; Chekanova, J.A.; Hamilton, D.A.; Belostotsky, D.A. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA 2004, 10, 1200–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belostotsky, D.A.; Sieburth, L.E. Kill the messenger: mRNA decay and plant development. Curr. Opin. Plant Biol. 2009, 12, 96–102. [Google Scholar] [CrossRef]
- Temme, C.; Zaessinger, S.; Meyer, S.; Simonelig, M.; Wahle, E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004, 23, 2862–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchese, F.P.; Aubareda, A.; Tudor, C.; Saklatvala, J.; Clark, A.R.; Dean, J.L. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J. Biol. Chem. 2010, 285, 27590–27600. [Google Scholar] [CrossRef] [Green Version]
- Meyer, S.; Temme, C.; Wahle, E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.; Green, P.J. mRNA Degradation Machinery in Plants. J. Plant Biol. 2009, 52, 114–124. [Google Scholar] [CrossRef]
- Zhao, T.; Huan, Q.; Sun, J.; Liu, C.; Hou, X.; Yu, X.; Silverman, I.M.; Zhang, Y.; Gregory, B.D.; Liu, C.M.; et al. Impact of poly(A)-tail G-content on Arabidopsis PAB binding and their role in enhancing translational efficiency. Genome Biol. 2019, 20, 189. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.J.; Zhang, X.H.; Hu, L.D.; Zhang, J.Q.; Jiang, Y.; Yang, Y.; Yan, Y.B. DsCaf1 is involved in environmental stress response of Dunaliella salina. Int. J. Biol. Macromol. 2016, 82, 369–374. [Google Scholar] [CrossRef]
- Arae, T.; Morita, K.; Imahori, R.; Suzuki, Y.; Yasuda, S.; Sato, T.; Yamaguchi, J.; Chiba, Y. Identification of Arabidopsis CCR4-NOT Complexes with Pumilio RNA-Binding Proteins, APUM5 and APUM2. Plant Cell Physiol. 2019, 60, 2015–2025. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.; Valencia-Sanchez, M.A.; Staples, R.R.; Chen, J.; Denis, C.L.; Parker, R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 2001, 104, 377–386. [Google Scholar] [CrossRef] [Green Version]
- Garapaty, S.; Mahajan, M.A.; Samuels, H.H. Components of the CCR4-NOT complex function as nuclear hormone receptor coactivators via association with the NRC-interacting Factor NIF-1. J. Biol. Chem. 2008, 283, 6806–6816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.J.; Gao, Q.; Fang, X.D.; Ding, Z.H.; Gao, D.M.; Xu, W.Y.; Cao, Q.; Qiao, J.H.; Yang, Y.Z.; Han, C.; et al. CCR4, a RNA decay factor, is hijacked by a plant cytorhabdovirus phosphoprotein to facilitate virus replication. eLife 2020, 9. [Google Scholar] [CrossRef]
- Balu, B.; Maher, S.P.; Pance, A.; Chauhan, C.; Naumov, A.V.; Andrews, R.M.; Ellis, P.D.; Khan, S.M.; Lin, J.W.; Janse, C.J.; et al. CCR4-associated factor 1 coordinates the expression of Plasmodium falciparum egress and invasion proteins. Eukaryot Cell 2011, 10, 1257–1263. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Bai, Y.; Zhang, A.; Zhang, Q.; Bartlam, M.G. Insights into the structure and architecture of the CCR4-NOT complex. Front. Genet. 2014, 5, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buschauer, R.; Matsuo, Y.; Sugiyama, T.; Chen, Y.H.; Alhusaini, N.; Sweet, T.; Ikeuchi, K.; Cheng, J.; Matsuki, Y.; Nobuta, R.; et al. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 2020, 368, 6488–6912. [Google Scholar] [CrossRef] [PubMed]
