Validation of Reference Genes for Studying Different Abiotic Stresses in Oat (Avena sativa L.) by RT-qPCR
Abstract
:1. Introduction
2. Results
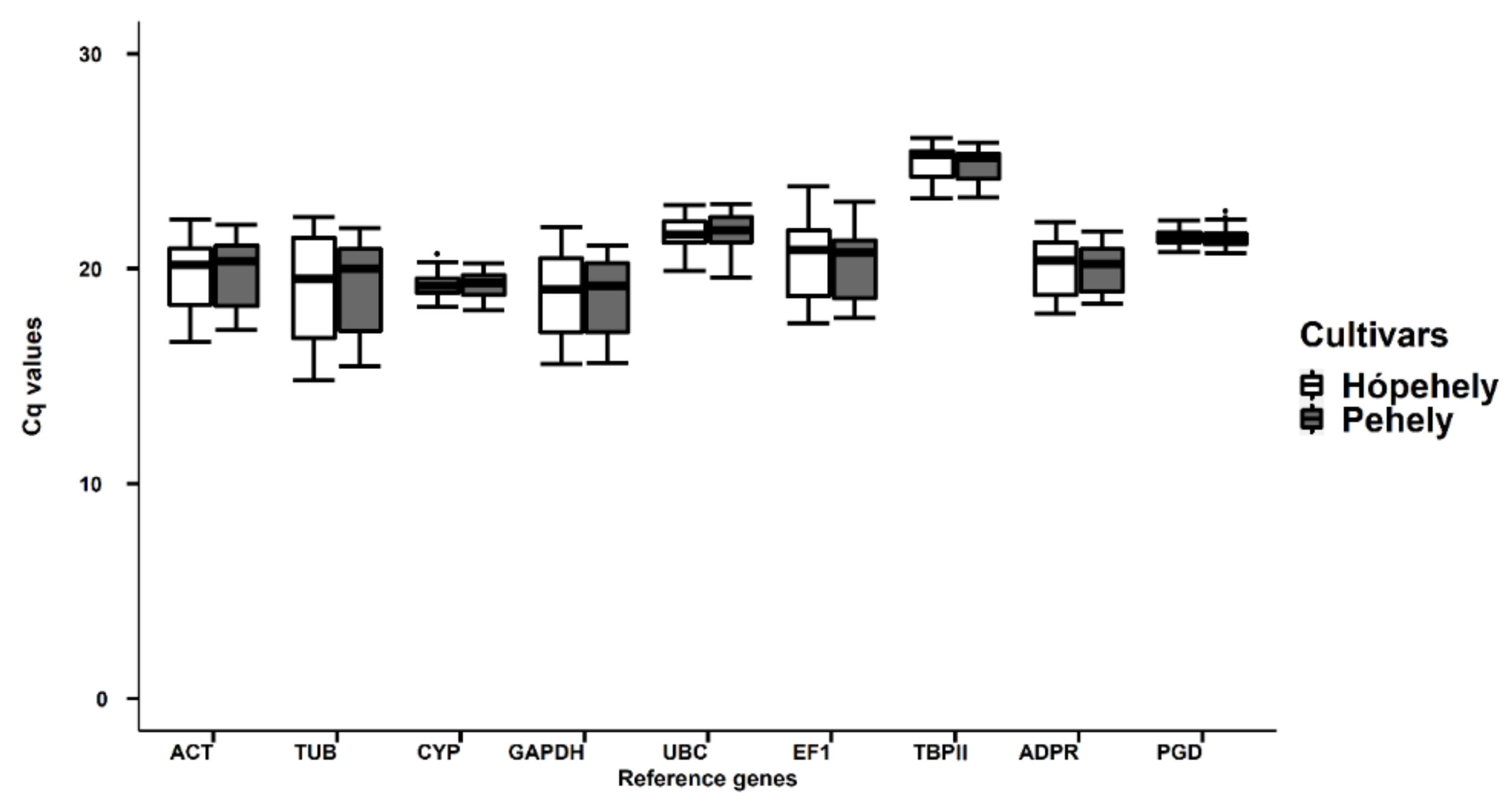
2.1. Verification of Amplification Products, Primer Specificity and Amplification Specificity
2.2. Evaluation of Stability Ranking for Candidate Reference Genes
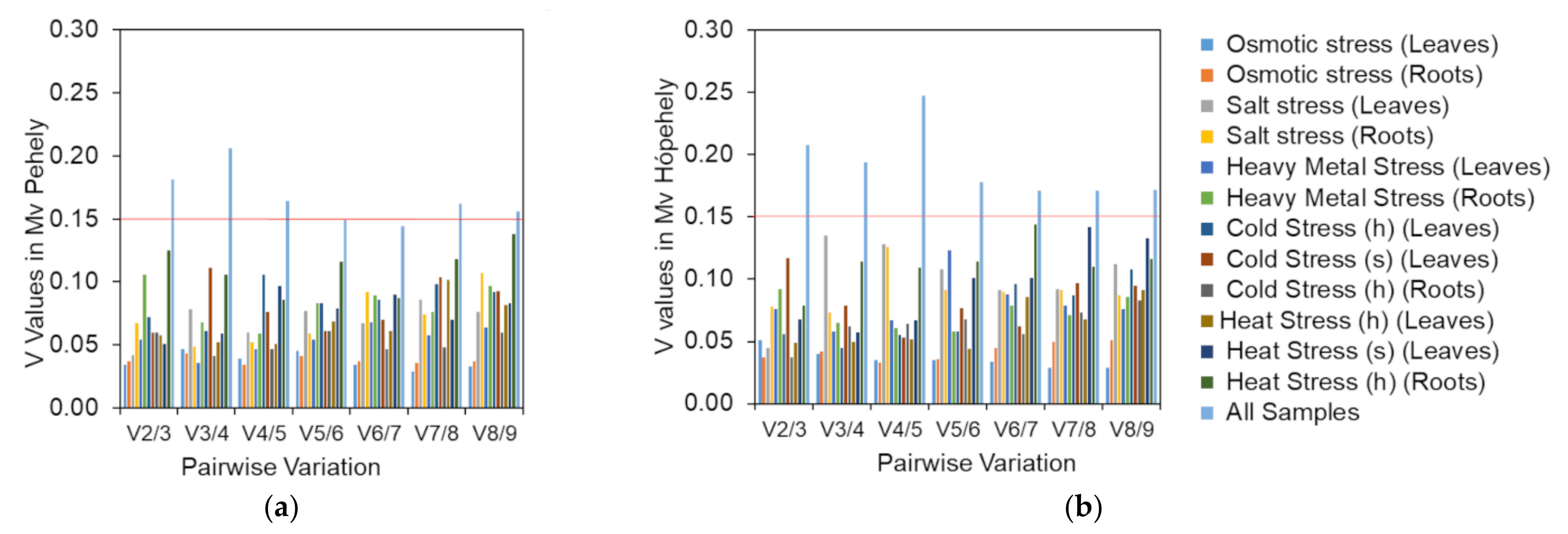
2.2.1. GeNorm Analysis
2.2.2. NormFinder Analysis
2.2.3. Bestkeeper Analysis
2.2.4. RefFinder Analysis
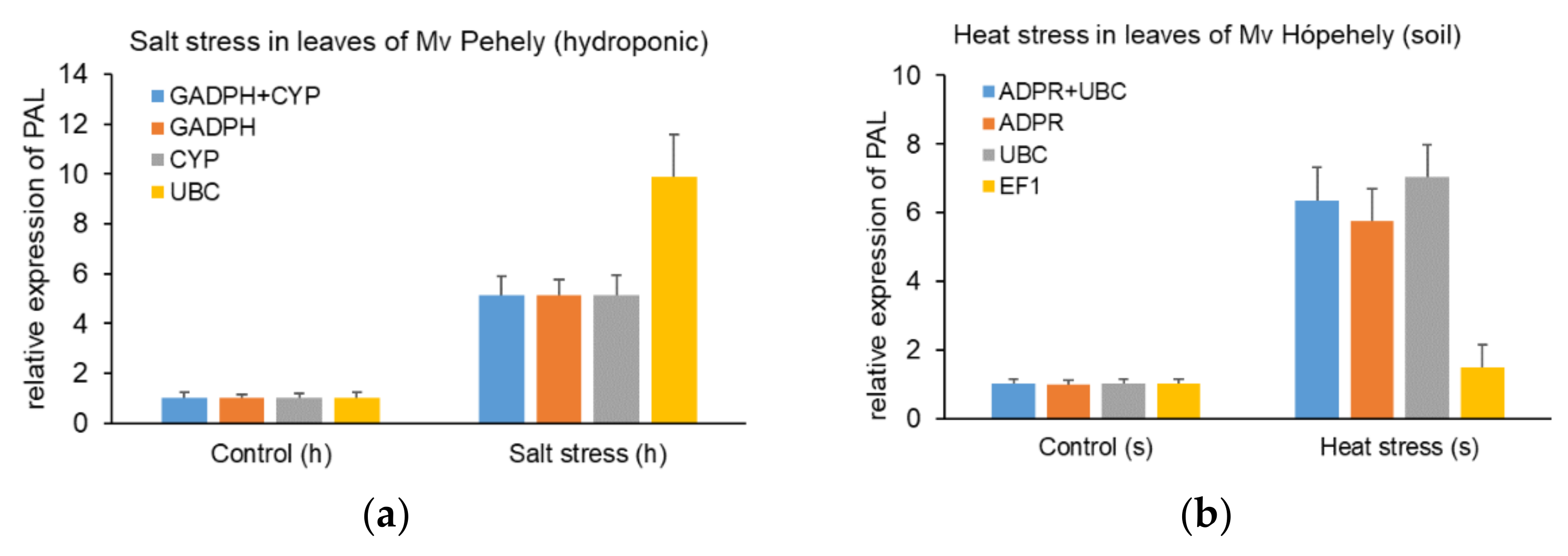
2.3. Validation of the Reference Gene Candidates
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Materials, Growth Conditions, and Stress Treatments
5.2. Plant Materials, Growth Conditions, and Stress Treatments Isolation of RNA and cDNA Synthesis
5.3. Reference Gene Selection and PCR Primer Design
5.4. Real Time Quantitative PCR and Amplification Efficiency Determination
5.5. Stability Ranking of Reference Genes
5.6. Validation of Reference Genes by Expression Analysis of PAL under Different Abiotic Stresses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prates, L.L.; Yu, P. Recent research on inherent molecular structure, physiochemical properties, and bio-functions of food and feed-type Avena sativa oats and processing-induced changes revealed with molecular microspectroscopic techniques. Appl. Spectrosc. Rev. 2017, 52, 850–867. [Google Scholar] [CrossRef]
- Butt, M.S.; Tahir-Nadeem, M.; Khan, M.K.I.; Shabir, R.; Butt, M.S. Oat: Unique among the cereals. Eur. J. Nutr. 2008, 47, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Yan, W.; Wang, Y.; Yin, Q.; Liu, J.; Wight, C.; Ma, B. Screening Oat Genotypes for Tolerance to Salinity and Alkalinity. Front. Plant Sci. 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Heuschele, D.J.; Case, A.; Smith, K.P. Evaluation of Fast Generation Cycling in Oat (Avena sativa). Cereal Res. Commun. 2019, 47, 626–635. [Google Scholar] [CrossRef]
- Goff, S.A.; Ricke, D.; Lan, T.H.; Presting, G.; Wang, R.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.S.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [Green Version]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Wong, M.L.; Medrano, J.F. One-Step Versus Two-Step Real- Time PCR. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Jarošová, J.; Kundu, J.K. Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol. 2010, 10, 146. [Google Scholar] [CrossRef] [Green Version]
- Akbarabadi, A.; Ismaili, A.; Kahrizi, D.; Nazarian Firouzabadi, F. Validation of expression stability of reference genes in response to herbicide stress in wild oat (Avena ludoviciana). Cell. Mol. Biol. 2018, 64, 113. [Google Scholar] [CrossRef]
- Liu, J.; Li, P.; Lu, L.; Xie, L.; Chen, X.; Zhang, B. Selection and evaluation of potential reference genes for gene expression analysis in avena fatua linn. Plant Prot. Sci. 2019, 55, 61–71. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Wang, K.; Aziz, U.; Zhao, C.; Zhang, M. Evaluation of duplicated reference genes for quantitative real-time PCR analysis in genome unknown hexaploid oat (Avena sativa L.). Plant Methods 2020, 16, 138. [Google Scholar] [CrossRef]
