Integrative Taxonomy Reveals Hidden Cryptic Diversity within Pin Nematodes of the Genus Paratylenchus (Nematoda: Tylenchulidae)
Abstract
:1. Introduction
2. Results
2.1. Systematics
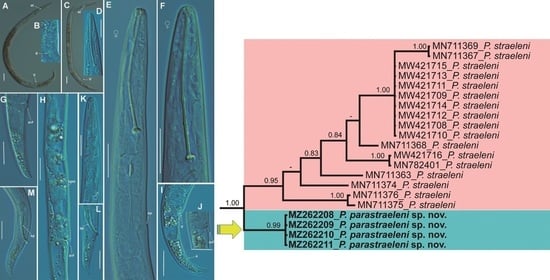
2.1.1. Description of Paratylenchus parastraeleni sp. nov.
Diagnosis and Relationships
Molecular Characterization
Type Habitat and Locality
Etymology
Type Material
2.1.2. Remarks of Paratylenchus aciculus Brown, 1959
Molecular Characterization
2.1.3. Remarks of Paratylenchus amundseni Bernard, 1982
Molecular Characterization
2.1.4. Remarks on Paratylenchus baldaccii (Oostenbrink, 1953) Raski, 1962, Paratylenchus enigmaticus Munawar, Yevtushenko, Palomares-Rius and Castillo, 2021, Paratylenchus holdemani Raski, 1975, Paratylenchus neoamblycephalus Geraert, 1965, Paratylenchus pedrami Clavero-Camacho, Cantalapiedra-Navarrete, Archidona-Yuste, Castillo and Palomares-Rius, 2021, and Paratylenchus veruculatus Wu, 1962
Molecular Characterization
2.1.5. Remarks on Paratylenchus goodeyi Oostenbrink, 1953
Molecular Characterization
2.1.6. Remarks on Paratylenchus macrodorus Brzeski, 1963
Molecular Characterization
2.1.7. Remarks on Paratylenchus pandatus (Raski, 1976) Siddiqi, 1986
Molecular Characterization
2.1.8. Remarks on Paratylenchus recisus Siddiqi, 1996
Molecular Characterization
2.1.9. Remarks on Paratylenchus sheri (Raski, 1973) Siddiqi, 1986
Molecular Characterization
2.1.10. Remarks on Paratylenchus variabilis Raski, 1975
Molecular Characterization
2.1.11. Remarks on Paratylenchus verus (Brzeski, 1995) Brzeski, 1998
Molecular Characterization
2.1.12. Remarks on Paratylenchus vitecus (Pramodini et al., 2006) Ghaderi et al., 2014
Molecular Characterization
2.2. Distribution of Paratylenchus spp. in Spain
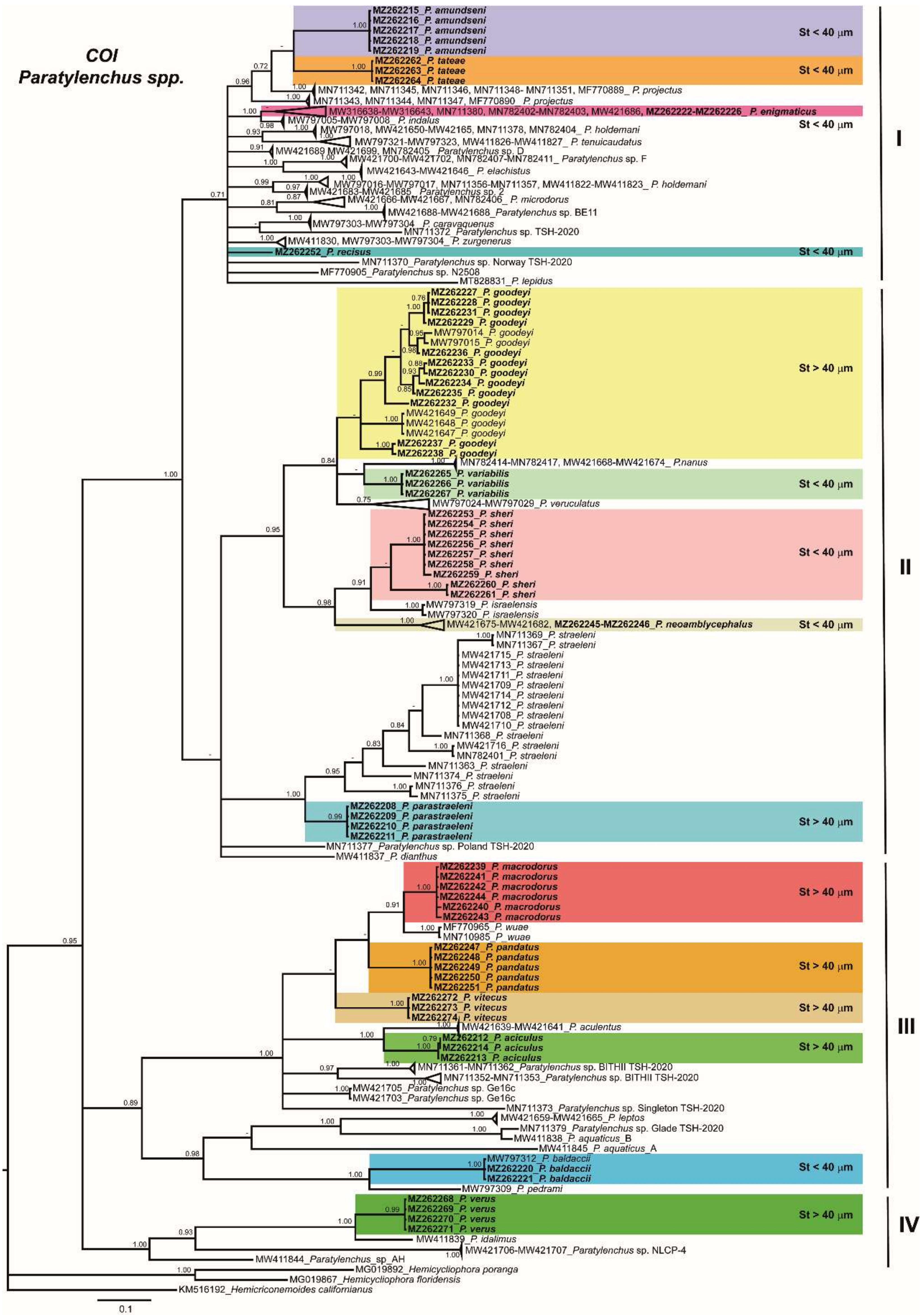
2.3. Phylogenetic Analyses of Paratylenchus spp.
3. Discussion
4. Materials and Methods
4.1. Nematode Sampling and Morphological Identification
4.2. Nematode Molecular Characterization
4.3. Phylogenetic Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Micoletzky, H. Die freilebenden Erd-Nematoden. Arch. Für Nat. 1922, 87, 1–650. [Google Scholar]
- Ghaderi, R.; Geraert, E.; Karegar, A. The Tylenchulidae of the World, Identification of the Family Tylenchulidae (Nematoda: Tylenchida); Academia Press: Ghent, Belgium, 2016. [Google Scholar]
- Singh, R.; Karssen, G.; Coureur, M.; Subbotin, S.; Bert, W. Integrative taxonomy and molecular phylogeny of the plant-parasitic nematode genus Paratylenchus (Nematoda: Paratylenchinae): Linking species with molecular barcodes. Plants 2021, 10, 408. [Google Scholar] [CrossRef] [PubMed]
- Clavero-Camacho, I.; Cantalapiedra-Navarrete, C.; Archidona-Yuste, A.; Castillo, P.; Palomares-Rius, J.E. Remarkable cryptic diversity of Paratylenchus spp. (Nematoda: Tylenchulidae) in Spain. Animals 2021, 11, 1161. [Google Scholar] [CrossRef] [PubMed]
- Raski, D.J. Paratylenchidae n.fam. with descriptions of five new species of Gracilacus n.g. and an emendation of Cacopaurus Thorne, 1943, Paratylenchus Micoletzky, 1922 and Criconematidae Thorne, 1943. Proc. Helminthol. Soc. Wash. 1962, 29, 189–207. [Google Scholar]
- Van den Berg, E.; Tiedt, L.R.; Subbotin, S.A. Morphological and molecular characterisation of several Paratylenchus Micoletzky, 1922 (Tylenchida: Paratylenchidae) species from South Africa and USA, together with some taxonomic notes. Nematology 2014, 16, 323–358. [Google Scholar] [CrossRef] [Green Version]
- Munawar, M.; Miao, W.; Castillo, P.; Zheng, J.-W. A new pin nematode, Paratylenchus sinensis n. sp. (Nematoda: Paratylenchinae) in the rhizosphere of white mulberry from Zhejiang Province, China. Eur. J. Plant Pathol. 2020, 156, 1023–1029. [Google Scholar]
- Munawar, M.; Yevtushenko, D.P.; Palomares-Rius, J.E.; Castillo, P. Species diversity of pin nematodes (Paratylenchus spp.) from potato growing regions of southern Alberta, Canada. Plants 2021, 10, 188. [Google Scholar] [CrossRef]
- Castillo, P.; Gómez-Barcina, A. Some species of Tylenchida from natural habitats in southeastern Spain. Nematol. Medit. 1988, 16, 75–86. [Google Scholar]
- Akyazi, F.; Felek, A.F.; Čermák, V.; Čudejková, M.; Foit, J.; Yildiz, S.; Háněl, L. Description of Paratylenchus (Gracilacus) straeleni (De Coninck, 1931) Oostenbrink, 1960 (Nematoda: Criconematoidea, Tylenchulidae) from hazelnut in Turkey and its comparison with other world populations. Helminthologia 2015, 52, 270–279. [Google Scholar] [CrossRef] [Green Version]
- Powers, T.O.; Harris, T.S.; Higgins, R.S.; Mullin, P.G.; Powers, K.S. Nematode biodiversity assessments need vouchered databases: A BOLD reference library for plant-parasitic nematodes in the superfamily Criconematoidea. Genome 2020, 64, 232–241. [Google Scholar] [CrossRef]
- Hernandez, M.; Mateo, M.D.; Jordana, R. Estudio comparativo entre grupos tróficos de Nematodos del suelo de cinco bosques de Navarra (tres naturales y dos de repoblación). Actas II Congr. Mund. Vasco. Sec. Biol. Ambient. 1988, 2, 323–335. [Google Scholar]
- Brzeski, M.; Hanel, L.; Nico, A.; Castillo, P. Paratylenchinae: Redescription of Paratylenchus arculatus Luc & de Guiran, 1962, a new senior synonym of P. nainianus Edward & Misra, 1963 (Nematoda: Tylenchulidae). Nematology 1999, 1, 375–380. [Google Scholar]
- Nico, A.I.; Rapoport, H.F.; Jiménez-Díaz, R.M.; Castillo, P. Incidence and population density of plant-parasitic nematodes associated with olive planting stocks at nurseries in southern Spain. Plant Dis. 2002, 86, 1075–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peña-Santiago, R. Plant-parasitic nematodes associated with olive (Olea europea L.) in the province of Jaén, Spain. Rev. Nématol. 1990, 13, 113–115. [Google Scholar]
- Archidona-Yuste, A.; Wiegand, T.; Castillo, P.; Navas-Cortés, J.A. Dataset on the diversity of plant-parasitic nematodes in cultivated olive trees in southern Spain. Data Brief 2019, 27, 104658. [Google Scholar] [CrossRef]
- Gomez-Barcina, A.; Castillo, P.; Pais, M.A.G. Four species of the genus Paratylenchus Micoletzky from southeasthern Spain. Nematol. Medit. 1990, 18, 169–177. [Google Scholar]
- Talavera, M.; Navas, A. Incidence of plant-parasitic nematodes in natural and semi-natural mountain grassland and the host status of some common grass species. Nematology 2002, 4, 541–552. [Google Scholar]
- Escuer, M. Els Nematodes. In El Patrimoni Biològic del Montseny. Catàleg de Flora i Fauna; Servei de Parcs Naturals, Diputació de Barcelona: Barcelona, Spain, 1995; Volume 2, pp. 17–22. [Google Scholar]
- Castillo, P.; González-País, M.A.; Gómez-Barcina, A. El género Gracilacus Raski, 1962 en España (Paratylenchinae: Tylenchida). Rev. Ibér. Parasitol. 1989, 49, 321–328. [Google Scholar]
- Gomez-Barcina, A.; Castillo, P.; González-País, M.A. Nematodos fitoparásitos de la subfamilia Criconematinae Taylor, 1936 en la Sierra de Cazorla. Rev. Ibér. Parasitol. 1989, 49, 241–255. [Google Scholar]
- Talavera, M.; Tobar Jimenez, A. Plant parasitic nematodes from unirrigated fields in Alhama, southeastern Spain. Nematol. Medit. 1997, 25, 73–81. [Google Scholar]
- Imaz, A.; Hernández, M.A.; Ariño, A.H.; Armendáriz, I.; Jordana, R. Diversity of soil nematodes across a Mediterranean ecotone. Appl. Soil Ecol. 2002, 20, 191–198. [Google Scholar] [CrossRef]
- Peña Santiago, R.; Geraert, E. New data on Aorolaimus perscitus (Doucet, 1980) and Gracilacus teres Raski, 1976 (Nematoda: Tylenchida) associated with olive (Olea europea L.) in the province of Jaén, Spain. Nematologica 1990, 36, 408–416. [Google Scholar]