- Prevot, D.; Morel, A.P.; Voeltzel, T.; Rostan, M.C.; Rimokh, R.; Magaud, J.P.; Corbo, L. Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: Involvement in estrogen receptor alpha signaling pathway. J. Biol. Chem. 2001, 276, 9640–9648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berthet, C.; Morera, A.M.; Asensio, M.J.; Chauvin, M.A.; Morel, A.P.; Dijoud, F.; Magaud, J.P.; Durand, P.; Rouault, J.P. CCR4-associated factor CAF1 is an essential factor for spermatogenesis. Mol. Cell Biol. 2004, 24, 5808–5820. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.X.; Li, J.S.; Hu, R.; Li, X.M.; Wang, H. CAF1-knockout mice are more susceptive to lipopolysaccharide-induced acute lung injury. J. Inflamm. Res. 2016, 9, 115–121. [Google Scholar] [CrossRef] [Green Version]
- Ahuja, I.; de Vos, R.C.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef]
- Chou, W.L.; Huang, L.F.; Fang, J.C.; Yeh, C.H.; Hong, C.Y.; Wu, S.J.; Lu, C.A. Divergence of the expression and subcellular localization of CCR4-associated factor 1 (CAF1) deadenylase proteins in Oryza sativa. Plant Mol. Biol. 2014, 85, 443–458. [Google Scholar] [CrossRef]
- Chou, W.L.; Chung, Y.L.; Fang, J.C.; Lu, C.A. Novel interaction between CCR4 and CAF1 in rice CCR4-NOT deadenylase complex. Plant Mol. Biol. 2017, 93, 79–96. [Google Scholar] [CrossRef]
- Hart, K.J.; Oberstaller, J.; Walker, M.P.; Minns, A.M.; Kennedy, M.F.; Padykula, I.; Adams, J.H.; Lindner, S.E. Plasmodium male gametocyte development and transmission are critically regulated by the two putative deadenylases of the CAF1/CCR4/NOT complex. PLoS Pathog. 2019, 15, e1007164. [Google Scholar] [CrossRef] [PubMed]
- Walley, J.W.; Kelley, D.R.; Nestorova, G.; Hirschberg, D.L.; Dehesh, K. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol. 2010, 152, 866–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarowar, S.; Oh, H.W.; Cho, H.S.; Baek, K.H.; Seong, E.S.; Joung, Y.H.; Choi, G.J.; Lee, S.; Choi, D. Capsicum annuum CCR4-associated factor CaCAF1 is necessary for plant development and defence response. Plant J. 2007, 51, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Shimo, H.M.; Terassi, C.; Lima Silva, C.C.; Zanella, J.L.; Mercaldi, G.F.; Rocco, S.A.; Benedetti, C.E. Role of the Citrus sinensis RNA deadenylase CsCAF1 in citrus canker resistance. Mol. Plant Pathol. 2019, 20, 1105–1118. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.; Li, C.; Liu, F.; Jiang, H.; Li, S.; Sun, J.; Wu, X.; Li, C. The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res. 2009, 19, 307–316. [Google Scholar] [CrossRef] [Green Version]
- Kwon, T.-M.; Yi, Y.-B.; Nam, J.-S. Overexpression of AtCAF1, CCR4-associated factor 1 homologue in Arabidopsis thaliana, negatively regulates wounding-mediated disease resistance. J. Plant Biotechnol. 2011, 38, 278–284. [Google Scholar] [CrossRef]
- Hirayama, T.; Matsuura, T.; Ushiyama, S.; Narusaka, M.; Kurihara, Y.; Yasuda, M.; Ohtani, M.; Seki, M.; Demura, T.; Nakashita, H.; et al. A poly(A)-specific ribonuclease directly regulates the poly(A) status of mitochondrial mRNA in Arabidopsis. Nat. Commun. 2013, 4, 2247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, J.C.; Liu, H.Y.; Tsai, Y.C.; Chou, W.L.; Chang, C.C.; Lu, C.A. A CCR4 Association Factor 1, OsCAF1B, Participates in the alphaAmy3 mRNA Poly(A) Tail Shortening and Plays a Role in Germination and Seedling Growth. Plant Cell Physiol. 2020, 61, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.C.; Tsai, Y.C.; Chou, W.L.; Liu, H.Y.; Chang, C.C.; Wu, S.J.; Lu, C.A. A CCR4-associated factor 1, OsCAF1B, confers tolerance of low-temperature stress to rice seedlings. Plant Mol. Biol. 2021, 105, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip. Rev. RNA 2016, 7, 438–454. [Google Scholar] [CrossRef] [Green Version]