- Duan, Z.L.; Han, W.H.; Yan, L.; Wu, B. Reference gene selections for real time quantitative PCR analysis of gene expression in different oat tissues and under salt stress. Biol. Plant. 2020, 64, 838–844. [Google Scholar] [CrossRef]
- Ding, P.; Ding, Y. Stories of Salicylic Acid: A Plant Defense Hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Pawlak-Sprada, S.; Arasimowicz-Jelonek, M.; Podgórska, M.; Deckert, J. Activation of phenylpropanoid pathway in legume plants exposed to heavy metals. Part I. Effects of cadmium and lead on phenylalanine ammonia-lyase gene expression, enzyme activity and lignin content. Acta Biochim. Pol. 2011, 58, 211–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandurska, H.; Cieślak, M. The interactive effect of water deficit and UV-B radiation on salicylic acid accumulation in barley roots and leaves. Environ. Exp. Bot. 2013, 94, 9–18. [Google Scholar] [CrossRef]
- Li, G.; Wang, H.; Cheng, X.; Su, X.; Zhao, Y.; Jiang, T.; Jin, Q.; Lin, Y.; Cai, Y. Comparative genomic analysis of the PAL genes in five Rosaceae species and functional identification of Chinese white pear. PeerJ 2019, 7, e8064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auler, P.A.; Benitez, L.C.; do Amaral, M.N.; Vighi, I.L.; dos Santos Rodrigues, G.; da Maia, L.C.; Braga, E.J.B. Evaluation of stability and validation of reference genes for RT-qPCR expression studies in rice plants under water deficit. J. Appl. Genet. 2017, 58, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Paolacci, A.R.; Tanzarella, O.A.; Porceddu, E.; Ciaffi, M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 2009, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Pu, Q.; Li, Z.; Nie, G.; Zhou, J.; Liu, L.; Peng, Y. Selection and Validation of Reference Genes for Quantitative Real-Time PCR in White Clover (Trifolium repens L.) Involved in Five Abiotic Stresses. Plants 2020, 9, 996. [Google Scholar] [CrossRef]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [Green Version]
- Joseph, J.T.; Poolakkalody, N.J.; Shah, J.M. Screening internal controls for expression analyses involving numerous treatments by combining statistical methods with reference gene selection tools. Physiol. Mol. Biol. Plants 2019, 25, 289–301. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, M.; Meng, Y. Identification and validation of reference genes for rt-qpcr analysis in switchgrass under heavy metal stresses. Genes 2020, 11, 502. [Google Scholar] [CrossRef]
- Janská, A.; Hodek, J.; Svoboda, P.; Zámečník, J.; Prášil, I.T.; Vlasáková, E.; Milella, L.; Ovesná, J. The choice of reference gene set for assessing gene expression in barley (Hordeum vulgare L.) under low temperature and drought stress. Mol. Genet. Genom. 2013, 288, 639–649. [Google Scholar] [CrossRef]
- Kiarash, J.G.; Wilde, H.D.; Amirmahani, F.; Moemeni, M.M.; Zaboli, M.; Nazari, M.; Moosavi, S.S.; Jamalvandi, M. Selection and validation of reference genes for normalization of qRT-PCR gene expression in wheat (Triticum durum L.) under drought and salt stresses. J. Genet. 2018, 97, 1433–1444. [Google Scholar] [CrossRef]
