Biotechnological Methods for Buckwheat Breeding
Abstract
:1. Introduction
2. Species of the Genus Fagopyrum as a Possible Source of Germplasm for Breeding and Their Reproductive Properties
| Species | Nuclear DNA Amount (pg) * | C-Value ** | Ploidy/Chromosome Number | References |
|---|---|---|---|---|
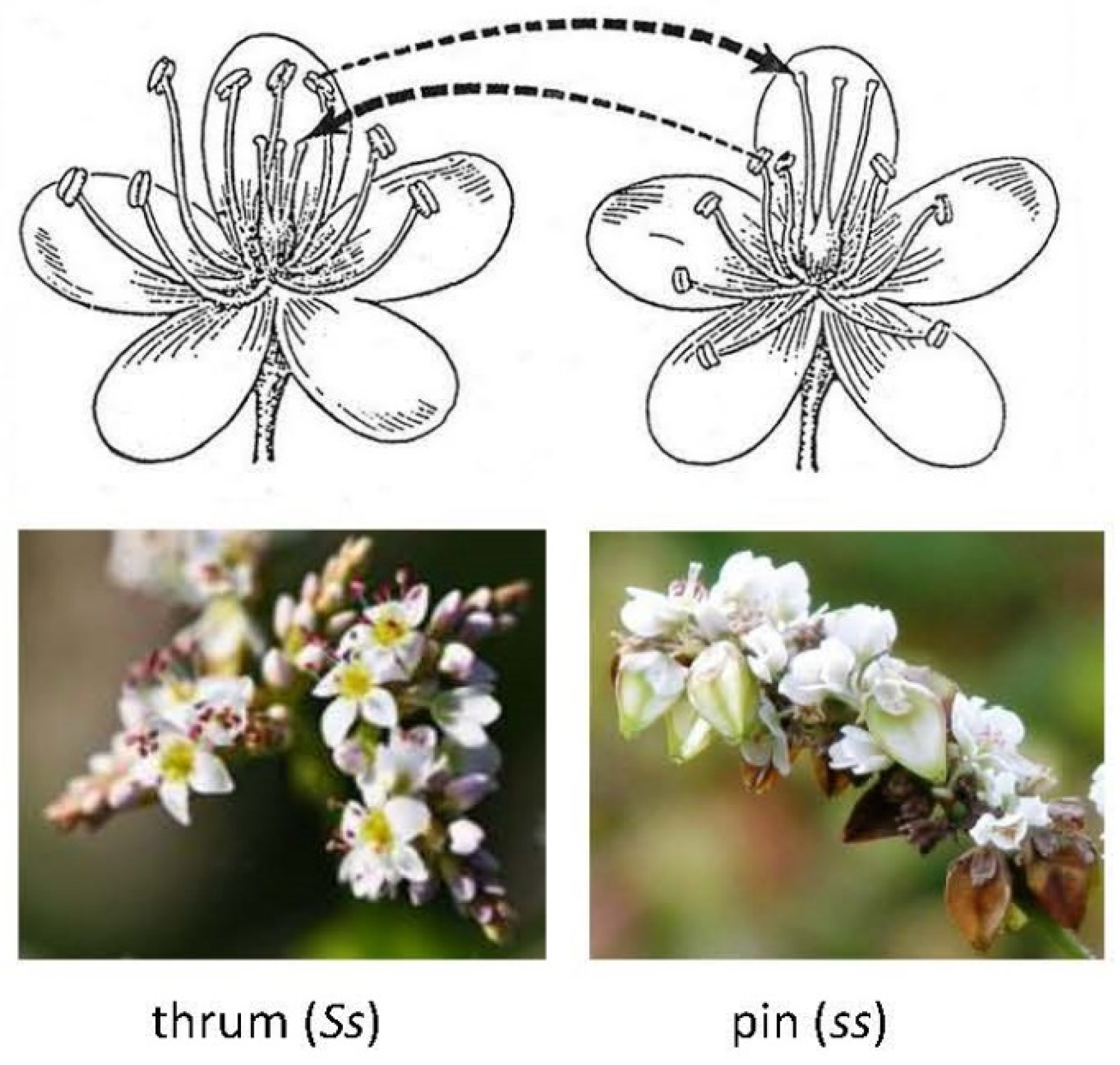
| Mating system: hetero-styled species, thrum/pin flower, self-incompatibility (SI) | ||||
| F. esculentum | 2.77 5.49 | 1.39 1.37 | 2n/16 4n/32 | [30,53,54,55,56] |
| F. cymosum | 2.32 3.37 | 1.16 0.48 | 2n/16 4n/32 | [54,55,57] |
| F. lineare | 1.08 | / | 2n/16 | [58,59] |
| F. urophyllum | 3.83 | / | 2n/16 | [60,61] |
| F. statice | 1.35 | 0.68 | 2n 16 | [61] |
| F. leptopodum | 1.43 | 0.72 | 2n/16 | [62,63] |
| F. gilessii | 1.80 | / | 2n 16 | [63] |
| F. capillatum | 1.71 | 0,68 | 2n/16 | [30] |
| F. gracilipedoides | / | / | 2n/16 | [34] |
| F. jinshaense | / | / | 2n/16 | [34] |
| F. pilus | 1.52 | / | 4n/32 | [33] |
| F. megaspartanium | / | / | 4n/32 | [33] |
| F. densovillosum | / | / | 2n/16 | [64] |
| F. qiangcai | / | / | 2n/16 | [38] |
| F.luojishanense | / | / | 2n/16 | [39] |
| F. hailuogouense | / | / | 4n/32 | [40] |
| Mating system: hetero-styled species, self-compatibility (SC) | ||||
| F. pleioramosum | 3.05 | 1.53 | 2n/16 | [30] |
| F. callianthum | / | / | 2n/16 | [30] |
| F. macrocarpum | 2.32 | / | 2n/16 | [32] |
| F. wenchuanense | / | / | 2n/16 | [38] |
| F. longzhoushanense | / | / | 2n/16 | [42] |
| F. longistylum | / | / | 2n/16 | [4] |
| Mating system: homo-styled species, self-compatibility (SC) | ||||
| F. tataricum | 1.11 | 0.56 | 2n/16 | [53,65,66] |
| F. homotropicum | 2.46 5.20 | 1.23 1.30 | 2n/16 4n/32 | [30,66] |
| F. gracilipes | 3.35 | 0.84 | 4n/32 | [62,67] |
| F. crispatifolium | / | / | 4n/32 | [35] |
| F. pugense | / | / | 2n/16 | [36,39,41] |
| F. rubifolium | 3.31 | / | 4n/32 | [32] |
| F. zuogongense | / | / | 2n/16 | [33] |
| Mating system: unknown | ||||
| F. tibeticum | / | / | 2n/48 | [68] |
| Interspecific hybrid F. cymosum × another species with hetero-styled flowers | ||||
| F. giganteum | 2.52 | / | [69] | |
3. Heteromorphic Self-Incompatibility in Buckwheat and Mode of Inheritance
4. Interspecific Hybridization
5. Tissue Culture and Plant Regeneration
6. Marker Systems for Property Studies
6.1. Random Amplified Polymorphic DNA
6.2. Amplified Fragment Length Polymorphism
| Year | Marker | Objectives of the Study | Results | Reference |
|---|---|---|---|---|
| 1987 | Morphological and allozyme | To analyze linkage relationship between morphological and allozyme marker | 30 morphological trait loci were identified, and first common buckwheat linkage map was constructed | [164] |