- Parsons, T.J.; Sinkar, V.P.; Stettler, R.F.; Nester, E.W.; Gordon, M.P. Transformation of Poplar by Agrobacterium tumefaciens. Bio/Technol. 1986, 4, 533–536. [Google Scholar] [CrossRef]
- Hu, J.; Yang, M.; Lu, M. Advances in biosafety studies on transgenic insect-resistant poplars in China. Biodivers. Sci. 2010, 18, 336–345. [Google Scholar]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Guo, Y.; Chen, Y.; Wu, D.; Jiang, L. Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in Brassica napus. BMC Plant Biol. 2020, 20, 543. [Google Scholar] [CrossRef]
- Movahedi, A.; Zhang, J.; Yin, T.; Qiang, Z. Functional Analysis of Two Orthologous NAC Genes, CarNAC3, and CarNAC6 from Cicer arietinum, Involved in Abiotic Stresses in Poplar. Plant Mol. Biol. Rep. 2015, 33, 1539–1551. [Google Scholar] [CrossRef]
- Hui, W.; Movahedi, A.; Xu, C.; Sun, W.; Li, L.; Li, D.; Qiang, Z. Characterization, expression profiling, and functional analysis of a Populus trichocarpa defensin gene and its potential as an anti-Agrobacterium rooting medium additive. Sci. Rep. 2019, 9, 15359–15375. [Google Scholar] [CrossRef] [Green Version]
- Wilusz, C.J.; Gao, M.; Jones, C.L.; Wilusz, J.; Peltz, S.W. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA 2001, 7, 1416–1424. [Google Scholar] [PubMed]
- Yang, X.; Morita, M.; Wang, H.; Suzuki, T.; Yang, W.; Luo, Y.; Zhao, C.; Yu, Y.; Bartlam, M.; Yamamoto, T.; et al. Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity. Nucleic Acids Res. 2008, 36, 6872–6881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, N.; Kitahata, N.; Seki, M.; Narusaka, Y.; Narusaka, M.; Kuromori, T.; Asami, T.; Shinozaki, K.; Hirayama, T. Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J. 2005, 44, 972–984. [Google Scholar] [CrossRef]
- Chong, L.; Guo, P.; Zhu, Y. Mediator Complex: A Pivotal Regulator of ABA Signaling Pathway and Abiotic Stress Response in Plants. Int. J. Mol. Sci 2020, 21, 7755. [Google Scholar] [CrossRef]
- Nishimura, N.; Okamoto, M.; Narusaka, M.; Yasuda, M.; Nakashita, H.; Shinozaki, K.; Narusaka, Y.; Hirayama, T. ABA hypersensitive germination2-1 causes the activation of both abscisic acid and salicylic acid responses in Arabidopsis. Plant Cell Physiol. 2009, 50, 2112–2122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otsuka, K.; Mamiya, A.; Konishi, M.; Nozaki, M.; Kinoshita, A.; Tamaki, H.; Arita, M.; Saito, M.; Yamamoto, K.; Hachiya, T.; et al. Temperature-dependent fasciation mutants provide a link between mitochondrial RNA processing and lateral root morphogenesis. eLife 2021, 10, 61611–61622. [Google Scholar] [CrossRef]