- Poli, M.; Salvi, S.; Li, M.; Varotto, C. Selection of reference genes suitable for normalization of qPCR data under abiotic stresses in bioenergy crop Arundo donax L. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yu, K.; Poysa, V.; Shi, C.; Zhou, Y. Selection of reference genes for normalization of qRT-PCR analysis of differentially expressed genes in soybean exposed to cadmium. Mol. Biol. Rep. 2012, 39, 1585–1594. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Q.; Jiang, Y.; Li, Y.; Zhang, H.; Li, R. Reference genes identification for normalization of qPCR under multiple stresses in Hordeum brevisubulatum. Plant Methods 2018, 14, 110. [Google Scholar] [CrossRef] [Green Version]
- Pál, M.; Horváth, E.; Janda, T.; Páldi, E.; Szalai, G. Cadmium stimulates the accumulation of salicylic acid and its putative precursors in maize (Zea mays) plants. Physiol. Plant. 2005, 125, 356–364. [Google Scholar] [CrossRef]
- Szalai, G.; Janda, K.; Darkó, É.; Janda, T.; Peeva, V.; Pál, M. Comparative analysis of polyamine metabolism in wheat and maize plants. Plant Physiol. Biochem. 2017, 112, 239–250. [Google Scholar] [CrossRef] [Green Version]
- Türkösi, E.; Darko, E.; Rakszegi, M.; Molnár, I.; Molnár-Láng, M.; Cseh, A. Development of a new 7BS.7HL winter wheat-winter barley robertsonian translocation line conferring increased salt tolerance and (1,3;1,4)-β-D-glucan content. PLoS ONE 2018, 13, e0206248. [Google Scholar] [CrossRef] [Green Version]
- Tajti, J.; Németh, E.; Glatz, G.; Janda, T.; Pál, M. Pattern of changes in salicylic acid-induced protein kinase (SIPK) gene expression and salicylic acid accumulation in wheat under cadmium exposure. Plant Biol. 2019, 21, 1176–1180. [Google Scholar] [CrossRef]
- Hua, W.; Zhu, J.; Shang, Y.; Wang, J.; Jia, Q.; Yang, J. Identification of Suitable Reference Genes for Barley Gene Expression Under Abiotic Stresses and Hormonal Treatments. Plant Mol. Biol. Report. 2015, 33, 1002–1012. [Google Scholar] [CrossRef]
- Pál, M.; Tajti, J.; Szalai, G.; Peeva, V.; Végh, B.; Janda, T. Interaction of polyamines, abscisic acid and proline under osmotic stress in the leaves of wheat plants. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Tajti, J.; Janda, T.; Majláth, I.; Szalai, G.; Pál, M. Comparative study on the effects of putrescine and spermidine pre-treatment on cadmium stress in wheat. Ecotoxicol. Environ. Saf. 2018, 148, 546–554. [Google Scholar] [CrossRef] [Green Version]
- Szalai, G.; Tajti, J.; Hamow, K.Á.; Ildikó, D.; Khalil, R.; Vanková, R.; Dobrev, P.; Misheva, S.P.; Janda, T.; Pál, M. Molecular background of cadmium tolerance in Rht dwarf wheat mutant is related to a metabolic shift from proline and polyamine to phytochelatin synthesis. Environ. Sci. Pollut. Res. 2020, 27, 23664–23676. [Google Scholar] [CrossRef] [Green Version]
- PepsiCo OT3098 Hexaploid Oat Genome Assembly and Annotation Release in Collaboration with GrainGenes. Available online: https://web.archive.org/web/20210616152750/https://wheat.pw.usda.gov/GG3/node/922 (accessed on 16 June 2021).