- Brzeski, M.W. Seasonal dynamics of Paratylenchus bukowinensis Micol. and some other nematodes. Rocz. Nauk Rol. Ser. E 1977, 7, 67–74. [Google Scholar]
- Brzeski, M.W.; Hanel, L. Paratylenchinae: Postembryonic developmental stages of Paratylenchus straeleni (De Coninck, 1931) and P. steineri Golden, 1961 (Nematoda: Tylenchulidae). Nematology 1999, 1, 673–680. [Google Scholar] [CrossRef]
- De Coninck, L.A.P. Sur trois espèces nouvelles de nématodes libres trouvés en Belgique. Bull. Musée R. d’Hist. Nat. Belg. 1931, 7, 1–15. [Google Scholar]
- Brzeski, M.W. Paratylenchinae: Morphology of some known species and descriptions of Gracilacus Bilineata sp. n. and G. Vera sp. n. (Nematoda: Tylenchulidae). Nematologica 1995, 41, 535–565. [Google Scholar] [CrossRef]
- Brzeski, M.W. Nematodes of Tylenchina in Poland and Temperate Europe; Muzeum i Instytutu Zoologii, PAN: Warsaw, Poland, 1998; p. 397. [Google Scholar]
- Ghaderi, R.; Karegar, A. Some species of Paratylenchus (Nematoda: Tylenchulidae) from Iran. Iran. J. Plant Pathol. 2013, 49, 137–156. [Google Scholar]
- Brown, G.L. Three new species of the genus Paratylenchus from Canada (Nematoda: Criconematidae). Proc. Helminthol. Soc. Wash. 1959, 26, 1–8. [Google Scholar]
- Bernard, E.C. Criconematina (nematoda: Tylenchida) from the aleutian islands. J. Nematol. 1982, 14, 323–331. [Google Scholar]
- Castillo, P.; Gomez-Barcina, A. Plant-parasitic nematodes associated with tropical and subtropical crops in southern Spain. Nematol. Medit. 1993, 21, 45–47. [Google Scholar]
- Escuer, M.; Cano, A.; Bello, A. Nematodos fitoparásitos de la Región de Murcia y alternativas de control. In Desinfección de Suelos en Invernaderos de Pimientos; Serie Jornadas y Congresos, 16; Consejería de Agricultura, Agua y Medioambiente: Murcia, Spain, 2004; pp. 27–57. [Google Scholar]
- Geraert, E. The Genus Paratylenchus. Nematologica 1965, 11, 301–334. [Google Scholar] [CrossRef]
- Oostenbrink, M. A note on Paratylenchus in the Netherlands with the description of P. goodeyi n. sp. (Nematoda, Criconematidae). Tijdschr. Plantenziekten 1953, 59, 207–216. [Google Scholar] [CrossRef]
- Raski, D.J. Revision of the genus Paratylenchus Micoletzky, 1922 and descriptions of new species. Part III of three parts—Gracilacus. J. Nematol. 1976, 8, 97–115. [Google Scholar] [PubMed]
- Yu, Q.; Ye, W.; Powers, T. Morphological and molecular characterization of Gracilacus wuae n. sp. (Nematoda: Criconematoidea) associated with cow parsnip (Heracleum maximum) in Ontario, Canada. J. Nematol. 2016, 48, 203–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brzeski, M. Paratylenchus macrodorus n. sp. (Nematoda, Paratylenchidae), a new plant parasitic nematode from Poland. Bull. L’Acad. Pol. Sci. 1963, 11, 277–280. [Google Scholar]
- Van den Berg, E.; Quénéhervé, P.; Tiedt, L. Two Paratylenchus species (Nemata: Tylenchulidae) from Martinique and New Caledonia. J. Nematode Morphol. Syst. 2006, 9, 1–10. [Google Scholar]
- Nguyen, C.N.; Baldwin, J.G.; Choi, Y.E. New records of Paratylenchus Micoletzky, 1922 (Nematoda: Paratylenchinae) from Viet Nam with description of Paratylenchus lapcaiensis sp. n. J. Nematode Morphol. Syst. 2004, 7, 51–75. [Google Scholar]
- Van Den Berg, E.; Mekete, T.; Tiedt, L.R. New records of Criconematidae (Nemata) from Ethiopia. J. Nematode Morphol. Syst. 2004, 6, 161–174. [Google Scholar]
- Siddiqi, M. Paratylenchus recisus sp.n. and P. perminimus sp.n. (Criconematina: Paratylenchidae). Afro-Asian J. Nematol. 1996, 6, 55–58. [Google Scholar]
- Raski, D.J. Paratylenchoides gen. n. and two new species (Nematoda: Paratyleiichidae). Proc. Helminthol. Soc. Wash. 1973, 40, 230–233. [Google Scholar]
- Mirbabaei, H.; Eskandari, A.; Ghaderi, R.; Karegar, A. On the synonymy of Trophotylenchulus asoensis and T. okamotoi with T. arenarius, and intra-generic structure of Paratylenchus (Nematoda: Tylenchulidae). J. Nematol. 2019, 51, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Raski, D.J. Revision of the genus Paratylenchus Micoletzky, 1922 and descriptions of new species. Part I of three parts. J. Nematol. 1975, 7, 15–34. [Google Scholar] [PubMed]
- Pramodini, M.; Mohilal, N.; Dhanachand, C. Gracilacus vitecus sp.n. and record of G. raskii Phukan & Sanwal from Manipur, India. Ind. J. Nematol. 2006, 36, 272–276. [Google Scholar]
- Siddiqi, M.R. Tylenchida: Parasites of Plants and Insects; Commonwealth Institute of Parasitology: Slough, UK, 1986; p. 645. [Google Scholar]
- Raski, D.; Luc, M. A reappraisal of Tylenchina (Nemata): 10. The superfamily Criconematoidea Taylor, 1936. Rev. Nématol. 1987, 10, 409–444. [Google Scholar]
- Derycke, S.; Backeljau, T.; Moens, T. Dispersal and gene flow in free-living marine nematodes. Front. Zool. 2013, 10, 109–152. [Google Scholar] [CrossRef] [Green Version]
- Jex, A.R.; Littlewood, D.T.; Gasser, R.B. Toward next-generation sequencing of mitochondrial genomes-focus on parasitic worms of animals and biotechnological implications. Biotechnol. Adv. 2010, 28, 151–159. [Google Scholar] [CrossRef]
- Coolen, W.A. Methods for extraction of Meloidogyne spp. and other nematodes from roots and soil. In Root-Knot Nematodes (Meloidogyne Species): Systematics, Biology and Control; Lamberti, F., Taylor, C.E., Eds.; Academic Press: New York, NY, USA, 1979; pp. 317–329. [Google Scholar]
- Seinhorst, J.W. Killing nematodes for taxonomic study with hot F.A. 4:1. Nematologica 1966, 12, 178. [Google Scholar] [CrossRef]
- De Grisse, A.T. Redescription ou modifications de quelques techniques utilisées dans l’étude de nématodes phytoparasitaires. Meded. Rijksfac. Landbouwwet. Gent 1969, 34, 315–359. [Google Scholar]
- Hunt, D.J.; Palomares-Rius, J.E. General morphology and morphometries of plant-parasitic nematodes. In Practical Plant Nematology; Biblioteca Basica de Agricultura: Texcoco, Mexico, 2012; pp. 25–64. [Google Scholar]
- Wergin, W.P. Scanning electron microscopic techniques and applications for use in nematology. In Plant Parasitic Nematodes; Zuckerman, B.M., Rohde, R.A., Eds.; Academic Press: New York, NY, USA; London, UK, 1981; Volume 3, pp. 175–204. [Google Scholar]
- Palomares-Rius, J.E.; Clavero-Camacho, I.; Archidona-Yuste, A.; Cantalapiedra-Navarrete, C.; León-Ropero, G.; Braun Miyara, S.; Karssen, G.; Castillo, P. Global distribution of the reniform nematode genus Rotylenchulus with the synonymy of Rotylenchulus macrosoma with Rotylenchulus borealis. Plants 2021, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- De Ley, P.; Felix, M.A.; Frisse, L.; Nadler, S.; Sternberg, P.; Thomas, W.K. Molecular and morphological characterisation of two reproductively isolated species with mirror-image anatomy (Nematoda: Cephalobidae). Nematology 1999, 1, 591–612. [Google Scholar] [CrossRef]
- Subbotin, S.A.; Vierstraete, A.; De Ley, P.; Rowe, J.; Waeyenberge, L.; Moens, M.; Vanfleteren, J.R. Phylogenetic relationships within the cyst-forming nematodes (Nematoda, Heteroderidae) based on analysis of sequences from the ITS regions of ribosomal DNA. Mol. Phylogenet. Evol. 2001, 21, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowles, J.; Blair, D.; McManus, D.P. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol. Biochem. Parasitol. 1992, 54, 165–173. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Subbotin, S.A.; Yan, G.; Kantor, M.; Handoo, Z. On the molecular identity of Paratylenchus nanus Cobb, 1923 (Nematoda: Tylenchida). J. Nematol. 2020, 52, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. FigTree v1.4.2, A Graphical Viewer of Phylogenetic Trees. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 11 June 2021).

















