OseIF3h Regulates Plant Growth and Pollen Development at Translational Level Presumably through Interaction with OsMTA2
Abstract
:1. Introduction
2. Results
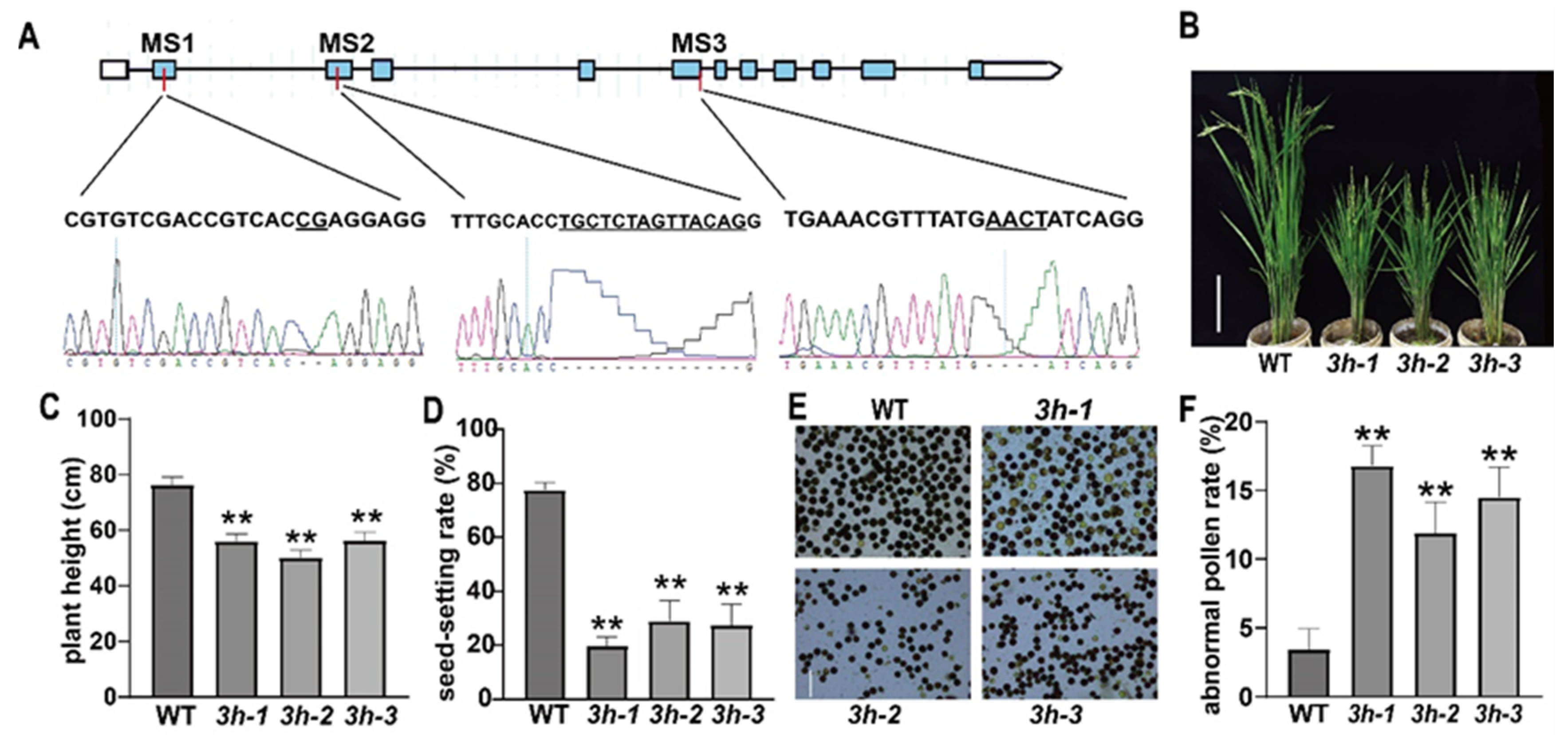
2.1. Generation and Characterization of oseif3h Mutants
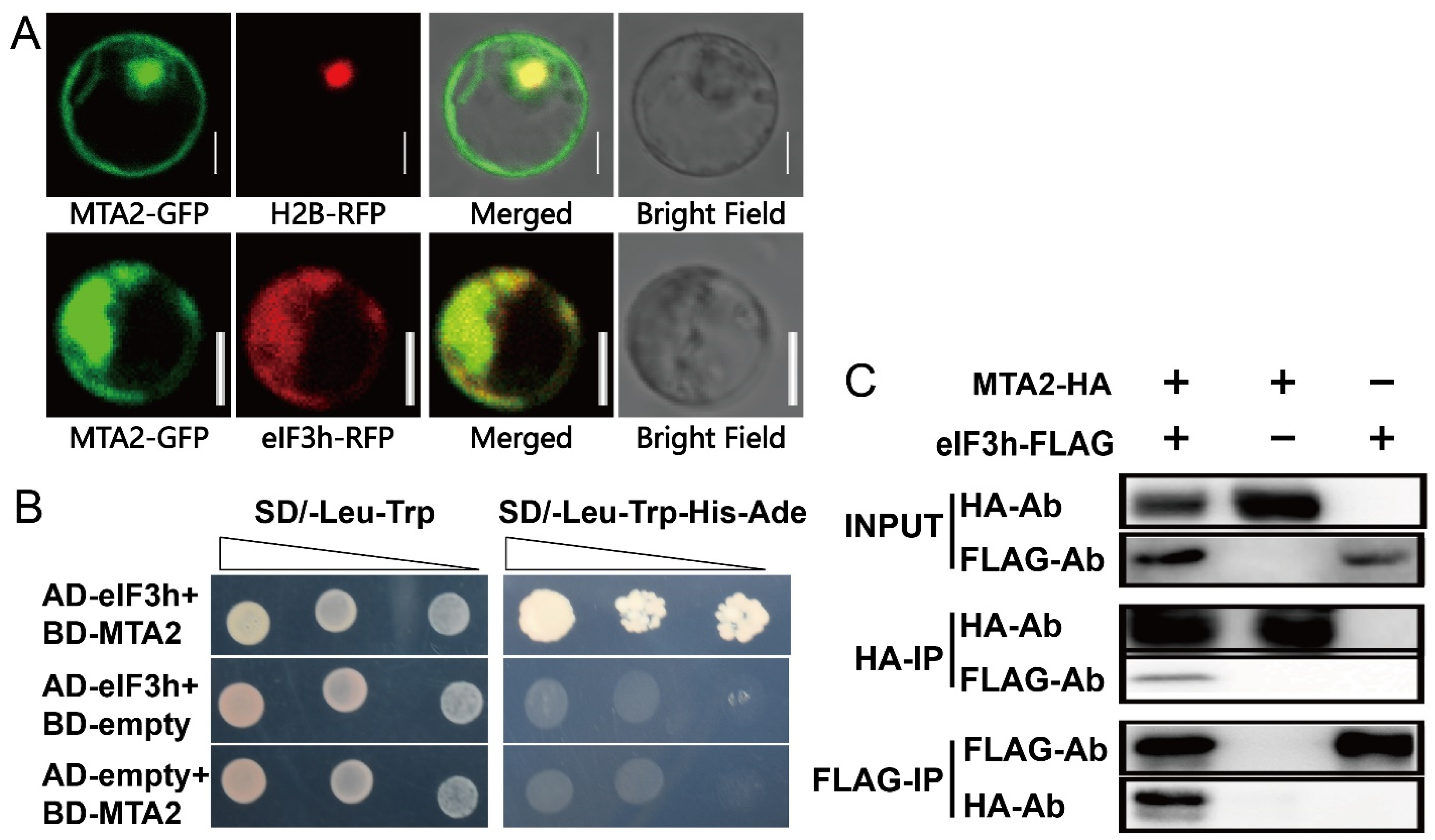
2.2. Subcellular Localization of OseIF3h and Its Expression and Polysome Profile Pattern
2.3. OseIF3h Interacts with OsMTA2
2.4. RIP-Seq Data Mapping and Character of OseIF3h Associated Peaks
3. Discussion
3.1. OseIF3h Is Required for Plant Growth and Pollen Development in Rice
3.2. OseIF3h Involves in the Regulation of Protein Translation Presumably through mRNA Methylation
3.3. OseIF3h Is Required for Photosynthetic Performance
4. Materials and Methods
4.1. Genetic Material
4.2. Generation of oseif3h Mutants
4.3. Yeast Two-Hybrid Assays
4.4. Transient Expression in Protoplast and Co-IP Assay
4.5. Polysome Profiling Assays
4.6. RIP-Seq and Data Analysis
4.7. I2-KI Staining for Pollen Fertility
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Sandercock, A.M.; Fraser, C.S.; Ridlova, G.; Stephens, E.; Schenauer, M.R.; Yokoi-Fong, T.; Barsky, D.; Leary, J.A.; Hershey, J.W.; et al. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl. Acad. Sci. USA 2008, 105, 18139–18144. [Google Scholar] [CrossRef] [Green Version]
- Burks, E.A.; Bezerra, P.P.; Le, H.; Gallie, D.R.; Browning, K.S. Plant initiation factor 3 subunit composition resembles mammalian initiation factor 3 and has a novel subunit. J. Biol. Chem. 2001, 276, 2122–2131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voigts-Hoffmann, F.; Klinge, S.; Ban, N. Structural insights into eukaryotic ribosomes and the initiation of translation. Curr. Opin. Struct. Biol. 2012, 22, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Valášek, L.S.; Zeman, J.; Wagner, S.; Beznosková, P.; Pavlíková, Z.; Mohammad, M.P.; Hronová, V.; Herrmannová, A.; Hashem, Y.; Gunišová, S. Embraced by eIF3: Structural and functional insights into the roles of eIF3 across the translation cycle. Nucleic Acids Res. 2017, 45, 10948–10968. [Google Scholar] [CrossRef] [PubMed]