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Owczarzy, R.; Tataurov, A.V.; Wu, Y.; Manthey, J.A.; McQuisten, K.A.; Almabrazi, H.G.; Pedersen, K.F.; Lin, Y.; Garretson, J.; McEntaggart, N.O.; et al. IDT SciTools: A suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008, 36, 163–169. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Rank | Osmotic Stress | Salt Stress | H. Metal Stress | Cold Stress | Heat Stress | All Samples | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | ||
| 1 | ADPR | GAPD | GAPD | UBC | ACT | GAPD | ACT | ACT | GAPD | ACT | TUB | ADPR | TBPII |
| 0.101 | 0.079 | 0.106 | 0.193 | 0.126 | 0.182 | 0.175 | 0.151 | 0.145 | 0.188 | 0.145 | 0.286 | 0.503 | |
| 2 | ACT | ADPR | CYP | ADPR | CYP | ACT | ADPR | ADPR | ADPR | ADPR | CYP | EF1 | UBC |
| 0.102 | 0.081 | 0.110 | 0.195 | 0.133 | 0.199 | 0.181 | 0.159 | 0.150 | 0.189 | 0.156 | 0.298 | 0.533 | |
| 3 | TBPII | ACT | PGD | CYP | ADPR | CYP | UBC | CYP | UBC | GAPD | ACT | PGD | ADPR |
| 0.106 | 0.092 | 0.117 | 0.203 | 0.145 | 0.239 | 0.196 | 0.166 | 0.163 | 0.192 | 0.159 | 0.328 | 0.552 | |
| 4 | CYP | UBC | ACT | TBPII | GAPD | PGD | CYP | TBPII | PGD | UBC | EF1 | ACT | ACT |
| 0.150 | 0.134 | 0.219 | 0.215 | 0.154 | 0.269 | 0.230 | 0.309 | 0.175 | 0.209 | 0.204 | 0.391 | 0.706 | |
| 5 | PGD | PGD | ADPR | EF1 | PGD | EF1 | TUB | TUB | TBPII | CYP | ADPR | TUB | EF1 |
| 0.177 | 0.156 | 0.253 | 0.244 | 0.192 | 0.298 | 0.355 | 0.352 | 0.206 | 0.236 | 0.319 | 0.428 | 0.785 | |
| 6 | GAPD | TBPII | EF1 | PGD | EF1 | ADPR | GAPD | PGD | ACT | PGD | UBC | CYP | GAPD |
| 0.212 | 0.188 | 0.325 | 0.290 | 0.241 | 0.373 | 0.415 | 0.371 | 0.264 | 0.300 | 0.373 | 0.524 | 0.861 | |
| 7 | EF1 | CYP | TUB | GAPD | TBPII | TBPII | EF1 | UBC | EF1 | TBPII | TBPII | TBPII | TUB |
| 0.228 | 0.216 | 0.370 | 0.396 | 0.311 | 0.450 | 0.476 | 0.410 | 0.294 | 0.340 | 0.448 | 0.563 | 0.934 | |
| 8 | TUB | TUB | TBPII | ACT | UBC | UBC | PGD | GAPD | TUB | EF1 | GAPD | UBC | CYP |
| 0.236 | 0.242 | 0.452 | 0.449 | 0.360 | 0.497 | 0.565 | 0.517 | 0.322 | 0.460 | 0.481 | 0.664 | 1.048 | |
| 9 | UBC | EF1 | UBC | TUB | TUB | TUB | TBPII | EF1 | CYP | TUB | PGD | GAPD | PGD |
| 0.254 | 0.271 | 0.507 | 0.565 | 0.413 | 0.583 | 0.632 | 0.591 | 0.378 | 0.528 | 0.544 | 0.795 | 1.143 | |
| Rank | Osmotic Stress | Salt Stress | H. Metal Stress | Cold Stress | Heat Stress | All Samples | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | ||
| 1 | UBC | UBC | PGD | UBC | ADPR | GAPD | ADPR | ADPR | ADPR | UBC | UBC/ADPR | EF1 | ACT |
| 0.105 | 0.078 | 0.116 | 0.134 | 0.146 | 0.177 | 0.111 | 0.218 | 0.121 | 0.129 | 0.197 | 0.183 | 0.517 | |
| 2 | ACT | ADPR | UBC | ADPR | ACT | PGD | UBC | CYP | PGD | ACT | CYP | ADPR | ADPR |
| 0.113 | 0.079 | 0.130 | 0.135 | 0.150 | 0.208 | 0.125 | 0.229 | 0.123 | 0.140 | 0.206 | 0.191 | 0.572 | |
| 3 | ADPR | GAPD | CYP | PGD | EF1 | ADPR | ACT | PGD | UBC | ADPR | ACT | PGD | EF1 |
| 0.127 | 0.090 | 0.134 | 0.167 | 0.177 | 0.232 | 0.140 | 0.271 | 0.124 | 0.146 | 0.227 | 0.21 | 0.601 | |
| 4 | PGD | EF1 | GAPD | EF1 | TUB | CYP | CYP | TBPII | GAPD | GAPD | GAPD | TUB | GAPD |
| 0.149 | 0.131 | 0.339 | 0.232 | 0.217 | 0.262 | 0.170 | 0.304 | 0.190 | 0.181 | 0.276 | 0.339 | 0.723 | |
| 5 | TBPII | PGD | ADPR | GAPD | GAPD | EF1 | EF1 | TUB | CYP | CYP | TBPII | ACT | TBPII |
| 0.168 | 0.151 | 0.465 | 0.394 | 0.272 | 0.294 | 0.217 | 0.314 | 0.250 | 0.220 | 0.388 | 0.431 | 0.949 | |
| 6 | TUB | ACT | ACT | ACT | CYP | ACT | TUB | ACT | EF1 | TUB | PGD | CYP | UBC |