| Species | Sample Code | Locality, Province, Host | D2-D3 | ITS | COI |
|---|---|---|---|---|---|
| 1. P. parastraeleni sp. nov. | CAZ_05 | Arroyo Frío, Jaén, Quercus faginea Lam. | MZ265064-MZ265070 | MZ265004-MZ265007 | MZ262208-MZ262211 |
| 2. P. aciculus Brown, 1959 | CAZ_07 | Coto Ríos, Jaén, Pinus halepensis Mill. | MZ265071-MZ265075 | MZ265008-MZ265011 | MZ262212-MZ262214 |
| 3. P. amundseni Bernard, 1982 | CAZ_02 | La Iruela, Jaén, Pinus halepensis Mill. | MZ265076-MZ265078 | MZ265012-MZ265014 | MZ262215-MZ262219 |
| 4. P. baldaccii (Oostenbrink, 1953) Raski, 1962 | CAZ_04 | Arroyo Frío, Jaén, grasses | MZ265079 | MZ265015-MZ265016 | MZ262220-MZ262221 |
| 5. P. enigmaticus Munawar et al., 2021 | IAS_21 | Córdoba, Córdoba, grasses | MZ265080-MZ265083 | MZ265017-MZ265019 | MZ262222-MZ262226 |
| 6. P. goodeyi Oostenbrink, 1953 | EP_ACA | Córdoba, Córdoba, wild olive | MZ265084-MZ265091 | MZ265020-MZ265024 | MZ262227-MZ262230 |
| AR_097 | Santa Mª de Trasierra, Córdoba, wild olive | MZ265092-MZ265097 | MZ265025-MZ265026 | MZ262231-MZ262233 | |
| PR_050 | Montalbán, Córdoba, almond | MZ265098-MZ265099 | MZ265027 | MZ262234-MZ262235 | |
| PR_017 | Córdoba, Córdoba, almond | MZ265100 | MZ265028 | MZ262236 | |
| PR_076 | Marmolejo, Jaén, almond | MZ265101-MZ265102 | MZ265029-MZ265031 | - * | |
| PR_019 | Córdoba, Córdoba, almond | MZ265103-MZ265105 | MZ265032-MZ265033 | MZ262237-MZ262238 | |
| 7. P. holdemani Raski, 1975 | AR_102 | Santa Mª de Trasierra, Córdoba, wild olive | MZ265106-MZ265107 | - | - |
| 8. P. macrodorus Brzeski, 1963 | AR_102 | Santa Mª de Trasierra, Córdoba, wild olive | MZ265108-MZ265113 | MZ265034-MZ265038 | MZ262239-MZ262244 |
| 9. P. neoamblycephalus Geraert, 1965 | CAZ_05 | Arroyo Frío, Jaén, Quercus faginea Lam. | MZ265114-MZ265115 | MZ265039-MZ265040 | MZ262245-MZ262246 |
| 10. P. pandatus (Raski, 1976) Siddiqi, 1986 | PIN_AR | Caravaca, Murcia, Pinus halepensis Mill. | MZ265116-MZ265117 | MZ265041-MZ265042 | MZ262247-MZ262251 |
| 11. P. pedrami Clavero-Camacho et al., 2021 | AR_102 | Santa Mª de Trasierra Córdoba, wild olive | MZ265118 | - | - |
| 12. P. recisus Siddiqi, 1996 | CAZ_06 | Arroyo Frío, Jaén, Quercus faginea Lam. | MZ265119-MZ265120 | MZ265043 | MZ262252 |
| 13. P. sheri (Raski, 1973) Siddiqi, 1986 | CAZ_04 | Arroyo Frío, Jaén, grasses | MZ265121-MZ265124 | MZ265044-MZ265048 | MZ262253-MZ262259 |
| CAZ_07 | Coto Ríos, Jaén, Pinus halepensis Mill. | MZ265125-MZ265126 | MZ265049-MZ265050 | MZ262260-MZ262261 | |
| 14. P. tateae Wu and Townshend (1973) | PR_187 | Ariza, Zaragoza, almond | MW282754-MW282759 | MW282766-MW282771 | MZ262262-MZ262264 |
| 15. P. variabilis Raski, 1975 | EP_ACA | Córdoba, Córdoba, wild olive | MZ265127-MZ265129 | MZ265051-MZ265053 | MZ262265-MZ262267 |
| 16. P. veruculatus Wu, 1962 | AR_102 | Santa Mª de Trasierra Córdoba, wild olive | MZ265134-MZ265135 | - | - |
| 17. P. verus (Brzeski, 1995) Brzeski, 1998 | AR_097 | Santa Mª de Trasierra, Córdoba, wild olive | MZ265130-MZ265133 | MZ265054-MZ265058 | MZ262268-MZ262271 |
| 18. P. vitecus (Pramodini et al., 2006) Ghaderi et al., 2014 | EP_ACA | Córdoba, Córdoba, wild olive | MZ265136-MZ265141 | MZ265059-MZ265062 | MZ262272-MZ262274 |
| Holotype | Paratypes | |||
|---|---|---|---|---|
| Female | Females | Males | Juveniles (J4) | |
| Sample Code | CAZ_05 | CAZ_05 | CAZ_05 | CAZ_05 |
| Locality | Arroyo Frío, Jaén | |||
| n | 1 | 19 | 4 | 5 |
| L | 425 | 417.7 ± 35.2 (363–467) | 389.5 ± 19.3 (369–414) | 337.6 ± 37.5 (302–382) |
| a * | 23.6 | 20.6 ± 3.4 (16.3–26.4) | 28.9 ± 1.3 (27.3–30.3) | 22.2 ± 1.2 (20.8–23.9) |
| b | 3.5 | 3.6 ± 0.3 (3.1–4.2) | 4.2 ± 0.7 (3.6–5.2) | 3.3 ± 0.4 (2.9–3.8) |
| c | 14.7 | 13.4 ± 1.5 (11.4–16.5) | 12.7 ± 1.2 (11.9–14.5) | 12.9 ± 1.9 (10.7–15.9) |
| c’ | 2.6 | 2.9 ± 0.2 (2.5–3.4) | 2.9 ± 0.1 (2.7–3.0) | 2.8 ± 0.3 (2.4–3.2) |
| V or T | 80.9 | 82.1 ± 0.9 (80.2–83.5) | - | - |
| G1 | 35.5 | 44.4 ± 4.4 (35.5–50.3) | - | - |
| Stylet length | 54.0 | 53.5 ± 1.5 (52.0–56.0) | - | 45.8 ± 2.2 (43.0–48.0) |
| (Stylet length/body length) × 100 | 12.7 | 12.9 ± 0.9 (11.3–14.6) | - | 13.7 ± 1.0 (12.6–14.7) |
| Conus length | 43.0 | 41.2 ± 1.4 (38.0–43.0) | - | 36.0 ± 3.2 (31.0–39.0) |
| m | 79.6 | 77.1 ± 1.9 (73.1–79.6) | - | 78.6 ± 6.3 (72.1–88.6) |
| DGO | 5.5 | 5.4 ± 0.5 (4.5–6.0) | - | 4.0 ± 0.7 (3.0–5.0) |
| O | 10.2 | 10.2 ± 1.0 (8.0–11.5) | - | 8.7 ± 1.4 (6.8–10.6) |
| Lip width | 5.5 | 4.9 ± 0.4 (4.0–5.5) | 4.1 ± 0.3 (4.0–4.5) | 4.8 ± 0.4 (4.5–5.0) |
| Median bulb length | 24.0 | 24.0 ± 2.7 (19.0–29.0) | - | 23.8 ± 2.8 (19.0–26.0) |
| Median bulb width | 11.0 | 11.1 ± 0.9 (9.0–13.0) | - | 9.4 ± 1.0 (8.5–11.0) |
| Anterior end to center median bulb | 78 | 74.0 ± 3.8 (67.0–81.0) | - | 62.4 ± 4.0 (58.0–68.0) |
| MB | 65.0 | 64.2 ± 2.5 (58.5–70.0) | - | 61.3 ± 4.0 (56.9–67.3) |
| Nerve ring to anterior end | 88.0 | 89.4 ± 5.1 (77.0–98.0) | - | 78.8 ± 4.2 (74.0–84.0) |
| Excretory pore to anterior end | 94.0 | 98.2 ± 6.6 (87.0–114.0) | 81.5 ± 5.7 (74.0–88.0) | 86.4 ± 6.1 (77.0–94.0) |
| Pharynx length | 120.0 | 115.2 ± 4.9 (107.0–123.0) | 95.0 ± 11.1 (80.0–106.0) | 101.8 ± 0.8 (101.0–103.0) |
| Maximum body diam. | 18.0 | 20.9 ± 4.3 (14.0–28.0) | 13.2 ± 0.3 (13.0–13.5) | 15.2 ± 1.8 (14.0–18.0) |