| 1998; 1999 | RAPD, SCAR | To identify RAPD markers linked to the homostylar (Ho) gene | 3 RAPD markers (one successfully converted to SCAR) linked to the Ho gene were developed | [82,148] |
| 2000 | RAPD | To study phylogenetic relationship among wild and cultivated Tartary buckwheat | Phylogenetic tree was constructed; north-western Yunnan is most likely the origin of cultivated Tartary buckwheat | [149] |
| 2004 | RAPD | Characterization of interspecific hybridization between F. esculentum and F. homotropicum | RAPD markers were able to successfully determine F1 hybrids between F. esculentum and F. homotropicum | [156] |
| 2004 | AFLP | To perform linkage analysis of F. esculentum and F. homotropicum | First high-density genetic map with genome-wide AFLP markers was constructed, and three morphological trait genes were mapped | [88] |
| 2005 | AFLP | To study genetic relationship among cultivated and wild common buckwheat | Phylogenetic tree was constructed; cultivated common buckwheat probably originated from Sanjiang area | [161] |
| 2012 | AFLP | To characterize Tartary buckwheat for rutin content variation | AFLP fingerprinting can be used to identify high rutin content accessions | [163] |
| 2006 | SSR | To develop SSR markers for common buckwheat | 54 SSR markers were developed, and transferability in closely related species was demonstrated | [165] |
| 2006 | SSR, AFLP | To construct linkage map for common buckwheat with SSR and AFLP markers | Female and male linkage map with 12 linkage groups (8 large) was constructed. 19 SSR markers also worked in Tartary buckwheat | [166] |
6.3. Genome Studies by Next-Generation Sequencing Methods
7. Genetic Transformation and Genome Editing
7.1. In Vitro Transformations
7.2. In Planta Transformations
7.3. Genome Editing
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ohnishi, O. Discovery of the wild ancestor of common buckwheat. Fagopyrum 1990, 11, 5–10. [Google Scholar]
- Ohsako, T.; Li, C. Classification and systematics of the Fagopyrum species. Breed. Sci. 2020, 70, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Chrungoo, N.K.; Chettry, U. Buckwheat: A critical approach towards assessment of its potential as a super crop. Indian J. Genet. Plant Breed. 2021, 81, 1–23. [Google Scholar] [CrossRef]
- Zhang, K.; Fan, Y.; Weng, W.; Tang, Y.; Zhou, M. Fagopyrum longistylum (Polygonaceae), a new species from Sichuan, China. Phytotaxa 2021, 482, 173–182. [Google Scholar] [CrossRef]
- De Arcangelis, E.; Cuomo, F.; Trivisonno, M.C.; Marconi, E.; Messia, M.C. Gelatinization and pasta making conditions for buckwheat gluten-free pasta. J. Cereal Sci. 2020, 95, 103073. [Google Scholar] [CrossRef]
- Eggum, B.O. The Protein Quality of Buckwheat in Comparison with other Protein Sources of Plant or Animal Origin. In Buckwheat. Genetics, Plant Breeding, Utilization; Kreft, I., Javornik, B., Dolinšek, B., Eds.; Biotechnical Faculty: Ljubljana, Slovenia, 1980; pp. 115–120. [Google Scholar]