- Scheel, H.; Hofmann, K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinform. 2005, 6, 71. [Google Scholar] [CrossRef] [Green Version]
- Nupponen, N.N.; Porkka, K.; Kakkola, L.; Tanner, M.; Persson, K.; Borg, A.; Isola, J.; Visakorpi, T. Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am. J. Pathol. 1999, 154, 1777–1783. [Google Scholar] [CrossRef] [Green Version]
- Savinainen, K.J.; Linja, M.J.; Saramäki, O.R.; Tammela, T.L.; Chang, G.T.; Brinkmann, A.O.; Visakorpi, T. Expression and copy number analysis of TRPS1, EIF3S3 and MYC genes in breast and prostate cancer. Br. J. Cancer 2004, 90, 1041–1046. [Google Scholar] [CrossRef] [Green Version]
- Choudhuri, A.; Maitra, U.; Evans, T. Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 9818–9823. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.H.; Kim, B.H.; Yahalom, A.; Chamovitz, D.A.; von Arnim, A.G. Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 2004, 16, 3341–3356. [Google Scholar] [CrossRef]
- Ray, A.; Bandyopadhyay, A.; Matsumoto, T.; Deng, H.; Maitra, U. Fission yeast translation initiation factor 3 subunit eIF3h is not essential for global translation initiation, but deletion of eif3h+ affects spore formation. Yeast 2008, 25, 809–823. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [Green Version]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Zhao, X.; Wu, Y.S.; Li, M.M.; Wang, X.J.; Yang, Y.G. N6-methyl-adenosine (m6A) in RNA: An old modification with a novel epigenetic function. Genom. Proteom. Bioinform. 2013, 11, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Vespa, L.; Vachon, G.; Berger, F.; Perazza, D.; Faure, J.D.; Herzog, M. The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 2004, 134, 1283–1292. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Zhang, Y.C.; Liao, J.Y.; Yu, Y.; Zhou, Y.F.; Feng, Y.Z.; Yang, Y.W.; Lei, M.Q.; Bai, M.; Wu, H.; et al. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 2019, 15, e1008120. [Google Scholar] [CrossRef] [Green Version]
- Bodi, Z.; Zhong, S.; Mehra, S.; Song, J.; Graham, N.; Li, H.; May, S.; Fray, R.G. Adenosine Methylation in Arabidopsis mRNA is Associated with the 3′ End and Reduced Levels Cause Developmental Defects. Front. Plant Sci. 2012, 3, 48. [Google Scholar] [CrossRef] [Green Version]