| Tail length | 29.0 | 31.4 ± 4.1 (25.5–39.0) | 30.9 ± 3.7 (25.5–34.0) | 26.8 ± 5.9 (19.5–35.0) |
| Anal body diam. | 11.0 | 11.0 ± 1.4 (8.5–14.0) | 10.6 ± 0.9 (9.5–11.5) | 9.7 ± 1.7 (8.0–12.0) |
| Spicules | - | - | 22.0 ± 1.1 (21.0–23.5) | - |
| Gubernaculum | - | - | 4.0 ± 0.4 (3.5–4.5) | - |
| Species | P. aciculus | P. aciculus | P. aculentus |
|---|---|---|---|
| Life Stage | Females | Females | Females |
| Sample Code | CAZ_07 | Type | Belgium |
| Locality, Province | Coto Ríos, Jaén | Population [31] | Singh et al. [3] |
| n | 12 | 25 | 12 |
| L | 309.3 ± 16.2 (285–339) | 240–310 | 266 ± 20.1 (233–03) |
| a * | 19.1 ± 1.7 (17.0–21.5) | 18–24 | 19.6 ± 2.0 (16.3–23.2) |
| b | 2.5 ± 0.2 (2.2–2.8) | 2.4–2.7 | 2.6 ± 0.1 (2.4–2.8) |
| c | 12.4 ± 1.4 (10.3–14.8) | 10–16 | 12.4 ± 1.5 (10.8–15.2) |
| c’ | 2.9 ± 0.2 (2.6–3.5) | 3.0 | 2.8 ± 0.3 (2.4–3.1) |
| V | 73.4 ± 0.8 (72.3–74.7) | 68–74 | 72.5 ± 1.5 (70.8–75.7) |
| G1 | 34.8 ± 7.6 (23.5–42.5) | - | - |
| Stylet length | 71.4 ± 2.8 (67.5–75.0) | 61–69 | 56.0 ± 3.3 (52.4–61.2) |
| (Stylet length/body length) × 100 | 23.1 ± 1.0 (21.8–24.6) | - | - |
| Conus length | 64.7 ± 3.5 (58.0–69.0) | - | 49.1 ± 3.6 (43.0–54.9) |
| m | 90.6 ± 2.7 (84.1–93.2) | - | - |
| DGO | 5.3 ± 0.6 (4.5–6.5) | - | - |
| O | 7.4 ± 0.7 (6.3–8.7) | - | - |
| Lip width | 5.4 ± 0.5 (5.5–6.5) | - | - |
| Median bulb length | 24.3 ± 1.9 (22.0–27.0) | - | - |
| Median bulb width | 11.5 ± 0.5 (11.0–12.0) | - | - |
| Anterior end to center median bulb | 85.0 ± 3.3 (79.0–89.0) | - | - |
| MB | 66.2 ± 2.5 (62.3–69.0) | - | - |
| Nerve ring to anterior end | 103.2 ± 4.7 (94.0–100.0) | - | - |
| Excretory pore to anterior end | 83.8 ± 5.9 (72.5–91.0) | 70 | 66.7 ± 5.2 (54.3–74.4) |
| Pharynx length | 125.5 ± 8.2 (109.0–138.0) | 101 ± 8.3 (87.0–113) | |
| Maximum body diam. | 16.4 ± 2.1 (14.0–20.0) | 15 | 13.6 ± 1.3 (11.6–15.5) |
| Tail length | 25.3 ± 3.4 (20.5–33.0) | 50 | 20.9 ± 2.3 (18.1–25.1) |
| Anal body diam. | 8.6 ± 0.7 (7.5–10.0) | - | 7.6 ± 0.5 (7.0–8.3) |
| Life Stage | Females | Fourth-Stage Juveniles | Females |
|---|---|---|---|
| Sample Code | CAZ_02 | CAZ_02 | Type |
| Locality, Province | La Iruela, Jaén | La Iruela, Jaén | Population [32] |
| n | 17 | 6 | 16 |
| L | 397.1 ± 35.0 (335–450) | 358.7 ± 10.8 (340–369) | 320–370 |
| a * | 18.7 ± 1.7 (16.0–22.5) | 19.0 ± 2.7 (15.6–21.7) | 19–25 |
| b | 4.2 ± 0.4 (3.6–5.2) | 3.8 ± 0.2 (3.6–4.1) | 3.6–4.6 |
| c | 11.5 ± 2.0 (7.9–16.0) | 13.5 ± 2.0 (11.7–17.3) | 9–14 |
| c’ | 3.5 ± 0.4 (2.8–4.4) | 2.7 ± 0.4 (2.3–3.2) | 4.5 |
| V | 80.5 ± 1.4 (78.6–82.8) | - | 76–80 |
| G1 | 45.1 ± 5.6 (35.2–56.7) | - | - |
| Stylet length | 17.0 ± 0.5 (16.0–18.0) | 15.7 ± 0.8 (14.0–16.0) | 17–19 |
| (Stylet length/body length) × 100 | 4.3 ± 0.4 (3.8–5.1) | 4.4 ± 0.1 (4.1–4.5) | - |
| Conus length | 10.8 ± 0.7 (10.0–12.0) | 10.5 ± 0.6 (9.5–11.0) | - |
| m | 63.7 ± 3.4 (58.8–70.6) | 67.0 ± 2.5 (62.5–68.8) | - |
| DGO | 4.9 ± 0.6 (4.0–6.0) | 4.3 ± 0.5 (3.5–5.0) | - |
| O | 28.5 ± 3.2 (23.5–36.4) | 27.6 ± 2.4 (25.0–31.3) | - |
| Lip width | 4.4 ± 0.5 (4.0–5.5) | 5.0 ± 0.6 (4.0–5.5) | - |
| Median bulb length | 22.8 ± 2.0 (19.0–25.0) | 20.4 ± 0.5 (20.0–21.0) | - |
| Median bulb width | 11.1 ± 1.6 (9.5–14.0) | 8.1 ± 0.2 (8.0–8.5) | - |
| Anterior end to center median bulb | 51.1 ± 4.2 (43.0–59.0) | 46.8 ± 1.4 (44.0–48.0) | - |
| MB | 53.8 ± 3.1 (46.2–59.8) | 50.1 ± 3.5 (44.9–53.9) | - |
| Nerve ring to anterior end | 69.8 ± 5.4 (57.0–81.0) | 67.5 ± 4.7 (62.0–74.0) | - |
| Excretory pore to anterior end | 82.8 ± 5.2 (72.0–94.0) | 85.0 ± 4.7 (79.0–91.0) | 75 |
| Pharynx length | 93.8 ± 6.6 (76.0–104.0) | 93.5 ± 5.0 (89.0–101.0) | - |
| Maximum body diam. | 21.5 ± 2.9 (16.5–26.0) | 19.2 ± 2.9 (16.0–23.0) | 18 |
| Tail length | 35.5 ± 6.0 (22.0–43.0) | 26.9 ± 3.1 (21.0–29.0) | 34 |
| Anal body diam. | 10.3 ± 1.5 (8.0–13.5) | 10.0 ± 1.5 (9.0–12.0) | - |
| P. baldaccii | P. enigmaticus | P. holdemani | P. neoamblycephalus | |
|---|---|---|---|---|
| Life Stage | Females | Females | Females | Female |
| Sample Code | CAZ_04 | IAS_21 | AR_102 | CAZ_05 |
| Locality, Province | Arroyo Frío, Jaén | Córdoba, Córdoba | St. Mª Trasierra, Córdoba | Arroyo Frío, Jaén |
| n | 4 | 4 | 4 | 1 |
| L | 270.5 ± 5.2 (267–278) | 367.0 ± 11.3 (358–383) | 392.3 ± 30.2 (364–435) | 363 |
| a * | 17.8 ± 2.1 (15.4–20.5) | 21.5 ± 1.5 (20.2–23.2) | 24.9 ± 1.6 (23.5–27.2) | 18.2 |
| b | 3.4 ± 0.1 (3.3–3.5) | 3.8 ± 0.2 (3.7–4.0) | 3.9 ± 0.3 (3.6–4.1) | 4.4 |
| c | 10.5 ± 1.3 (8.7–11.7) | 15.0 ± 1.2 (13.2–16.0) | 13.3 ± 1.0 (12.2–14.6) | 14.0 |
| c’ | 2.8 ± 0.1 (2.7–2.9) | 2.5 ± 0.2 (2.3–2.8) | 3.0 ± 0.4 (2.7–3.5) | 2.2 |
| V | 81.2 ± 1.5 (79.5–83.0) | 83.2 ± 1.2 (81.8–84.5) | 81.0 ± 0.8 (79.9–81.9) | 81.3 |
| G1 | 40.1 ± 5.4 (34.5–46.4) | 41.8 ± 9.3 (31.8–51.2) | 28.4 ± 0.5 (28.0–28.7) | 36.9 |
| Stylet length | 29.5 ± 1.2 (28.0–31.0) | 26.8 ± 1.0 (26.0–28.0) | 26.8 ± 0.6 (26.0–27.5) | 33.0 |
| (Stylet length/body length) × 100 | 10.9 ± 0.5 (10.5–11.6) | 7.3 ± 0.1 (7.2–7.4) | 6.8 ± 0.5 (6.3–7.4) | 9.1 |
| Conus length | 21.4 ± 1.1 (20.5–23.0) | 18.3 ± 0.5 (18.0–19.0) | 16.0 ± 0.7 (15.0–16.5) | 23 |
| m | 72.5 ± 4.8 (67.4–78.0) | 68.3 ± 3.8 (64.3–73.1) | 59.8 ± 1.5 (57.7–61.1) | 69.7 |