- Corbin, C.; Drouet, S.; Markulin, L.; Auguin, D.; Laine, E.; Davin, L.B.; Cort, J.R.; Lewis, N.G.; Hano, C. A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: From gene identification and evolution to differential regulation. Plant Mol. Biol. 2018, 97, 73–101. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Sun, W.; Ma, Z.; Zheng, T.; Huang, L.; Wu, Q.; Zhao, G.; Tang, Z.; Bu, T.; Li, C.; et al. Genome-wide investigation of the AP2/ERF gene family in tartary buckwheat (Fagopyum Tataricum). BMC Plant Biol. 2019, 19, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maestrini, P.; Cavallini, A.; Rizzo, M.; Giordani, T.; Bernardi, R.; Durante, M.; Natali, L. Isolation and expression analysis of low temperature-induced genes in white poplar (Populus alba). J. Plant Physiol. 2009, 166, 1544–1556. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cui, X.; Meng, Z.; Huang, X.; Xie, Q.; Wu, H.; Jin, H.; Zhang, D.; Liang, W. Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012, 158, 1279–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Lee, J.H.; Poindexter, M.R.; Shao, Y.; Liu, W.; Lenaghan, S.C.; Ahkami, A.H.; Blumwald, E.; Stewart, C.N., Jr. Rational design and testing of abiotic stress-inducible synthetic promoters from poplar cis-regulatory elements. Plant Biotechnol. J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lin, Y.J.; Wang, P.; Zhang, B.; Li, M.; Chen, S.; Shi, R.; Tunlaya-Anukit, S.; Liu, X.; Wang, Z.; et al. The AREB1 Transcription Factor Influences Histone Acetylation to Regulate Drought Responses and Tolerance in Populus trichocarpa. Plant Cell 2019, 31, 663–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Gao, S.; Tang, Y.; Li, L.; Zhang, F.; Feng, B.; Fang, Z.; Ma, L.; Zhao, C. Genome-wide identification and evolutionary analyses of bZIP transcription factors in wheat and its relatives and expression profiles of anther development related TabZIP genes. BMC Genom. 2015, 16, 976. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yang, J.; Wang, W.; Li, D.; Hu, X.; Wang, H.; Wei, M.; Liu, Q.; Wang, Z.; Li, C. Genome-wide identification and expression profiling of the copper transporter gene family in Populus trichocarpa. Plant Physiol. Biochem. 2015, 97, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Jiang, M.; Li, P.; Chu, Z. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium distachyon. BMC Genom. 2016, 17, 175. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Liu, P.; Yu, J.; Wang, L.; Dossa, K.; Zhang, Y.; Zhou, R.; Wei, X.; Zhang, X. Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biol. 2017, 17, 152. [Google Scholar] [CrossRef] [PubMed]
- Faraji, S.; Filiz, E.; Kazemitabar, S.K.; Vannozzi, A.; Palumbo, F.; Barcaccia, G.; Heidari, P. The AP2/ERF Gene Family in Triticum durum: Genome-Wide Identification and Expression Analysis under Drought and Salinity Stresses. Genes 2020, 11, 1464. [Google Scholar] [CrossRef] [PubMed]
- Makova, K.D.; Li, W.-H. Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 2003, 13, 1638–1645. [Google Scholar] [CrossRef] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.H.; Yang, J.; Gu, X. Expression divergence between duplicate genes. Trends Genet. Tig 2005, 21, 602–607. [Google Scholar] [CrossRef]