- Choe, J.; Lin, S.; Zhang, W.; Liu, Q.; Wang, L.; Ramirez-Moya, J.; Du, P.; Kim, W.; Tang, S.; Sliz, P.; et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556–560. [Google Scholar] [CrossRef] [Green Version]
- Pisarev, A.V.; Hellen, C.U.; Pestova, T.V. Recycling of eukaryotic posttermination ribosomal complexes. Cell 2007, 131, 286–299. [Google Scholar] [CrossRef] [Green Version]
- Skabkin, M.A.; Skabkina, O.V.; Hellen, C.U.; Pestova, T.V. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol. Cell 2013, 51, 249–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinnebusch, A.G. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006, 31, 553–562. [Google Scholar] [CrossRef]
- Liu, S.; Zhuo, L.; Wang, J.; Zhang, Q.; Li, Q.; Li, G.; Yan, L.; Jin, T.; Pan, T.; Sui, X.; et al. METTL3 plays multiple functions in biological processes. Am. J. Cancer Res. 2020, 10, 1631–1646. [Google Scholar]
- McCormick, S. Male Gametophyte Development. Plant Cell 1993, 5, 1265. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, J.P. The biochemistry of angiosperm pollen development. Bot. Rev. 1975, 41, 259–314. [Google Scholar] [CrossRef]
- Zhang, D.; Luo, X.; Zhu, L. Cytological analysis and genetic control of rice anther development. J. Genet. Genom. 2011, 38, 379–390. [Google Scholar] [CrossRef]
- Virmani, S.S.; Shinjyo, C. Current status of analysis and symbols for male-sterile cytoplasms and fertility-restoring genes. Rice Genet. Newsl. 1988, 5, 9–15. [Google Scholar]
- Chassé, H.; Boulben, S.; Glippa, V.; Pontheaux, F.; Cormier, P.; Morales, J. In vivo analysis of protein translation activity in sea urchin eggs and embryos. Methods Cell Biol. 2019, 151, 335–352. [Google Scholar]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, M.C.; Gregory, B.D. Using Protein Interaction Profile Sequencing (PIP-seq) to Identify RNA Secondary Structure and RNA-Protein Interaction Sites of Long Noncoding RNAs in Plants. Methods Mol. Biol. 2019, 1933, 343–361. [Google Scholar] [PubMed]
- Köster, T.; Staiger, D. RNA-binding protein immunoprecipitation from whole-cell extracts. Methods Mol. Biol. 2014, 1062, 679–695. [Google Scholar]