| DGO | 5.0 ± 0.7 (4.0–5.5) | 5.8 ± 0.9 (4.5–6.5) | 6.3 ± 0.6 (5.5–7.0) | 5.5 |
| O | 17.0 ± 2.5 (13.6–19.6) | 21.5 ± 3.6 (16.7–25.0) | 23.3 ± 2.0 (21.2–25.5) | 16.7 |
| Lip width | 3.9 ± 0.3 (3.5–4.0) | 6.3 ± 0.3 (6.0–6.5) | 6.6 ± 0.5 (6.0–7.0) | 6 |
| Median bulb length | 17.2 ± 0.8 (16.5–18.0) | 23.3 ± 3.8 (20.0–27.0) | 19.4 ± 3.1 (16.5–23.5) | 26 |
| Median bulb width | 8.5 ± 0.5 (8.0–9.0) | 10.0 ± 0.7 (9.5–11.0) | 10.0 ± 0.4 (9.5–10.5) | 11 |
| Anterior end to center median bulb | 44.8 ± 1.9 (42.0–46.0) | 53.5 ± 1.3 (52.0–55.0) | 56.5 ± 3.3 (52.5–60.5) | 57 |
| MB | 56.4 ± 3.7 (52.1–59.7) | 55.5 ± 1.1 (54.6–57.1) | 56.4 ± 1.0 (55.2–57.6) | 69.5 |
| Nerve ring to anterior end | 58.3 ± 1.5 (57.0–60.0) | 73.3 ± 3.4 (70.0–78.0) | 72.8 ± 3.6 (68.0–75.5) | 66.0 |
| Excretory pore to anterior end | 68.3 ± 3.6 (63.0–71.0) | 82.3 ± 4.3 (76.0–83.0) | 85.5 ± 5.2 (79.5–82.0) | 74.0 |
| Pharynx length | 79.5 ± 2.1 (77.0–82.0) | 96.5 ± 3.9 (91.0–100.0) | 100.3 ± 5.1 (93.0–105.0) | 82.0 |
| Maximum body diam. | 15.4 ± 2.1 (13.0–18.0) | 17.1 ± 1.7 (15.5–19.0) | 15.8 ± 0.6 (15.0–16.5) | 20.0 |
| Tail length | 26.3 ± 4.0 (23.0–32.0) | 24.6 ± 2.9 (23.0–29.0) | 29.6 ± 3.4 (26.5–33.5) | 26.0 |
| Anal body diam. | 9.4 ± 1.1 (8.5–11.0) | 9.8 ± 0.6 (9.0–10.5) | 9.8 ± 0.3 (9.5–10.0) | 12.0 |
| Locality, Province | Córdoba, Córdoba | Sta. Mª Trasierra, Córdoba | Montalbán, Córdoba | Córdoba, Córdoba | ||||
|---|---|---|---|---|---|---|---|---|
| Life Stage | Females | Fourth-Stage Juveniles | Females | Fourth-Stage Juveniles | Females | Fourth-Stage Juveniles | Females | Fourth-Stage Juveniles |
| Sample Code | EP_ACA | EP_ACA | AR_097 | AR_097 | PR_050 | PR_050 | PR_017 | PR_017 |
| n | 14 | 10 | 5 | 5 | 4 | 4 | 5 | 5 |
| L | 433.8 ± 38.3 (396–513) | 440.8 ± 25.3 (409–486) | 466.4 ± 35.1 (408–495) | 432.8 ± 19.0 (412–461) | 460.0 ± 34.6 (411–490) | 395.8 ± 15.0 (375–409) | 413.3 ± 8.2 (403–422) | 395.8 ± 15.0 (375–409) |
| a * | 22.6 ± 2.5 (18.8–26.8) | 23.9 ± 1.5 (21.6–25.8) | 22.3 ± 2.2 (19.6–24.3) | 24.1 ± 1.6 (21.9–25.8) | 22.5 ± 1.9 (20.6–24.3) | 22.4 ± 1.9 (20.2–24.7) | 20.3 ± 2.0 (17.5–22.2) | 22.4 ± 1.8 (20.2–24.7) |
| b | 3.9 ± 0.2 (3.5–4.3) | 4.9 ± 0.3 (4.4–5.4) | 3.9 ± 0.2 (3.6–4.3) | 4.9 ± 0.3 (4.5–5.2) | 4.0 ± 0.3 (3.6–4.3) | 4.5 ± 0.4 (4.0–4.8) | 3.6 ± 0.4 (3.3–4.2) | 4.5 ± 0.4 (4.0–4.8) |
| c | 12.6 ± 2.6 (10.7–20.9) | 11.2 ± 1.0 (10.0–13.5) | 13.7 ± 3.4 (11.3–19.3) | 11.4 ± 1.1 (10.1–13.1) | 13.4 ± 3.2 (11.6–18.1) | 10.8 ± 1.3 (9.4–12.4) | 12.0 ± 2.0 (10.6–14.9) | 11.1 ± 1.5 (9.9–13.2) |
| c’ | 3.5 ± 0.5 (2.1–4.4) | 3.2 ± 0.3 (2.6–3.5) | 3.3 ± 0.7 (2.3–4.3) | 3.0 ± 0.2 (2.7–3.3) | 3.3 ± 0.4 (2.7–3.6) | 3.1 ± 0.2 (2.8–3.3) | 3.3 ± 0.4 (2.7–3.6) | 3.0 ± 0.3 (2.6–3.3) |
| V | 80.8 ± 1.3 (78.2–82.4) | - | 79.7 ± 1.5 (77.8–81.2) | - | 79.2 ± 1.4 (77.8–81.1) | - | 80.9 ± 0.9 (79.7–81.8) | - |
| G1 | 30.6 ± 2.5 (25.7–33.9) | - | 30.8 ± 3.4 (26.1–34.7) | - | 29.8 ± 3.0 (26.1–33.1) | - | 33.0 ± 2.3 (31.1–36.3) | - |
| Stylet length | 50.9 ± 2.9 (46.0–56.0) | 17.4 ± 1.2 (15.0–18.5) | 52.8 ± 2.5 (51.0–56.0) | 16.9 ± 0.9 (16.0–18.0) | 51.8 ± 2.2 (50.0–55.0) | 17.1 ± 0.9 (16.0–18.0) | 51.8 ± 2.2 (50.0–55.0) | 16.9 ± 0.6 (16.0–17.5) |
| (Stylet length/body length) × 100 | 11.8 ± 0.7 (10.4–12.7) | 4.0 ± 0.3 (3.5–4.4) | 11.4 ± 0.8 (10.4–12.5) | 3.9 ± 0.3 (3.5–4.4) | 11.3 ± 0.9 (10.2–12.4) | 4.3 ± 0.3 (3.9–4.7) | 12.5 ± 0.5 (12.1–13.2) | 4.3 ± 0.3 (3.9–4.7) |
| Conus length | 40.8 ± 2.4 (37.0–45.0) | 12.4 ± 1.1 (10.0–14.0) | 41.8 ± 1.9 (40.0–45.0) | 12.2 ± 1.3 (10.0–13.0) | 41.5 ± 1.7 (40.0–44.0) | 12.8 ± 0.5 (12.0–13.0) | 41.8 ± 2.2 (40.0–45.0) | 12.8 ± 0.5 (12.0–13.0) |
| m | 80.2 ± 2.1 (75.0–83.0) | 71.3 ± 4.6 (62.5–80.0) | 79.2 ± 2.6 (75.0–81.8) | 72.1 ± 5.6 (62.5–76.5) | 80.7 ± 0.8 (80.0–81.8) | 74.4 ± 1.8 (72.2–76.5) | 80.7 ± 0.8 (80.0–81.8) | 75.6 ± 1.1 (74.3–76.5) |
| DGO | 5.3 ± 0.4 (4.5–6.0) | 4.1 ± 0.4 (3.5–5.0) | 5.5 ± 0.5 (5.0–6.0) | 4.2 ± 0.4 (4.0–5.0) | 5.1 ± 0.5 (4.5–5.5) | 4.3 ± 0.5 (4.0–5.0) | 4.8 ± 0.3 (3.5–4.0) | 4.1 ± 0.3 (4.0–4.5) |
| O | 10.5 ± 1.0 (8.8–12.0) | 23.6 ± 2.4 (20.0–27.8) | 10.4 ± 1.0 (9.1–11.7) | 24.9 ± 2.8 (22.2–29.4) | 9.9 ± 1.1 (8.8–11.0) | 24.9 ± 3.3 (22.2–24.9) | 9.2 ± 0.6 (8.8–10.0) | 24.5 ± 1.6 (22.9–26.5) |
| Lip width | 4.8 ± 0.6 (4.0–5.5) | 4.0 ± 0.2 (3.5–4.5) | 4.2 ± 0.4 (4.0–5.0) | 3.7 ± 0.3 (3.5–4.0) | 4.1 ± 0.3 (4.0–4.5) | 3.8 ± 0.3 (3.5–4.0) | 4.1 ± 0.3 (4.0–4.5) | 3.8 ± 0.3 (3.5–4.0) |
| Median bulb length | 22.6 ± 2.9 (17.0–26.0) | 24.3 ± 1.3 (22.0–27.0) | 25.4 ± 0.5 (25.0–26.0) | 24.4 ± 0.9 (24.0–26.0) | 25.5 ± 0.6 (25.0–26.0) | 24.5 ± 1.0 (24.0–26.0) | 25.5 ± 0.6 (25.0–26.0) | 23.8 ± 0.5 (23.0–24.0) |
| Median bulb width | 11.2 ± 0.8 (10.0–13.0) | 7.9 ± 0.4 (7.0–8.5) | 11.3 ± 1.0 (10.0–12.5) | 7.8 ± 0.6 (7.0–8.5) | 11.3 ± 0.8 (10.2–12.4) | 7.8 ± 0.6 (7.0–8.5) | 11.3 ± 1.0 (10.0–12.0) | 7.5 ± 0.4 (7.0–8.0) |
| Anterior end to center median bulb | 74.8 ± 3.1 (70.0–81.0) | 48.4 ± 1.5 (46.0–51.0) | 77.0 ± 3.4 (73.0–81.0) | 47.8 ± 0.8 (47.0–49.0) | 76.3 ± 3.4 (73.0–81.0) | 47.8 ± 1.0 (47.0–49.0) | 75.8 ± 2.5 (73.0–79.0) | 47.8 ± 1.0 (47.0–49.0) |
| MB | 65.9 ± 4.8 (59.5–74.0) | 53.5 ± 3.2 (48.5–57.5) | 64.5 ± 6.6 (59.5–76.0) | 53.7 ± 3.2 (53.9–56.6) | 65.5 ± 8.2 (59.5–77.5) | 54.2 ± 3.2 (50.0–57.3) | 65.5 ± 8.2 (59.5–77.6) | 54.2 ± 3.2 (51.0–57.3) |