- Walley, J.W.; Kelley, D.R.; Savchenko, T.; Dehesh, K. Investigating the function of CAF1 deadenylases during plant stress responses. Plant Signal. Behav. 2010, 5, 802–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meinke, D.W. Genome-wide identification of EMBRYO-DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 2020, 226, 306–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.-H.; Liu, Y.-X.; Deng, R.; Lei, T.-T.; Tian, A.-J.; Ren, H.-H.; Wang, S.-F.; Wang, X.-F. Genome-wide identification and expression analysis of the GSK gene family in Solanum tuberosum L. under abiotic stress and phytohormone treatments and functional characterization of StSK21 involvement in salt stress. Gene 2021, 766, 145156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yin, T. Analysis of topology properties in different tissues of poplar based on gene co-expression networks. Tree Genet. Genomes 2019, 16, 178–189. [Google Scholar] [CrossRef]
- Jia, H.F.; Chai, Y.M.; Li, C.L.; Lu, D.; Luo, J.J.; Qin, L.; Shen, Y.Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milborrow, B.V. The Metabolism of Abscisic Acid. J. Exp. Bot. 1970, 21, 17–29. [Google Scholar] [CrossRef]
- Joshi-Saha, A.; Valon, C.; Leung, J. Chapter 7—Molecular Mechanisms of Abscisic Acid Action in Plants and Its Potential Applications to Human Health. In Advances in Botanical Research; Turkan, I., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 57, pp. 249–292. [Google Scholar]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Knight, H.; Knight, M.R. Abiotic stress signalling pathways: Specificity and cross-talk. Trends Plant Sci. 2001, 6, 262–267. [Google Scholar] [CrossRef]
- Yi, L.; Li, B.; Korpelainen, H.; Yu, F.; Wu, L.; Tong, L.; Liu, M. Mechanisms of drought response in Populus. South. For. J. For. Sci. 2020, 82, 359–366. [Google Scholar] [CrossRef]
- Thakur, A.K.; Kumar, P.; Parmar, N.; Shandil, R.K.; Aggarwal, G.; Gaur, A.; Srivastava, D.K. Achievements and prospects of genetic engineering in poplar: A review. New For. 2021, 2, 361–390. [Google Scholar] [CrossRef]
- Yan, C.; Wang, Y.; Lyu, T.; Hu, Z.; Ye, N.; Liu, W.; Li, J.; Yao, X.; Yin, H. Alternative Polyadenylation in response to temperature stress contributes to gene regulation in Populus trichocarpa. BMC Genom. 2021, 22, 53. [Google Scholar] [CrossRef]
- Wu, L.; Belasco, J.G. Let Me Count the Ways: Mechanisms of Gene Regulation by miRNAs and siRNAs. Mol. Cell 2008, 29, 1–7. [Google Scholar] [CrossRef]
- Wu, L.; Fan, J.; Belasco, J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Nat. Acad. Sci. USA 2006, 103, 4034–4039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Sun, G.; Shi, C.; Sun, D. Transcriptome analysis reveals new microRNAs-mediated pathway involved in anther development in male sterile wheat. BMC Genom. 2018, 19, 333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myburg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D.; et al. The genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Duan, W.; Song, X.; Tang, J.; Wu, P.; Zhang, B.; Hou, X. Retention, Molecular Evolution, and Expression Divergence of the Auxin/Indole Acetic Acid and Auxin Response Factor Gene Families in Brassica Rapa Shed Light on Their Evolution Patterns in Plants. Genome Biol. Evol. 2015, 8, 302–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]






| Name | Phytozome Gene ID | Ensembl Gene ID | CDS | AA | MW (kDa) | pI | Subcellular Location |
|---|---|---|---|---|---|---|---|