| 0.191 | 0.175 | 0.531 | 0.453 | 0.430 | 0.323 | 0.265 | 0.369 | 0.312 | 0.246 | 0.481 | 0.521 | 1.027 | |
| 7 | GAPD | CYP | TUB | TUB | PGD | UBC | PGD | UBC | ACT | TBPII | EF1 | UBC | CYP |
| 0.212 | 0.218 | 0.569 | 0.511 | 0.489 | 0.395 | 0.384 | 0.396 | 0.348 | 0.352 | 0.647 | 0.666 | 1.106 | |
| 8 | EF1 | TUB | TBPII | TBPII | UBC | TBPII | TBPII | GAPD | TUB | PGD | TUB | TBPII | PGD |
| 0.227 | 0.267 | 0.615 | 0.572 | 0.531 | 0.449 | 0.466 | 0.495 | 0.413 | 0.408 | 0.772 | 0.733 | 1.197 | |
| 9 | CYP | TBPII | EF1 | CYP | TBPII | TUB | GAPD | EF1 | TBPII | EF1 | GAPD | TUB | |
| 0.241 | 0.314 | 0.706 | 0.628 | 0.571 | 0.525 | 0.580 | 0.579 | 0.491 | 0.502 | 0.806 | 1.299 | ||
| Rank | Osmotic Stress | Salt Stress | H. Metal Stress | Cold Stress | Heat Stress | All Samples | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | ||
| 1 | PGD | ADPR | GAPD | PGD | CYP | EF1 | ADPR | ACT | ADPR | ADPR | ADPR | ADPR | ADPR |
| 0.096 | 0.032 | 0.049 | 0.090 | 0.120 | 0.103 | 0.121 | 0.070 | 0.117 | 0.091 | 0.079 | 0.091 | 0.178 | |
| 2 | TBPII | GAPD | CYP | ADPR | GAPD | PGD | UBC | CYP | EF1 | CYP | UBC | EF1 | TBPII |
| 0.112 | 0.053 | 0.196 | 0.216 | 0.126 | 0.108 | 0.148 | 0.089 | 0.142 | 0.133 | 0.113 | 0.169 | 0.357 | |
| 3 | CYP | PGD | PGD | TBPII | ADPR | CYP | CYP | ADPR | PGD | GAPD | ACT | PGD | UBC |
| 0.119 | 0.067 | 0.218 | 0.257 | 0.145 | 0.191 | 0.152 | 0.127 | 0.146 | 0.155 | 0.211 | 0.215 | 0.374 | |
| 4 | ADPR | UBC | ADPR | GAPD | ACT | GAPD | ACT | UBC | GAPD | UBC | TBPII | ACT | ACT |
| 0.124 | 0.074 | 0.251 | 0.268 | 0.171 | 0.243 | 0.216 | 0.166 | 0.173 | 0.173 | 0.279 | 0.337 | 0.400 | |
| 5 | ACT | ACT | ACT | ACT | EF1 | ADPR | TUB | TBPII | ACT | ACT | TUB | CYP | EF1 |
| 0.127 | 0.102 | 0.276 | 0.307 | 0.188 | 0.247 | 0.338 | 0.348 | 0.178 | 0.218 | 0.280 | 0.406 | 0.496 | |
| 6 | EF1 | TBPII | EF1 | UBC | PGD | ACT | GAPD | TUB | UBC | TBPII | GAPD | TBPII | GAPD |
| 0.137 | 0.120 | 0.330 | 0.308 | 0.213 | 0.284 | 0.357 | 0.392 | 0.219 | 0.383 | 0.293 | 0.413 | 0.588 | |
| 7 | GAPD | CYP | TUB | CYP | TBPII | TBPII | EF1 | PGD | TUB | PGD | CYP | TUB | CYP |
| 0.139 | 0.138 | 0.369 | 0.308 | 0.270 | 0.417 | 0.463 | 0.408 | 0.232 | 0.409 | 0.332 | 0.427 | 0.691 | |
| 8 | TUB | TUB | TBPII | EF1 | UBC | UBC | PGD | GAPD | TBPII | EF1 | EF1 | UBC | TUB |
| 0.151 | 0.153 | 0.382 | 0.349 | 0.302 | 0.444 | 0.478 | 0.450 | 0.241 | 0.437 | 0.380 | 0.694 | 0.743 | |
| 9 | UBC | EF1 | UBC | TUB | TUB | TUB | TBPII | EF1 | CYP | TUB | PGD | GAPD | PGD |
| 0.173 | 0.157 | 0.408 | 0.587 | 0.354 | 0.584 | 0.493 | 0.492 | 0.349 | 0.461 | 0.507 | 0.769 | 0.884 | |
| Rank | Osmotic Stress | Salt Stress | H. Metal Stress | Cold Stress | Heat Stress | All Samples | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | ||
| 1 | ACT | GAPD | GAPD | PGD | GAPD | GAPD | ADPR | ADPR | ADPR | ADPR | CYP | ADPR | ADPR |
| 0.034 | 0.008 | 0.137 | 0.081 | 0.184 | 0.055 | 0.057 | 0.102 | 0.090 | 0.034 | 0.087 | 0.041 | 0.207 | |
| 2 | UBC | UBC | ADPR | EF1 | ACT | ADPR | UBC | UBC | PGD | GAPD | ADPR | EF1 | ACT |
| 0.036 | 0.014 | 0.243 | 0.091 | 0.219 | 0.110 | 0.071 | 0.149 | 0.125 | 0.107 | 0.095 | 0.052 | 0.393 | |
| 3 | ADPR | ADPR | ACT | ADPR | ADPR | PGD | ACT | CYP | CYP | UBC | UBC | PGD | TBPII |
| 0.082 | 0.021 | 0.317 | 0.183 | 0.253 | 0.132 | 0.134 | 0.163 | 0.149 | 0.146 | 0.137 | 0.146 | 0.467 | |
| 4 | PGD | EF1 | PGD | UBC | PGD | EF1 | CYP | ACT | GAPD | ACT | ACT | TUB | UBC |