- Škrabanja, V.; Kreft, I. Nutritional Value of Buckwheat Proteins and Starch. In Molecular Breeding and Nutritional Aspects of Buckwheat; Zhou, M., Kreft, I., Woo, S.H., Chrungoo, N., Javornik, B., Dolinsek, B., Eds.; Elsevier BV: Amsterdam, The Netherlands, 2016; pp. 169–176. [Google Scholar]
- Krumina-Zemture, G.; Beitane, I.; Gramatina, I. Amino acid and dietary fibre content of pea and buckwheat flours. Res. Rural Dev. 2016, 1, 84–90. [Google Scholar]
- Sytar, O.; Chrenkova, M.; Ferencova, J.; Polacikova, M.; Rajsky, M.; Brestic, M. Nutrient capacity of amino acids from buckwheat seeds and sprouts. J. Food Nutr. Res. 2018, 57, 38–47. [Google Scholar]
- Skrabanja, V.; Kreft, I. Resistant starch formation following autoclaving of Buckwheat (Fagopyrum esculentum Moench) Groats. An In Vitro Study. J. Agric. Food Chem. 1998, 46, 2020–2023. [Google Scholar] [CrossRef]
- Skrabanja, V.; Lærke, H.N.; Kreft, I. Protein-polyphenol interactions and in vivo digestibility of buckwheat groat proteins. Pflügers Arch. Eur. J. Physiol. 2000, 440, R129–R131. [Google Scholar] [CrossRef] [PubMed]
- Annor, G.A.; Tyl, C.; Marcone, M.; Ragaee, S.; Marti, A. Why do millets have slower starch and protein digestibility than other cereals? Trends Food Sci. Technol. 2017, 66, 73–83. [Google Scholar] [CrossRef]
- Jin, J.; Okagu, O.D.; Yagoub, A.E.A.; Udenigwe, C.C. Effects of sonication on the in vitro digestibility and structural properties of buckwheat protein isolates. Ultrason. Sonochem. 2021, 70, 105348. [Google Scholar] [CrossRef] [PubMed]
- Holasova, M.; Fiedlerova, V.; Smrcinova, H.; Orsak, M.; Lachman, J.; Vavreinova, S. Buckwheat—The source of antioxidant activity in functional foods. Food Res. Int. 2002, 35, 207–211. [Google Scholar] [CrossRef]
- Fabjan, N.; Rode, J.; Košir, I.J.; Wang, Z.; Zhang, Z.; Kreft, I. Tartary Buckwheat (Fagopyrum tataricum Gaertn.) as a Source of Dietary Rutin and Quercitrin. J. Agric. Food Chem. 2003, 51, 6452–6455. [Google Scholar] [CrossRef] [PubMed]
- Kreft, M. Buckwheat phenolic metabolites in health and disease. Nutr. Res. Rev. 2016, 29, 30–39. [Google Scholar] [CrossRef] [Green Version]
- Germ, M.; Árvay, J.; Vollmannova, A.; Tóth, T.; Golob, A.; Luthar, Z.; Kreft, I. The temperature threshold for the transformation of rutin to quercetin in Tartary buckwheat dough. Food Chem. 2019, 283, 28–31. [Google Scholar] [CrossRef]
- Glavač, N.K.; Stojilkovski, K.; Kreft, S.; Park, C.H.; Kreft, I. Determination of fagopyrins, rutin, and quercetin in Tartary buckwheat products. LWT 2017, 79, 423–427. [Google Scholar] [CrossRef]