| Nerve ring to anterior end | 90.9 ± 7.1 (78.0–99.0) | 65.0 ± 3.2 (61.0–71.0) | 95.4 ± 2.7 (91.0–98.0) | 64.2 ± 2.3 (62.0–69.0) | 94.8 ± 2.6 (91.0–97.0) | 64.8 ± 2.2 (63.0–68.0) | 94.8 ± 2.6 (91.0–97.0) | 64.8 ± 2.2 (63.0–68.0) |
| Excretory pore to anterior end | 94.6 ± 6.8 (82.0–106.0) | 83.1 ± 6.0 (74.0–91.0) | 98.2 ± 3.3 (94.0–103.0) | 81.6 ± 5.7 (75.0–88.0) | 98.5 ± 3.7 (94.0–103.0) | 81.8 ± 5.6 (76.0–87.0) | 98.5 ± 3.7 (94.0–103.0) | 81.8 ± 5.6 (76.0–87.0) |
| Pharynx length | 112.2 ± 12.0 (97.0–132.0) | 90.8 ± 6.6 (82.0–101.0) | 119.4 ± 12.1 (100.0–129.0) | 89.2 ± 5.3 (83.0–97.0) | 116.5 ± 13.5 (98.0–127.0) | 88.3 ± 5.3 (82.0–94.0) | 116.5 ± 13.5 (98.0–127.0) | 88.3 ± 5.3 (82.0–94.0) |
| Maximum body diam. | 19.4 ± 2.8 (15.0–26.0) | 18.5 ± 1.3 (16.0–20.0) | 21.0 ± 2.3 (19.0–25.0) | 18.0 ± 1.6 (16.0–20.0) | 20.5 ± 1.7 (19.0–23.0) | 17.8 ± 1.7 (16.0–20.0) | 20.5 ± 1.7 (19.0–23.0) | 17.8 ± 1.7 (16.0–20.0) |
| Tail length | 35.2 ± 4.2 (23.5–42.0) | 39.6 ± 4.3 (31.0–44.0) | 35.2 ± 6.2 (25.0–41.0) | 38.2 ± 4.3 (32.0–43.0) | 35.3 ± 5.9 (27.0–40.0) | 37.0 ± 3.2 (33.0–40.0) | 35.3 ± 5.9 (27.0–40.0) | 36.0 ± 3.6 (31.0–39.0) |
| Anal body diam. | 10.2 ± 0.8 (9.0–11.0) | 12.5 ± 0.9 (11.5–14.0) | 10.7 ± 0.7 (9.5–11.0) | 12.6 ± 0.8 (11.5–13.5) | 10.8 ± 0.5 (10.0–11.0) | 12.1 ± 0.9 (11.0–13.0) | 10.8 ± 0.5 (10.0–11.0) | 12.1 ± 0.9 (11.0–13.0) |
| Life Stage | Females | Male | Fourth-Stage Juveniles |
|---|---|---|---|
| Sample Code | AR_102 | ||
| Locality, province | St. Mª Trasierra, Córdoba | ||
| n | 10 | 1 | 3 |
| L | 365.8.3 ± 26.6 (317–410) | 395 | 304.3 ± 6.1 (299–311) |
| a * | 23.0 ± 2.6 (19.9–28.3) | 31.6 | 20.8 ± 0.6 (20.2–21.4) |
| b | 2.8 ± 0.2 (2.5–3.0) | 4.0 | 2.8 ± 0.1 (2.7–2.8) |
| c | 8.8 ± 1.1 (7.4–11.1) | 12.3 | 13.6 ± 0.5 (13.2–14.1) |
| c’ | 4.2 ± 0.5 (3.5–4.9) | 2.9 | 2.7 ± 0.03 (2.7–2.8) |
| V or T | 75.1 ± 0.8 (73.9–76.3) | 48.1 | - |
| G1 | 29.7 ± 3.1 (24.1–33.8) | - | - |
| Stylet length | 76.2 ± 3.9 (70.0–84.0) | - | 62.3 ± 1.5 (61.0–64.0) |
| (Stylet length/body length) × 100 | 20.9 ± 1.6 (18.8–24.0) | - | 20.5 ± 0.3 (20.1–20.7) |
| Conus length | 68.9 ± 4.3 (61.5–77.0) | - | 53.0 ± 1.0 (52.0–54.0) |
| m | 90.4 ± 1.6 (87.9–93.7) | - | 85.0 ± 1.6 (83.9–86.9) |
| DGO | 5.6 ± 0.6 (5.0–6.5) | - | 5.5 ± 0.5 (5.0–6.0) |
| O | 7.4 ± 0.8 (6.4–9.0) | - | 8.8 ± 0.8 (8.2–8.7) |
| Lip width | 4.7 ± 0.2 (4.5–5.0) | 3.5 | 4.7 ± 0.6 (4.0–5.0) |
| Median bulb length | 26.7 ± 2.4 (24.0–31.0) | - | 18.7 ± 0.6 (18.0–19.0) |
| Median bulb width | 10.3 ± 1.2 (9.0–13.0) | - | 8.8 ± 0.3 (8.5–9.0) |
| Anterior end to center median bulb | 92.9 ± 6.4 (83.0–102.0) | - | 72.0 ± 1.0 (71.0–73.0) |
| MB | 69.3 ± 3.2 (61.5–72.2) | - | 66.1 ± 0.3 (65.8–66.4) |
| Nerve ring to anterior end | 109.7 ± 10.0 (91.0–123.0) | 84 | 85.7 ± 1.5 (84.0–87.0) |
| Excretory pore to anterior end | 94.1 ± 9.0 (82.0–109.0) | 91 | 73.0 ± 2.0 (71.0–75.0) |
| Pharynx length | 133.1 ± 9.4 (115.0–143.0) | 98 | 109.0 ± 2.0 (107.0–111.0) |
| Maximum body diam. | 16.0 ± 1.1 (14.5–18.0) | 12.5 | 14.7 ± 0.6 (14.0–15.0) |
| Tail length | 42.0 ± 4.4 (33.5–49.0) | 32 | 22.3 ± 0.6 (22.0–23.0) |
| Anal body diam. | 10.0 ± 1.1 (8.5–12.0) | 11 | 8.2 ± 0.3 (8.0–8.5) |
| Spicules | - | 19.5 | - |
| Gubernaculum | - | 5.5 | - |
| Life Stage | Females | Fourth-Stage Juveniles | Females |
|---|---|---|---|
| Sample Code | PIN_AR | PIN_AR | Type |
| Locality, Province | Caravaca, Murcia | Caravaca, Murcia | Population [37] |
| n | 12 | 3 | 10 |
| L | 317.2 ± 15.9 (290–339) | 277.3 ± 22.7 (252–296) | 330–420 |
| a * | 18.7 ± 1.8 (15.7–21.5) | 18.1 ± 1.5 (16.8–19.7) | 23–32 |
| b | 2.7 ± 0.1 (2.5–2.9) | 3.8 ± 0.3 (3.5–4.0) | 2.8–3.2 |
| c | 12.2 ± 2.2 (9.2–16.6) | 13.2 ± 1.6 (11.5–14.2) | 9–12 |
| c’ | 2.5 ± 0.2 (2.2–3.0) | 2.2 ± 0.1 (2.1–2.3) | 4.7 |
| V | 75.8 ± 0.9 (74.5–77.7) | - | 70–76 |
| G1 | 32.0 ± 2.9 (28.6–38.6) | - | - |
| Stylet length | 61.3 ± 3.8 (57.0–68.5) | - | 63–70 |
| (Stylet length/body length) × 100 | 19.3 ± 0.8 (17.8–21.1) | - | - |
| Conus length | 53.2 ± 3.1 (49.0–59.0) | - | - |
| m | 86.9 ± 1.1 (85.7–89.4) | - | - |
| DGO | 5.5 ± 0.5 (5.0–6.5) | - | - |
| O | 9.1 ± 0.9 (7.9–10.5) | - | - |
| Lip width | 5.9 ± 0.7 (5.0–7.0) | 4.7 ± 0.3 (4.5–5.0) | - |
| Median bulb length | 23.7 ± 2.1 (19.0–26.5) | - | - |
| Median bulb width | 11.0 ± 1.1 (10.0–14.0) | - | - |
| Anterior end to center median bulb | 79.5 ± 6.7 (64.0–88.0) | - | - |
| MB | 67.0 ± 2.5 (62.8–71.3) | - | - |
| Nerve ring to anterior end | 97.7 ± 6.3 (82.0–106.0) | 55.0 ± 1.0 (54.0–56.0) | - |
| Excretory pore to anterior end | 84.9 ± 8.9 (71.0–111.0) | 63.0 ± 1.0 (62.0–64.0) | 94–119 |
| Pharynx length | 118.6 ± 7.6 (102.0–128.0) | 73.0 ± 1.0 (72.0–74.0) | - |
| Maximum body diam. | 17.1 ± 1.4 (15.0–19.5) | 15.3 ± 0.6 (15.0–16.0) | 17 |
| Tail length | 26.7 ± 4.6 (18.0–32.0) | 21.0 ± 1.0 (20.0–22.0) | 38 |
| Anal body diam. | 10.6 ± 1.5 (8.0–13.0) | 9.3 ± 0.3 (9.0–9.5) | - |
| Life Stage | Females | Fourth-Stage Juveniles | Females |
|---|---|---|---|
| Sample Code | CAZ_06 | CAZ_06 | Type |
| Locality, Province | Arroyo Frío, Jaén | Arroyo Frío, Jaén | Population [43] |
| n | 4 | 3 | 20 |