| 0.094 | 0.025 | 0.342 | 0.253 | 0.278 | 0.161 | 0.184 | 0.174 | 0.171 | 0.148 | 0.208 | 0.394 | 0.527 | |
| 5 | GAPD | PGD | CYP | GAPD | CYP | CYP | TUB | PGD | UBC | CYP | GAPD | CYP | EF1 |
| 0.100 | 0.041 | 0.344 | 0.337 | 0.304 | 0.223 | 0.192 | 0.339 | 0.188 | 0.159 | 0.237 | 0.401 | 0.581 | |
| 6 | TUB | ACT | UBC | ACT | UBC | ACT | EF1 | TUB | EF1 | TUB | TBPII | ACT | GAPD |
| 0.113 | 0.072 | 0.354 | 0.441 | 0.336 | 0.270 | 0.283 | 0.370 | 0.248 | 0.224 | 0.512 | 0.479 | 0.662 | |
| 7 | TBPII | CYP | TUB | TBPII | EF1 | UBC | PGD | TBPII | ACT | PGD | PGD | UBC | CYP |
| 0.120 | 0.117 | 0.385 | 0.518 | 0.367 | 0.349 | 0.410 | 0.385 | 0.282 | 0.399 | 0.575 | 0.583 | 0.714 | |
| 8 | EF1 | TUB/TBPII | TBPII | CYP | TUB | TBPII | TBPII | GAPD | TUB | TBPII | EF1 | GAPD | PGD |
| 0.124 | 0.175 | 0.570 | 0.551 | 0.431 | 0.431 | 0.538 | 0.395 | 0.418 | 0.413 | 0.609 | 0.619 | 0.890 | |
| 9 | CYP | EF1 | TUB | TBPII | TUB | GAPD | EF1 | TBPII | EF1 | TUB | TBPII | TUB | |
| 0.132 | 0.615 | 0.638 | 0.452 | 0.493 | 0.617 | 0.561 | 0.457 | 0.528 | 0.795 | 0.662 | 0.945 | ||
| Rank | Osmotic Stress | Salt Stress | H. Metal Stress | Cold Stress | Heat Stress | All Samples | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | ||
| 1 | PGD | TBPII | CYP | CYP | UBC | PGD | PGD | TUB | PGD | PGD | TBPII | ADPR | PGD |
| 0.36 ± 0.08 | 0.23 ± 0.06 | 0.58 ± 0.11 | 0.99 ± 0.19 | 0.60 ± 0.13 | 0.66 ± 0.14 | 0.80 ± 0.17 | 0.40 ± 0.08 | 0.67 ± 0.14 | 0.49 ± 0.11 | 0.22 ± 0.06 | 1.16 ± 0.22 | 1.49 ± 0.32 | |
| 2 | TUB | PGD | TBPII | UBC | TBPII | CYP | TBPII | TBPII | TBPII | TBPII | GAPD | PGD | CYP |
| 0.52 ± 0.11 | 0.43 ± 0.09 | 0.72 ± 0.18 | 1.00 ± 0.22 | 0.78 ± 0.19 | 0.98 ± 0.19 | 1.06 ± 0.27 | 0.47 ± 0.12 | 0.94 ± 0.23 | 0.75 ± 0.19 | 0.46 ± 0.09 | 1.23 ± 0.27 | 2.44 ± 0.47 | |
| 3 | CYP | UBC | PGD | TBPII | PGD | EF1 | UBC | PGD | UBC | UBC | UBC | EF1 | TBPII |
| 0.64 ± 0.13 | 0.76 ± 0.16 | 0.79 ± 0.17 | 1.10 ± 0.27 | 1.09 ± 0.23 | 1.15 ± 0.21 | 1.35 ± 0.30 | 0.69 ± 0.15 | 1.14 ± 0.24 | 1.40 ± 0.31 | 1.02 ± 0.23 | 1.49 ± 0.27 | 2.78 ± 0.69 | |
| 4 | EF1 | ADPR | UBC | EF1 | CYP | ADPR | ACT | ADPR | EF1 | ADPR | PGD | TBPII | UBC |
| 0.64 ± 0.14 | 1.03 ± 0.19 | 1.15 ± 0.26 | 1.23 ± 0.23 | 1.67 ± 0.32 | 1.40 ± 0.26 | 1.57 ± 0.33 | 1.25 ± 0.26 | 1.49 ± 0.28 | 1.95 ± 0.41 | 1.13 ± 0.24 | 1.71 ± 0.41 | 3.12 ± 0.68 | |
| 5 | TBPII | CYP | GAPD | ADPR | ACT | TBPII | ADPR | ACT | ADPR | GAPD | ADPR | CYP | ADPR |
| 0.67 ± 0.17 | 1.32 ± 0.25 | 1.17 ± 0.24 | 1.57 ± 0.30 | 2.33 ± 0.49 | 1.69 ± 0.40 | 1.80 ± 0.38 | 1.66 ± 0.35 | 1.52 ± 0.29 | 2.00 ± 0.41 | 1.40 ± 0.29 | 2.11 ± 0.40 | 4.75 ± 0.95 | |
| 6 | GAPD | GAPD | ACT | PGD | EF1 | UBC | CYP | CYP | GAPD | CYP | ACT | ACT | ACT |
| 0.80 ± 0.16 | 1.35 ± 0.22 | 2.72 ± 0.57 | 2.20 ± 0.48 | 2.40 ± 0.53 | 2.36 ± 0.50 | 1.90 ± 0.37 | 1.97 ± 0.38 | 1.83 ± 0.30 | 2.63 ± 0.51 | 2.93 ± 0.62 | 2.78 ± 0.50 | 6.91 ± 1.36 | |
| 7 | ADPR | ACT | ADPR | ACT | ADPR | GAPD | TUB | UBC | CYP | ACT | TUB | TUB | EF1 |
| 1.01 ± 0.22 | 1.37 ± 0.24 | 2.77 ± 0.58 | 4.51 ± 0.83 | 2.55 ± 0.53 | 2.43 ± 0.41 | 3.36 ± 0.71 | 2.56 ± 0.57 | 1.85 ± 0.36 | 2.65 ± 0.56 | 3.13 ± 0.64 | 3.11 ± 0.51 | 7.32 ± 1.48 | |
| 8 | ACT | TUB | EF1 | GAPD | GAPD | ACT | GAPD | EF1 | ACT | TUB | EF1 | UBC | GAPD |
| 1.05 ± 0.23 | 1.71 ± 0.28 | 3.25 ± 0.71 | 4.68 ± 0.80 | 2.74 ± 0.56 | 2.49 ± 0.44 | 4.31 ± 0.86 | 4.77 ± 1.04 | 2.15 ± 0.38 | 4.33 ± 0.91 | 4.06 ± 0.90 | 3.98 ± 0.82 | 7.88 ± 1.48 | |
| 9 | UBC | EF1 | TUB | TUB | TUB | TUB | EF1 | GAPD | TUB | EF1 | CYP | GAPD | TUB |