- Germ, M.; Árvay, J.; Vollmannová, A.; Tóth, T.; Kreft, I.; Golob, A. Hydrothermal Treatments Affecting the Concentration of Neochlorogenic Acid in Dough of Tartary Buckwheat. Agriculture 2020, 10, 601. [Google Scholar] [CrossRef]
- Zhou, M.; Sun, Z.; Ding, M.; Logacheva, M.D.; Kreft, I.; Wang, D.; Yan, M.; Shao, J.; Tang, Y.; Wu, Y.; et al. FtSAD2 and FtJAZ1 regulate activity of the FtMYB11 transcription repressor of the phenylpropanoid pathway in Fagopyrum tataricum. New Phytol. 2017, 216, 814–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dziedzic, K.; Kurek, S.; Mildner–Szkudlarz, S.; Kreft, I.; Walkowiak, J. Fatty acids profile, sterols, tocopherol and squalene content in Fagopyrum tataricum seed milling fractions. J. Cereal Sci. 2020, 96, 103118. [Google Scholar] [CrossRef]
- Golob, A.; Stibilj, V.; Kreft, I.; Germ, M. The Feasibility of Using Tartary Buckwheat as a Se-Containing Food Material. J. Chem. 2015, 2015, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Golob, A.; Germ, M.; Kreft, I.; Zelnik, I.; Kristan, U.; Stibilj, V. Selenium uptake and Se compounds in Se-treated buckwheat. Acta Bot. Croat. 2016, 75, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Germ, M.; Stibilj, V.; Šircelj, H.; Jerše, A.; Kroflič, A.; Golob, A.; Maršić, N.K. Biofortification of common buckwheat microgreens and seeds with different forms of selenium and iodine. J. Sci. Food Agric. 2019, 99, 4353–4362. [Google Scholar] [CrossRef]
- Ožbolt, L.; Kreft, S.; Kreft, I.; Germ, M.; Stibilj, V. Distribution of selenium and phenolics in buckwheat plants grown from seeds soaked in Se solution and under different levels of UV-B radiation. Food Chem. 2008, 110, 691–696. [Google Scholar] [CrossRef]
- Stojilkovski, K.; Glavač, N.K.; Kreft, S.; Kreft, I. Fagopyrin and flavonoid contents in common, Tartary, and cymosum buckwheat. J. Food Compos. Anal. 2013, 32, 126–130. [Google Scholar] [CrossRef]
- Golob, A.; Stibilj, V.; Kreft, I.; Vogel-Mikuš, K.; Gaberščik, A.; Germ, M. Selenium treatment alters the effects of UV radiation on chemical and production parameters in hybrid buckwheat. Acta Agric. Scand. Sect. B Plant Soil Sci. 2017, 68, 1–11. [Google Scholar] [CrossRef]
- Wang, K.-J.; Zhang, Y.-J.; Yang, C.-R. Antioxidant phenolic constituents from Fagopyrum dibotrys. J. Ethnopharmacol. 2005, 99, 259–264. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Yan, J.; Zhou, M.; Woo, S.-H.; Weng, W.; Cheng, J.; Zhang, K. Tartary Buckwheat: An Under-utilized Edible and Medicinal Herb for Food and Nutritional Security. Food Rev. Int. 2020, 1–15. [Google Scholar] [CrossRef]
- Ohnishi, O. Search for the wild ancestor of buckwheat I. Description of new Fagopyrum (Polygonaceae) species and theirdistribution in China and the Himalayan hills. Fagopyrum 1998, 15, 18–28. [Google Scholar]