| L | 397.0 ± 36.5 (363–448) | 353.0 ± 32.5 (329–390) | 270–390 |
| a * | 18.4 ± 1.3 (17.2–20.2) | 16.2 ± 1.0 (15.5–17.3) | 16–27 |
| b | 4.6 ± 0.7 (3.8–5.4) | 4.9 ± 0.3 (4.7–5.3) | 3.8–5.0 |
| c | 12.6 ± 1.0 (11.2–13.6) | 14.7 ± 0.8 (14.2–15.6) | 13–16 |
| c’ | 3.3 ± 0.4 (2.8–3.5) | 2.4 ± 0.1 (2.4–2.6) | 2.7–3.3 |
| V | 81.2 ± 0.7 (80.4–82.0) | - | 78–83 |
| G1 | 44.3 ± 1.6 (42.1–45.8) | - | - |
| Stylet length | 15.1 ± 0.6 (14.5–16.0) | - | 15–17 |
| (Stylet length/body length) × 100 | 3.8 ± 0.4 (3.3–4.2) | - | - |
| Conus length | 9.8 ± 0.5 (9.0–10.0) | - | - |
| m | 64.5 ± 2.5 (62.1–66.7) | - | - |
| DGO | 5.3 ± 0.5 (5.0–6.0) | - | - |
| O | 34.8 ± 4.5 (31.3–41.4) | - | - |
| Lip width | 5.6 ± 0.8 (5.0–6.5) | 4.5 ± 0.5 (4.0–5.0) | - |
| Median bulb length | 23.0 ± 2.2 (20.0–25.0) | 14.0 ± 0.5 (13.5–14.5) | - |
| Median bulb width | 10.4 ± 1.5 (9.0–12.5) | 7.2 ± 0.3 (7.0–7.5) | - |
| Anterior end to center median bulb | 47.5 ± 7.4 (39.0–57.0) | 33.7 ± 1.5 (32.0–35.0) | - |
| MB | 54.0 ± 4.9 (47.0–58.0) | 46.7 ± 0.9 (45.7–47.3) | - |
| Nerve ring to anterior end | 66.3 ± 7.1 (59.0–76.0) | 62.0 ± 2.0 (60.0–64.0) | - |
| Excretory pore to anterior end | 77.5 ± 9.7 (71.0–92.0) | 68.7 ± 1.5 (67.0–70.0) | 58–70 |
| Pharynx length | 87.8 ± 9.1 (81.0–101.0) | 72.0 ± 2.0 (70.0–74.0) | - |
| Maximum body diam. | 21.8 ± 3.3 (18.0–26.0) | 21.8 ± 0.8 (21.0–22.5) | 12–16 |
| Tail length | 31.5 ± 2.6 (28.0–34.0) | 24.0 ± 1.0 (23.0–25.0) | 18–29 |
| Anal body diam. | 9.8 ± 1.7 (8.0–12.0) | 9.8 ± 0.8 (9.0–10.5) | - |
| Life Stage | Females | Fourth-Stage Juveniles | Females |
|---|---|---|---|
| Sample Code | CAZ_04 | CAZ_04 | CAZ_07 |
| Locality, Province | Arroyo Frío, Jaén | Arroyo Frío, Jaén | Coto Ríos, Jaén |
| n | 14 | 5 | 4 |
| L | 548.6 ± 53.2 (459–626) | 523.6 ± 11.6 (514–543) | 540.0 ± 45.4 (492–595) |
| a * | 20.5 ± 2.5 (15.8–24.6) | 23.6 ± 1.3 (21.5–24.8) | 19.9 ± 2.3 (16.6–22.0) |
| b | 4.8 ± 0.5 (3.8–5.4) | 4.9 ± 0.2 (4.6–5.1) | 4.8 ± 0.3 (4.6–5.2) |
| c | 11.1 ± 1.6 (9.2–14.6) | 14.3 ± 2.9 (11.1–17.9) | 11.5 ± 1.5 (10.3–13.5) |
| c’ | 3.8 ± 0.3 (3.2–4.2) | 3.0 ± 0.2 (2.8–3.3) | 3.6 ± 0.3 (3.2–3.9) |
| V | 78.9 ± 1.7 (75.8–81.7) | - | 79.5 ± 1.7 (77.0–80.9) |
| G1 | 49.7 ± 5.6 (40.7–56.7) | - | 48.5 ± 7.0 (41.3–55.0) |
| Stylet length | 23.8 ± 0.7 (22.5–25.0) | 20.0 ± 0.4 (19.5–20.5) | 23.6 ± 0.5 (23.0–24.0) |
| (Stylet length/body length) × 100 | 4.4 ± 0.4 (4.0–5.5) | 3.9 ± 0.1 (3.8–4.0) | 4.4 ± 0.4 (4.0–4.9) |
| Conus length | 15.4 ± 0.8 (14.0–17.0) | 12.3 ± 0.3 (12.0–12.5) | 14.8 ± 0.5 (14.0–15.0) |
| m | 64.5 ± 2.5 (62.0–70.8) | 61.5 ± 1.1 (60.0–62.5) | 62.4 ± 1.2 (60.9–63.8) |
| DGO | 6.0 ± 0.5 (5.0–7.0) | 4.2 ± 0.4 (4.0–5.0) | 6.3 ± 0.5 (6.0–7.0) |
| O | 25.3 ± 2.1 (20.8–30.4) | 21.0 ± 1.9 (20.0–24.4) | 26.5 ± 2.6 (25.0–30.4) |
| Lip width | 5.1 ± 0.6 (4.5–6.0) | 3.9 ± 0.4 (3.5–4.5) | 5.0 ± 0.7 (4.5–6.0) |
| Median bulb length | 27.2 ± 1.9 (24.0–31.0) | - | 25.8 ± 1.3 (24.0–27.0) |
| Median bulb width | 14.3 ± 1.9 (12.0–18.5) | - | 13.3 ± 1.2 (12.0–14.0) |
| Anterior end to center median bulb | 63.0 ± 3.8 (56.0–70.0) | - | 61.8 ± 3.9 (57.0–65.0) |
| MB | 55.6 ± 1.5 (52.5–58.1) | - | 55.2 ± 1.0 (54.3–56.5) |
| Nerve ring to anterior end | 86.0 ± 6.4 (76.0–99.0) | 82.4 ± 1.7 (81.0–85.0) | 83.3 ± 6.8 (76.0–90.0) |
| Excretory pore to anterior end | 101.9 ± 6.4 (93.0–116.0) | 99.2 ± 1.9 (97.0–102.0) | 100.8 ± 6.9 (95.0–110.0) |
| Pharynx length | 113.4 ± 6.3 (104.0–126.0) | 107.0 ± 5.0 (102.0–115.0) | 111.8 ± 6.4 (105.0–119.0) |
| Maximum body diam. | 27.1 ± 4.0 (21.0–33.5) | 22.2 ± 1.3 (21.0–24.0) | 27.3 ± 2.7 (24.5–31.0) |
| Tail length | 50.1 ± 7.0 (39.0–62.0) | 38.0 ± 8.3 (29.0–49.0) | 47.5 ± 5.5 (42.0–54.0) |
| Anal body diam. | 13.3 ± 1.2 (11.5–15.5) | 12.8 ± 2.1 (10.5–15.0) | 13.3 ± 1.0 (12.0–14.0) |
| Life Stage | Females | Fourth-Stage Juveniles | Females |
|---|---|---|---|
| Sample Code | EP_ACA | EP_ACA | Type |
| Locality, Province | Córdoba, Córdoba | Córdoba, Córdoba | Population [46] |
| n | 10 | 4 | 27 |
| L | 337.3 ± 30.5 (302–407) | 312.3 ± 38.2 (282–367) | 250–340 |
| a * | 21.7 ± 1.1 (20.1–23.5) | 21.2 ± 2.3 (19.4–24.5) | 19–25 |
| b | 3.9 ± 0.6 (2.8–4.8) | 3.9 ± 0.2 (3.7–4.2) | 3.6–4.7 |
| c | 15.3 ± 1.2 (13.1–17.2) | 14.7 ± 1.3 (13.1–16.3) | 12–18 |
| c’ | 2.6 ± 0.2 (2.3–2.9) | 2.2 ± 0.2 (1.9–2.4) | 2.7 |
| V | 84.5 ± 1.0 (83.1–86.5) | - | 82–85 |
| G1 | 39.4 ± 6.6 (28.6–52.8) | - | - |
| Stylet length | 14.9 ± 0.6 (14.0–16.0) | 11.6 ± 0.8 (11.0–12.5) | 13–16 |
| (Stylet length/body length) × 100 | 4.5 ± 0.4 (3.4–4.9) | 3.7 ± 0.3 (3.3–4.0) | - |
| Conus length | 10.5 ± 0.5 (10.0–11.0) | 7.5 ± 0.4 (7.0–8.0) | - |
| m | 70.5 ± 3.7 (64.5–75.9) | 64.6 ± 2.5 (62.5–68.2) | - |
| DGO | 3.5 ± 0.6 (2.5–4.5) | 2.6 ± 0.5 (2.0–3.0) | - |
| O | 23.5 ± 3.9 (17.9–30.0) | 22.6 ± 4.2 (18.2–27.3) | - |
| Lip width | 5.6 ± 0.6 (5.0–6.5) | 4.3 ± 0.3 (4.0–4.5) | - |
| Median bulb length | 18.9 ± 1.7 (17.0–23.0) | 18.0 ± 1.4 (17.0–20.0) | - |
| Median bulb width | 8.9 ± 1.4 (7.5–12.0) | 7.5 ± 0.6 (7.0–8.0) | - |
| Anterior end to center median bulb | 46.0 ± 3.6 (40.0–52.0) | 41.0 ± 0.8 (40.0–42.0) | - |
| MB | 53.1 ± 7.4 (39.5–63.6) | 51.9 ± 3.6 (46.6–54.5) | - |
| Nerve ring to anterior end | 62.6 ± 3.0 (58.0–68.0) | 57.8 ± 5.0 (51.0–63.0) | - |