| PtCAF1A | Potri.001G038500.1 | PNT52564 | 876 | 292 | 33.40 | 6.86 | cytosol |
| PtCAF1B | Potri.001G039000.1 | PNT52569 | 648 | 216 | 24.78 | 8.47 | cytosol |
| PtCAF1C | Potri.001G040400.1 | PNT52586 | 789 | 263 | 30.25 | 9.91 | cytosol |
| PtCAF1D | Potri.001G046700.1 | PNT52695 | 825 | 275 | 31.30 | 4.56 | cytosol |
| PtCAF1E | Potri.001G368400.1 | PNT58721 | 1425 | 475 | 53.50 | 7.85 | nucleus |
| PtCAF1F | Potri.003G181100.1 | PNT46250 | 825 | 275 | 31.22 | 4.49 | cytosol |
| PtCAF1G | Potri.003G186300.1 | PNT46340 | 915 | 305 | 34.53 | 5.21 | nucleus |
| PtCAF1H | Potri.004G048800.1 | PNT39610 | 828 | 276 | 31.98 | 6.37 | chloroplast |
| PtCAF1I | Potri.004G200400.1 | PNT42179 | 651 | 217 | 25.12 | 4.62 | cytosol |
| PtCAF1J | Potri.006G187200.1 | PNT32402 | 810 | 270 | 30.31 | 5.34 | cytosol |
| PtCAF1K | Potri.006G205600.1 | PNT32745 | 888 | 296 | 33.70 | 5.96 | chloroplast |
| PtCAF1L | Potri.006G262500.1 | PNT33885 | 834 | 278 | 31.49 | 4.46 | cytosol |
| PtCAF1M | Potri.009G161500.1 | PNT21675 | 834 | 278 | 31.71 | 5.08 | cytosol |
| PtCAF1N | Potri.014G018500.1 | PNT02530 | 1815 | 605 | 67.72 | 7.27 | chloroplast |
| PtCAF1O | Potri.014G177400.1 | PNT05527 | 1881 | 627 | 70.23 | 6.12 | chloroplast |
| PtCAF1P | Potri.016G073000.1 | PNS98364 | 891 | 297 | 33.43 | 5.8 | cytosol |
| PtCAF1Q | Potri.018G020900.1 | PNS92197 | 834 | 278 | 31.42 | 4.41 | cytosol |
| PtCAF1R | Potri.018G036100.1 | PNS92495 | 918 | 306 | 33.97 | 9.1 | cytosol |
| PtCAF1S | Potri.018G038700.1 | PNS92540 | 909 | 303 | 33.75 | 8.8 | cytosol |
| Sequence | Ka | Ks | Ka/Ks | p-Value (Fisher) | Length | Time (Mya) |
|---|---|---|---|---|---|---|
| PtCAF1D & PtCAF1F | 0.016708 | 0.306409 | 0.05453 | 1.36 × 10−22 | 822 | 102.1363 |
| PtCAF1A & PtCAF1C | 0.014891 | 0.019054 | 0.781482 | 0.429132 | 780 | 6.351433 |
| PtCAF1C & PtCAF1G | 1.01436 | 0.952687 | 1.06473 | 0.247271 | 723 | 317.5623 |
| PtCAF1A & PtCAF1G | 0.12254 | 0.316388 | 0.387308 | 5.67 × 10−8 | 873 | 105.4627 |
| PtCAF1F & PtCAF1L | 0.082084 | 2.14757 | 0.038222 | 4.1× 10−125 | 819 | 715.8567 |
| PtCAF1K & PtCAF1P | 0.049893 | 0.434185 | 0.114912 | 8.21× 10−28 | 858 | 144.7283 |
| PtCAF1N & PtCAF1O | 0.008309 | 0.01364 | 0.609174 | 0.239107 | 1797 | 4.546667 |
| PtCAF1L & PtCAF1Q | 0.034304 | 0.337114 | 0.101757 | 2.86× 10−20 | 831 | 112.3713 |
| PtCAF1R & PtCAF1S | 0.028137 | 0.084529 | 0.332862 | 0.000561 | 906 | 28.17643 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Li, L.; Wei, H.; Sun, W.; Zhou, P.; Zhu, S.; Li, D.; Zhuge, Q. Genome-Wide and Comprehensive Analysis of the Multiple Stress-Related CAF1 (CCR4-Associated Factor 1) Family and Its Expression in Poplar. Plants 2021, 10, 981. https://doi.org/10.3390/plants10050981
Wang P, Li L, Wei H, Sun W, Zhou P, Zhu S, Li D, Zhuge Q. Genome-Wide and Comprehensive Analysis of the Multiple Stress-Related CAF1 (CCR4-Associated Factor 1) Family and Its Expression in Poplar. Plants. 2021; 10(5):981. https://doi.org/10.3390/plants10050981
Chicago/Turabian StyleWang, Pu, Lingling Li, Hui Wei, Weibo Sun, Peijun Zhou, Sheng Zhu, Dawei Li, and Qiang Zhuge. 2021. "Genome-Wide and Comprehensive Analysis of the Multiple Stress-Related CAF1 (CCR4-Associated Factor 1) Family and Its Expression in Poplar" Plants 10, no. 5: 981. https://doi.org/10.3390/plants10050981