- Ohnishi, O. Search for the wild ancestor of buckwheat III. The wild ancestor of cultivated common buckwheat, and of tatary buckwheat. Econ. Bot. 1998, 52, 123–133. [Google Scholar] [CrossRef]
- Ohsako, T.; Ohnishi, O. New Fagopyrum species revealed by morphological and molecular analyses. Genes Genet. Syst. 1998, 73, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.-F. A study of resources of Fagopyrum (Polygonaceae) native to China. Bot. J. Linn. Soc. 1999, 130, 53–64. [Google Scholar] [CrossRef]
- Ohsako, T.; Yamane, K.; Ohnishi, O. Two new Fagopyrum(Polygonaceae) species, F. gracilipedoides and F. jinshaense from Yunnan, China. Genes Genet. Syst. 2002, 77, 399–408. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.L.; Tang, Y.; Xia, M.Z.; Shao, J.R.; Cai, G.Z.; Luo, Q.; Sun, J.X. Fagopyrum crispatifolium J. L. Liu, a new species of Polygonaceae from Sichuan, China. J. Syst. Evol. 2008, 46, 929–932. [Google Scholar] [CrossRef]
- Tang, Y.; Zhou, M.-L.; Bai, D.-Q.; Shao, J.-R.; Zhu, X.-M.; Wang, D.-Z.; Tang, Y.-X. Fagopyrum pugense (Polygonaceae), a New Species from Sichuan, China. Novon A J. Bot. Nomencl. 2010, 20, 239–242. [Google Scholar] [CrossRef]
- Tang, Y.; Shao, J.R.; Zhou, M.L. A Taxonomic Revision of Fagopyrum Mill. from China. J. Plant Genet. Resour. 2019, 20, 646–653. [Google Scholar]
- Shao, J.-R.; Zhou, M.-L.; Zhu, X.-M.; Wang, D.-Z.; Bai, D.-Q. Fagopyrum wenchuanenseandFagopyrum qiangcai, Two New Species of Polygonaceae from Sichuan, China. Novon A J. Bot. Nomencl. 2011, 21, 256–261. [Google Scholar] [CrossRef]
- Hou, L.-L.; Zhou, M.-L.; Zhang, Q.; Qi, L.-P.; Yang, X.-B.; Tang, Y.; Zhu, X.-M.; Shao, J.-R. Fagopyrum luojishanense, a New Species of Polygonaceae from Sichuan, China. Novon A J. Bot. Nomencl. 2015, 24, 22–26. [Google Scholar] [CrossRef]
- Zhou, M.-L.; Zhang, Q.; Zheng, Y.-D.; Tang, Y.; Li, F.-L.; Zhu, X.-M.; Shao, J.-R. Fagopyrum hailuogouense(Polygonaceae), One New Species from Sichuan, China. Novon A J. Bot. Nomencl. 2015, 24, 222–224. [Google Scholar] [CrossRef]
- Zhou, M.; Tang, Y.; Deng, X.; Ruan, C.; Tang, Y.; Wu, Y. Classification and Nomenclature of Buckwheat Plants. In Buckwheat Germplasm in the World; Elsevier BV: Amsterdam, The Netherlands, 2018; pp. 9–20. [Google Scholar]
- Wang, C.-L.; Li, Z.-Q.; Ding, M.-Q.; Tang, Y.; Zhu, X.-M.; Liu, J.-L.; Shao, J.-R.; Zhou, M.-L. Fagopyrum longzhoushanense, a new species of Polygonaceae from Sichuan, China. Phytotaxa 2017, 291, 73. [Google Scholar] [CrossRef]
- Jin, J.; Li, B.; Li, D.; Chen, S. A common Bistorta was misidentified as a novel species in Fagopyrum (Polygonaceae): The confirmation of the taxonomic identify of F. hailuogouense by morphological and molecular evidences. Phytotaxa 2018, 348, 221–228. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.-F.; Huang, X.-Y.; Li, H.-Y.; Yang, L.-J.; Cui, Y.-S. Recent Progress in Perennial Buckwheat Development. Sustainability 2018, 10, 536. [Google Scholar] [CrossRef] [Green Version]