- Kirchhoff, H. Chloroplast ultrastructure in plants. New Phytol. 2019, 223, 565–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Guo, T.; Li, X.M.; Zhang, Y.M.; Yang, Y.B.; Ye, W.W.; Dong, N.Q.; Shi, C.L.; Kan, Y.; Xiang, Y.H.; et al. Translational Regulation of Plant Response to High Temperature by a Dual-Function tRNA(His) Guanylyltransferase in Rice. Mol. Plant. 2019, 12, 1123–1142. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, K. A high-efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.). Methods Mol. Biol. 2012, 847, 51–57. [Google Scholar]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D.; et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant. Methods 2011, 7, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustroph, A.; Juntawong, P.; Bailey-Serres, J. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol. Biol. 2009, 553, 109–126. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Uren, P.J.; Bahrami-Samani, E.; Burns, S.C.; Qiao, M.; Karginov, F.V.; Hodges, E.; Hannon, G.J.; Sanford, J.R.; Penalva, L.O.; Smith, A.D. Site identification in high-throughput RNA-protein interaction data. Bioinformatics 2012, 28, 3013–3020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Zheng, P.; Liu, X.; Chen, H.; Tu, J. OseIF3h Regulates Plant Growth and Pollen Development at Translational Level Presumably through Interaction with OsMTA2. Plants 2021, 10, 1101. https://doi.org/10.3390/plants10061101
Huang Y, Zheng P, Liu X, Chen H, Tu J. OseIF3h Regulates Plant Growth and Pollen Development at Translational Level Presumably through Interaction with OsMTA2. Plants. 2021; 10(6):1101. https://doi.org/10.3390/plants10061101
Chicago/Turabian StyleHuang, Yuqing, Peng Zheng, Xuejiao Liu, Hao Chen, and Jumin Tu. 2021. "OseIF3h Regulates Plant Growth and Pollen Development at Translational Level Presumably through Interaction with OsMTA2" Plants 10, no. 6: 1101. https://doi.org/10.3390/plants10061101
APA StyleHuang, Y., Zheng, P., Liu, X., Chen, H., & Tu, J. (2021). OseIF3h Regulates Plant Growth and Pollen Development at Translational Level Presumably through Interaction with OsMTA2. Plants, 10(6), 1101. https://doi.org/10.3390/plants10061101