| 1.14 ± 0.26 | 2.04 ± 0.37 | 3.57 ± 0.75 | 7.20 ± 1.24 | 3.76 ± 0.79 | 4.46 ± 0.75 | 4.47 ± 0.96 | 4.86 ± 0.93 | 2.37 ± 0.38 | 4.36 ± 0.94 | 4.10 ± 0.78 | 6.04 ± 1.05 | 9.62 ± 1.84 | |
| Rank | Osmotic Stress | Salt Stress | H. Metal Stress | Cold Stress | Heat Stress | All Samples | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | ||
| 1 | ADPR | PGD | TBPII | TBPII | TBPII | CYP | PGD | TBPII | PGD | TBPII | TBPII | EF1 | PGD |
| 0.50 ± 0.11 | 1.21 ± 0.26 | 0.67 ± 0.17 | 1.04 ± 0.25 | 0.62 ± 0.16 | 0.78 ± 0.15 | 1.09 ± 0.23 | 0.30 ± 0.08 | 0.63 ± 0.13 | 0.70 ± 0.18 | 0.45 ± 0.11 | 0.90 ± 0.16 | 1.45 ± 0.31 | |
| 2 | TBPII | CYP | PGD | UBC | UBC | PGD | TBPII | PGD | UBC | PGD | PGD | ADPR | CYP |
| 0.51 ± 0.13 | 1.45 ± 0.28 | 0.81 ± 0.17 | 1.06 ± 0.23 | 0.99 ± 0.22 | 1.11 ± 0.24 | 1.10 ± 0.28 | 0.35 ± 0.07 | 1.01 ± 0.22 | 0.97 ± 0.21 | 0.64 ± 0.14 | 0.93 ± 0.17 | 2.31 ± 0.44 | |
| 3 | PGD | EF1 | UBC | PGD | PGD | ADPR | TUB | TUB | ADPR | CYP | GAPD | CYP | TBPII |
| 0.67 ± 0.14 | 1.46 ± 0.27 | 0.83 ± 0.19 | 1.18 ± 0.25 | 1.08 ± 0.23 | 1.36 ± 0.25 | 1.32 ± 0.28 | 0.97 ± 0.21 | 1.07 ± 0.20 | 1.32 ± 0.26 | 1.87 ± 0.38 | 1.11 ± 0.21 | 2.76 ± 0.69 | |
| 4 | UBC | UBC | CYP | ADPR | CYP | GAPD | UBC | ADPR | CYP | ADPR | UBC | PGD | UBC |
| 0.72 ± 0.16 | 1.51 ± 0.33 | 1.04 ± 0.20 | 1.28 ± 0.24 | 1.20 ± 0.23 | 1.40 ± 0.22 | 1.99 ± 0.44 | 1.64 ± 0.35 | 1.19 ± 0.23 | 1.61 ± 0.35 | 2.32 ± 0.51 | 1.46 ± 0.32 | 2.78 ± 0.60 | |
| 5 | TUB | GAPDH | GAPD | CYP | GAPD | TBPII | ADPR | CYP | TBPII | GAPD | ADPR | TBPII | ADPR |
| 0.97 ± 0.21 | 1.57 ± 0.25 | 3.01 ± 0.62 | 1.51 ± 0.28 | 3.53 ± 0.73 | 1.82 ± 0.44 | 2.25 ± 0.48 | 1.83 ± 0.36 | 1.40 ± 0.34 | 1.76 ± 0.37 | 2.93 ± 0.62 | 2.58 ± 0.62 | 5.92 ± 1.19 | |
| 6 | ACT | ADPR | ADPR | EF1 | ACT | EF1 | ACT | ACT | GAPD | UBC | CYP | UBC | ACT |
| 0.98 ± 0.22 | 1.80 ± 0.34 | 4.00 ± 0.85 | 2.21 ± 0.41 | 3.99 ± 0.84 | 1.89 ± 0.34 | 2.82 ± 0.60 | 2.66 ± 0.57 | 1.73 ± 0.28 | 1.94 ± 0.43 | 3.55 ± 0.68 | 3.21 ± 0.66 | 7.29 ± 1.44 | |
| 7 | EF1 | TBPII | ACT | GAPD | ADPR | UBC | CYP | UBC | EF1 | TUB | ACT | TUB | EF1 |
| 1.05 ± 0.25 | 1.94 ± 0.48 | 4.69 ± 0.98 | 4.64 ± 0.78 | 4.30 ± 0.91 | 1.91 ± 0.40 | 2.94 ± 0.57 | 2.90 ± 0.63 | 2.17 ± 0.40 | 2.31 ± 0.49 | 4.01 ± 0.84 | 3.32 ± 0.53 | 8.20 ± 1.69 | |
| 8 | GAPD | ACT | TUB | ACT | EF1 | ACT | EF1 | GAPD | ACT | ACT | EF1 | ACT | GAPD |
| 1.31 ± 0.28 | 2.34 ± 0.41 | 5.07 ± 1.05 | 4.72 ± 0.84 | 4.66 ± 1.04 | 1.97 ± 0.34 | 3.02 ± 0.69 | 5.11 ± 1.00 | 2.58 ± 0.45 | 2.34 ± 0.50 | 6.5 ± 1.44 | 4.16 ± 0.74 | 8.95 ± 1.69 | |
| 9 | CYP | TUB | EF1 | TUB | TUB | TUB | GAPD | EF1 | TUB | EF1 | TUB | GAPD | TUB |
| 1.71 ± 0.34 | 2.85 ± 0.45 | 6.38 ± 1.41 | 6.10 ± 1.00 | 5.24 ± 1.09 | 3.59 ± 0.57 | 6.06 ± 1.22 | 5.14 ± 1.15 | 3.97 ± 0.64 | 4.45 ± 1.00 | 8.19 ± 1.65 | 5.77 ± 0.98 | 11.43 ± 2.19 | |
| Rank | Osmotic Stress | Salt Stress | H. Metal Stress | Cold Stress | Heat Stress | All Samples | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | |||
| 1 | PGD | ADPR | GAPD | ADPR | GAPD | PGD | ADPR | ADPR | ADPR | ADPR | UBC | ADPR | ADPR | |
| 2 | TBPII | GAPD | CYP | PGD | ADPR | CYP | UBC | ACT | PGD | UBC | TBPII | EF1 | TBPII | |
| 3 | EF1 | PGD | PGD | CYP | CYP | EF1 | ACT | CYP | GAPD | GAPD | ADPR | PGD | UBC | |
| 4 | TUB | UBC | ADPR | UBC | ACT | GAPD | CYP | TUB | UBC | CYP | GAPD | ACT | ACT | |
| 5 | CYP | TBPII | ACT | TBPII | PGD | ACT | PGD | TBPII | EF1 | PGD | ACT | CYP | PGD | |