- Ohsako, T.; Ohnishi, O. Nucleotide sequence variation of the chloroplast trnK/matK region in two wild Fagopyrum(Polygonaceae) species, F. leptopodum and F. statice. Genes Genet. Syst. 2001, 76, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Ohsako, T.; Fukuoka, S.; Bimb, H.P.; Baniya, B.K.; Yasui, Y.; Ohnishi, O. Phylogenetic analysis of the genus Fagopyrum (Polygonaceae), including the Nepali species F. megacarpum, based on nucleotide sequence of the rbcL-accD region in chloroplast DNA. Fagopyrum 2001, 18, 9–14. [Google Scholar]
- Ohsako, T.; Li, C.; Tian, B. Evolutionary relationship between a wild ancestor of common buckwheat Fagopyrum esculentum subsp. ancestrale and a self-compatible relative F. homotropicum based on microsatellite variability. Genet. Resour. Crop. Evol. 2016, 64, 1595–1603. [Google Scholar] [CrossRef]
- Woo, S.H.; Roy, S.; Kwon, S.; Cho, S.-W.; Sarker, K.; Lee, M.-S.; Chung, K.-Y.; Kim, H.-H. Concepts, Prospects, and Potentiality in Buckwheat (Fagopyrum esculentum Moench): A Research Perspective. In Molecular Breeding and Nutritional Aspects of Buckwheat; Elsevier BV: Amsterdam, The Netherlands, 2016; pp. 21–49. [Google Scholar]
- Joshi, D.C.; Chaudhari, G.V.; Sood, S.; Kant, L.; Pattanayak, A.; Zhang, K.; Fan, Y.; Janovská, D.; Meglič, V.; Zhou, M. Revisiting the versatile buckwheat: Reinvigorating genetic gains through integrated breeding and genomics approach. Planta 2019, 250, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, O.; Matsuoka, Y. Search for the wild ancestor of buckwheat II. Taxonomy of Fagopyrum (Polygonaceae) species based on morphology, isozymes and cpDNA variability. Genes Genet. Syst. 1996, 71, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, O.; Asano, N. Genetic diversity of Fagopyrum homotropicum, a wild species related to common buckwheat. Genet. Resour. Crop. Evol. 1999, 46, 389–398. [Google Scholar] [CrossRef]
- Tian, X.M.; Liu, R.R.; Tian, B.; Liu, J.Q. Karyological studies of Parapteropyrum and Atraphaxis (Polygonaceae). Caryologia 2009, 62, 261–266. [Google Scholar]
- Linnaeus, C. Species Plantarum; Laurentius Salvius: Stockholm, Sweden, 1753; Volume 1. [Google Scholar]
- Treviranus, L.C. Horti Botanici Vratislaviensis Plantarum vel Novarum vel Minus Cognitarum Manipulus. Plantarum vel Novarum vel Minus Cognitarum Manipulus: Descripsit et Observationibus nec non Tabulis Tribus Illustravit. In Nova Acta Physico Academia Caesareae Leopoldino-Carolinae; Schlesische Friedrich-Wilhelms-Universität zu Breslau: Breslau, Poland, 1826; Volume 13. [Google Scholar]
- Meisner, C.D.F. Polygonaceae. In Plantae Asiaticae Rariores: Or, Descriptions and Figures of a Select Number of Unpublished East Indian Plants; Wallich, N., Ed.; Treuttel & Wuürtz: London, UK, 1832; Volume 3, pp. 51–65. [Google Scholar]