| Excretory pore to anterior end | 74.5 ± 7.8 (67.0–95.0) | 67.0 ± 6.2 (60.0–75.0) | 59–71 |
| Pharynx length | 87.9 ± 12.4 (77.0–119.0) | 79.3 ± 5.9 (76.0–88.0) | - |
| Maximum body diam. | 15.6 ± 1.6 (14.0–19.0) | 14.8 ± 0.6 (14.0–15.5) | 13 |
| Tail length | 22.2 ± 3.5 (20.0–31.0) | 21.4 ± 2.6 (19.0–25.0) | 21 |
| Anal body diam. | 8.8 ± 1.8 (7.0–13.0) | 9.8 ± 1.0 (9.0–11.0) | - |
| Females | Fourth-Stage Juveniles | Females | |
|---|---|---|---|
| Sample Code | AR_097 | AR_097 | [28] |
| Locality | Sta. Mª Trasierra, Córdoba | Sta. Mª Trasierra, Córdoba | Type Population |
| n | 14 | 7 | 11 |
| L | 324.4 ± 25.0 (265–355) | 276.9 ± 22.2 (253–319) | 270–320 |
| a * | 19.4 ± 3.7 (12.2–24.7) | 21.8 ± 2.3 (18.8–24.2) | 20–24 |
| b | 2.4 ± 0.2 (2.1–2.7) | 3.0 ± 0.5 (2.5–3.8) | 2.2–2.5 |
| c | 12.1 ± 2.0 (9.1–16.9) | 11.1 ± 1.1 (9.4–12.5) | 13–19 |
| c’ | 3.3 ± 0.3 (2.7–3.6) | 3.0 ± 0.3 (2.8–3.3) | 1.8–2.9 |
| V | 77.0 ± 1.8 (73.6–80.3) | - | 76–79 |
| G1 | 29.3 ± 4.1 (24.5–40.4) | - | - |
| Stylet length | 89.1 ± 5.8 (79.0–97.0) | 49.1 ± 1.2 (48.0–51.0) | 66–86 |
| (Stylet length/body length) × 100 | 27.6 ± 1.9 (24.8–30.2) | 17.8 ± 1.4 (15.8–20.2) | - |
| Conus length | 77.8 ± 5.3 (70.0–86.5) | 41.9 ± 1.2 (40.0–43.0) | - |
| m | 87.3 ± 1.9 (84.3–89.9) | 85.2 ± 1.1 (83.3–86.9) | - |
| DGO | 4.6 ± 0.7 (3.5–6.0) | 4.2 ± 0.3 (4.0–4.5) | - |
| O | 5.2 ± 0.8 (4.1–6.7) | 8.6 ± 0.6 (7.8–9.4) | - |
| Lip width | 4.4 ± 0.4 (4.0–5.0) | 3.7 ± 0.3 (3.5–4.0) | - |
| Median bulb length | 25.5 ± 3.3 (22.0–34.0) | 15.2 ± 1.2 (14.0–17.0) | - |
| Median bulb width | 10.6 ± 0.7 (9.5–12.0) | 8.3 ± 0.4 (8.0–9.0) | - |
| Anterior end to center median bulb | 101.7 ± 7.9 (87.0–115.0) | 69.3 ± 3.4 (66.0–74.0) | - |
| MB | 76.1 ± 4.0 (70.2–83.0) | 73.7 ± 5.3 (68.3–83.5) | - |
| Nerve ring to anterior end | 112.6 ± 11.3 (94.0–133.0) | 77.9 ± 0.9 (77.0–79.0) | - |
| Excretory pore to anterior end | 89.0 ± 5.2 (81.0–95.0) | 82.0 ± 2.5 (78.0–85.0) | 68–92 |
| Pharynx length | 133.9 ± 12.1 (112.0–161.0) | 94.4 ± 7.8 (85.0–105.0) | - |
| Maximum body diam. | 17.4 ± 4.2 (13.0–27.0) | 12.9 ± 2.3 (11.0–17.0) | 14 |
| Tail length | 27.1 ± 2.6 (21.0–30.0) | 25.3 ± 3.9 (23.0–34.0) | 18 |
| Anal body diam. | 8.1 ± 0.5 (7.0–9.0) | 8.4 ± 1.4 (7.5–11.5) | - |
| Females | Fourth-Stage Juveniles | Females | |
|---|---|---|---|
| Sample Code | EP_ACA | EP_ACA | Type Population |
| Locality | Córdoba, Córdoba | Córdoba, Córdoba | [47] |
| n | 3 | 7 | 7 |
| L | 341.2 ± 22.1 (323–366) | 271.6 ± 12.0 (259–288) | 220–350 |
| a * | 20.3 ± 1.7 (18.3–21.5) | 19.5 ± 1.4 (18.8–24.2) | 20–26 |
| b | 2.7 ± 0.1 (2.5–2.8) | 2.9 ± 0.2 (2.6–3.1) | 2.7–2.9 |
| c | 11.6 ± 1.6 (9.8–12.9) | 10.6 ± 0.6 (9.6–11.5) | 9–16 |
| c’ | 3.1 ± 0.4 (2.7–3.5) | 2.9 ± 0.2 (2.7–3.1) | 2.9 |
| V | 72.8.0 ± 4.1 (68.1–75.4) | - | 72–77 |
| G1 | 28.3 ± 3.8 (24.5–32.1) | - | - |
| Stylet length | 66.0 ± 4.0 (62.0–70.0) | 42.5 ± 1.6 (40.0–44.0) | 42–65 |
| (Stylet length/body length) × 100 | 19.4 ± 2.1 (16.9–20.8) | 15.7 ± 1.1 (13.9–16.7) | - |
| Conus length | 59.3 ± 1.5 (58.0–61.0) | 35.6 ± 1.0 (34.0–37.0) | - |
| m | 90.0 ± 3.2 (87.1–93.5) | 83.7 ± 1.5 (81.8–85.7) | - |
| DGO | 4.3 ± 0.6 (4.0–5.0) | 3.1 ± 0.4 (2.5–4.0) | - |
| O | 6.1 ± 0.8 (5.4–7.0) | 7.2 ± 1.1 (6.3–9.5) | - |
| Lip width | 4.5 ± 0.5 (4.0–5.0) | 4.5 ± 0.5 (4.0–5.0) | - |
| Median bulb length | 24.0 ± 2.0 (22.0–26.0) | 17.6 ± 3.8 (15.0–26.0) | - |
| Median bulb width | 9.3 ± 2.3 (8.0–12.0) | 8.1 ± 0.6 (7.5–9.0) | - |
| Anterior end to center median bulb | 88.3 ± 4.9 (85.0–94.0) | 57.1 ± 3.1 (53.0–61.0) | - |
| MB | 68.9 ± 1.8 (67.2–70.8) | 59.9 ± 0.9 (58.2–61.1) | - |
| Nerve ring to anterior end | 105.7 ± 7.2 (101.0–114.0) | 70.0 ± 3.7 (63.0–74.0) | - |
| Excretory pore to anterior end | 84.3 ± 6.8 (79.0–92.0) | 78.4 ± 6.6 (72.0–89.0) | 85–94 |
| Pharynx length | 128.3 ± 8.5 (120.0–137.0) | 95.4 ± 4.4 (91.0–102.0) | - |
| Maximum body diam. | 17.0 ± 2.6 (15.0–20.0) | 14.0 ± 1.6 (12.5–16.5) | 10–15 |
| Tail length | 29.8 ± 3.5 (26.0–33.0) | 25.6 ± 1.5 (24.0–27.5) | 14–39 |
| Anal body diam. | 9.8 ± 1.5 (8.5–11.5) | 8.8 ± 0.6 (8.0–9.5) | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clavero-Camacho, I.; Palomares-Rius, J.E.; Cantalapiedra-Navarrete, C.; León-Ropero, G.; Martín-Barbarroja, J.; Archidona-Yuste, A.; Castillo, P. Integrative Taxonomy Reveals Hidden Cryptic Diversity within Pin Nematodes of the Genus Paratylenchus (Nematoda: Tylenchulidae). Plants 2021, 10, 1454. https://doi.org/10.3390/plants10071454
Clavero-Camacho I, Palomares-Rius JE, Cantalapiedra-Navarrete C, León-Ropero G, Martín-Barbarroja J, Archidona-Yuste A, Castillo P. Integrative Taxonomy Reveals Hidden Cryptic Diversity within Pin Nematodes of the Genus Paratylenchus (Nematoda: Tylenchulidae). Plants. 2021; 10(7):1454. https://doi.org/10.3390/plants10071454
Chicago/Turabian StyleClavero-Camacho, Ilenia, Juan Emilio Palomares-Rius, Carolina Cantalapiedra-Navarrete, Guillermo León-Ropero, Jorge Martín-Barbarroja, Antonio Archidona-Yuste, and Pablo Castillo. 2021. "Integrative Taxonomy Reveals Hidden Cryptic Diversity within Pin Nematodes of the Genus Paratylenchus (Nematoda: Tylenchulidae)" Plants 10, no. 7: 1454. https://doi.org/10.3390/plants10071454