| 6 | ADPR | ACT | EF1 | EF1 | UBC | ADPR | TUB | UBC | TBPII | TBPII | TUB | TUB | CYP | |
| 7 | GAPD | CYP | TBPII | GAPD | TBPII | TBPII | TBPII | PGD | ACT | ACT | CYP | TBPII | EF1 | |
| 8 | ACT | TUB | TUB | ACT | EF1 | UBC | GAPD | GAPD | TUB | TUB | PGD | UBC | GAPD | |
| 9 | UBC | EF1 | UBC | TUB | TUB | TUB | EF1 | EF1 | CYP | EF1 | EF1 | GAPD | TUB | |
| Rank | Osmotic Stress | Salt Stress | H. Metal Stress | Cold Stress | Heat Stress | All Samples | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | Leaves (h) | Leaves (s) | Roots (h) | ||
| 1 | ACT | GAPD | GAPD | UBC | ACT | GAPD | ADPR | ADPR | ADPR | ADPR | UBC | ADPR | ADPR |
| 2 | UBC | UBC | CYP | PGD | GAPD | PGD | UBC | CYP | PGD | UBC | ADPR | EF1 | ACT |
| 3 | ADPR | ADPR | PGD | ADPR | ADPR | CYP | ACT | PGD | UBC | GAPD | CYP | PGD | TBPII |
| 4 | PGD | EF1 | ADPR | EF1 | PGD | ADPR | TUB | TBPII | GAPD | ACT | TBPII | CYP | UBC |
| 5 | TBPII | PGD | UBC | TBPII | UBC | EF1 | PGD | UBC | CYP | CYP | GAPD | TUB | PGD |
| 6 | TUB | CYP | ACT | GAPD | CYP | ACT | CYP | TUB | EF1 | TBPII | ACT | ACT | CYP |
| 7 | GAPD | ACT | TBPII | CYP | TBPII | UBC | EF1 | ACT | ACT | PGD | PGD | UBC | EF1 |
| 8 | EF1 | TUB | TUB | ACT | EF1 | TBPII | TBPII | GAPD | TBPII | TUB | EF1 | TBPII | GAPD |
| 9 | CYP | TBPII | EF1 | TUB | TUB | TUB | GAPD | EF1 | TUB | EF1 | TUB | GAPD | TUB |
| Gene Abb. | Gene Name | Primer Sequence Forward and Reverse | Amplicon Length (bp) | Efficiency | R2 | Accession No. |
|---|---|---|---|---|---|---|
| ACT | Actin1 | CGAGCGGGAAATTGTAAGGG | 191 | 92% | 0.994 | MF405765.1 |
| CGATCATGGATGGCTGGAAG | ||||||
| TUB | α-Tubulin | AGGTCTTCTCCCGCATCG | 90 | 98% | 0.995 | U76558.1 |
| CCTCCTCCATGCCCTCAC | ||||||
| CYP | Cyclophilin | AGTCCATCTACGGCGAGAAGT | 120 | 93% | 0.998 | EU035525.1 |
| GGGACGGTGCAGATGAAGAA | ||||||
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | GTTTGGCATCGTTGAGGGTT | 131 | 93% | 0.998 | KR029492.1 |
| TGCTGCTGGGAATGATGTTG | ||||||
| UBC | Ubiquitin conjugating enzyme | CAAGCTGACCCTGCAATTCA | 135 | 91% | 0.992 | M62720.1 |
| GGGCTCCACTGGTTCTGTA | ||||||
| EF1 | Elongation factor 1-α | AAGGAGGCAGCCAACTTCA | 122 | 96% | 0.999 | M90077.2 |
| AGCTCAGCAAACTTGACAGC | ||||||
| TBPII | TATA-binding protein II subunit | GATGAGGCAGCCGAAGATTG | 156 | 91% | 0.993 | L07604.1 |
| TCCAAAGTCAACCATCATTGCT | ||||||
| ADPR | ADP-ribosylation factor | CTCATGGTTGGTCTCGATGC | 143 | 94% | 0.998 | Ta2291 (Unigene cluster) |
| ACATCCCAAACAGTGAAGCT | ||||||
| PGD | Phosphogluconate dehydrogenase | GCAAAGATGAAACTGGTGGTCA | 90 | 93% | 0.995 | Ta30797 (Unigene cluster) |
| CAACCCACTTTTGTCCGCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tajti, J.; Pál, M.; Janda, T. Validation of Reference Genes for Studying Different Abiotic Stresses in Oat (Avena sativa L.) by RT-qPCR. Plants 2021, 10, 1272. https://doi.org/10.3390/plants10071272
Tajti J, Pál M, Janda T. Validation of Reference Genes for Studying Different Abiotic Stresses in Oat (Avena sativa L.) by RT-qPCR. Plants. 2021; 10(7):1272. https://doi.org/10.3390/plants10071272
Chicago/Turabian StyleTajti, Judit, Magda Pál, and Tibor Janda. 2021. "Validation of Reference Genes for Studying Different Abiotic Stresses in Oat (Avena sativa L.) by RT-qPCR" Plants 10, no. 7: 1272. https://doi.org/10.3390/plants10071272
APA StyleTajti, J., Pál, M., & Janda, T. (2021). Validation of Reference Genes for Studying Different Abiotic Stresses in Oat (Avena sativa L.) by RT-qPCR. Plants, 10(7), 1272. https://doi.org/10.3390/plants10071272