- Moench, C. Methodus plantas horti botanici et agri Marburgensis: A Staminum Situ Describendi/Auctore Conrado Moench. In Methodus Plantas Horti Botanici et Agri Marburgensis: A Staminum Situ Describendi/Auctore Conrado Moench; Smithsonian Institution: Washington, DC, USA, 1794. [Google Scholar]
- Meisner, C.D.F. Polygonaceae. In Prodromus Systematis Naturalis Regni Vegetabilis; de Candolle, A., Ed.; Victoris Masson: Paris, France, 1857; Volume 14, pp. 1–186. [Google Scholar]
- Samuelsson, G. Polygoneceae. In Symbolae Sinicae 7; Handel-Mazzetti, H., Ed.; Julius Springer: Wien, Austria, 1929; pp. 166–188. [Google Scholar]
- Haraldson, K. Anatomy and Taxonomy in Polygonaceae Subfam. Polygonoideae Meisn. Emend. Jaretzky (Symbolae Botanicae Upsalienses); Almqvist & Wiksell international: Uppsala, Sweden, 1978. [Google Scholar]
- Bureau, L.E.; Franchet, A.R. Plantes Nouvelles du Thibet et de la Chine Occidentale: Recueillies Pendant le Voyage de M. Bonvalot et du Prince Henri D’orléans en 1890; Nabu Press: Paris, France, 1891. [Google Scholar]
- Gross, M.H. Remarques sur les Polygonées de I’Asie Orientale. Bull. Géographie Bot. 1913, 23, 7–32. [Google Scholar]
- Diels, L. Die flora von Central China. Botanische Jahrbücher für Systematik. Pflanzengesch. Pflanzengeogr. 1901, 29, 169–659. [Google Scholar]
- Hedberg, O. Pollen morphology in the genus Polygonum L. s. lat. and its taxonomical significance. Sven. Bot. Tidskr. 1946, 40, 371–404. [Google Scholar]
- Liu, J.L.; Tang, Y.; Xia, M.Z.; Shao, J.R.; Cai, G.Z.; Luo, Q.; Sun, J.X. Fagopyrum densovillosum J. L. Liu, a New Species of Polygonaceae from Sichuan, China. Bull. Bot. Res. 2008, 28, 530–533. [Google Scholar] [CrossRef]
- Gaertner, J. De Fructibus et Seminibus Plantarum; Academiae Carolina: Stuttgart, Germany, 1791; Volume 2. [Google Scholar]
- Ohnishi, O. Discovery of new Fagopyrum species and its implication for the studies of evolution of Fagopyrum and of the origin of cultivated buckwheat. In Current Advances in Buckwheat Research: Proceedings of the 6th International Symposium on Buckwheat Shinsu, Japan, 24–29 August 1995; Matano, T., Ujihara, A., Eds.; Shinshu University Press: Shinshu, Japan, 1995; Volume I–III, pp. 175–190. [Google Scholar]
- Forbes, F.B.; Hemsley, W.B. An Enumeration of all the Plants known from China Proper, Formosa, Hainan, Corea, the Luchu Archipelago, and the Island of Hongkong, together with their Distribution and Synonymy-Part X. J. Linn. Soc. London, Bot. 1891, 26, 317–396. [Google Scholar] [CrossRef]
- Tian, X.; Luo, J.; Wang, A.; Mao, K.; Liu, J. On the origin of the woody buckwheat Fagopyrum tibeticum (=Parapteropyrum tibeticum) in the Qinghai–Tibetan Plateau. Mol. Phylogenetics Evol. 2011, 61, 515–520. [Google Scholar] [CrossRef]
- Krotov, A.S.; Dranenko, E.T. An Amphidiploid of Buckwheat Fagopyrum giganteum Krotov sp. nova. Byulleten Vsesoyuznogo Ordena Lenina Inst. Rastenievod. Im. N.I. Vavilova 1973, 30, 41–45. [Google Scholar]
- Nagano, M.; Aii, J.; Campbell, C.; Kawasakp, S.; Adachi, T. Genome size analysis of the genus Fagopyrum. Fagopyrum 2000, 17, 35–39. [Google Scholar]
- Nagatomo, T.; Adachi, T. Fagopyrum esculentum. In Handbook of Flowering; Halevy, A.H., Ed.; CRC Press: Boca Raton, FL, USA, 1985; Volume III, pp. 1–8. [Google Scholar]
- Campbell, C.G. Buckwheat, Fagopyrum esculentum Moench; International Plant Genetic Resources Institute: Rome, Italy, 1997. [Google Scholar]
- Quinet, M.; Cawoy, V.; Lefèvre, I.; Van Miegroet, F.; Jacquemart, A.-L.; Kinet, J.-M. Inflorescence structure and control of flowering time and duration by light in buckwheat (Fagopyrum esculentum Moench). J. Exp. Bot. 2004, 55, 1509–1517. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.H.; Kamal, A.H.M.; Tatsuro, S.; Campbell, C.G.; Adachi, T.; Yun, S.H.; Chung, K.Y.; Choi, J.S. Buckwheat (Fagopyrum esculentum Moench.): Concepts, Prospects and Potential. Eur. J. Plant Sci. Biotechnol. 2010, 4, 1–16. [Google Scholar]
- Yasui, Y.; Hirakawa, H.; Ueno, M.; Matsui, K.; Katsube-Tanaka, T.; Yang, S.J.; Aii, J.; Sato, S.; Mori, M. Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res. 2016, 23, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Ganders, F.R. The biology of heterostyly. N. Z. J. Bot. 1979, 17, 607–635. [Google Scholar] [CrossRef]
- Matsui, K.; Nishio, T.; Tetsuka, T. Genes Outside the S Supergene Suppress S Functions in Buckwheat (Fagopyrum esculentum). Ann. Bot. 2004, 94, 805–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garber, R.J.; Quisenberry, K.S. Self-fertilization in Buckwheat. J. Agric. Res. 1927, 34, 185–190. [Google Scholar]
- Lewis, D.; Jones, D.A. The Genetics of Heterostyly. Monogr. Theor. Appl. Genet. 1992, 15, 129–150. [Google Scholar] [CrossRef]
- Sharma, K.D.; Boyes, J.W. Modified incompatibility of buckwheat following irradiation. Can. J. Bot. 1961, 39, 1241–1246. [Google Scholar] [CrossRef]
- Yasui, Y.; Mori, M.; Aii, J.; Abe, T.; Matsumoto, D.; Sato, S.; Hayashi, Y.; Ohnishi, O.; Ota, T. S-LOCUS EARLY FLOWERING 3 Is Exclusively Present in the Genomes of Short-Styled Buckwheat Plants that Exhibit Heteromorphic Self-Incompatibility. PLoS ONE 2012, 7, e31264. [Google Scholar] [CrossRef]
- Ali, J.; Nagano, M.; Penner, G.A.; Campbell, C.G.; Adachi, T. Identification of RAPD markers linked to the Homostylar (Ho) gene in buckwheat. Breed. Sci. 1998, 48, 59–62. [Google Scholar]
- Woo, S.H.; Adachi, T.; Jong, S.K.; Campbell, C.G. Inheritance of self-compatibility and flower morphology in an inter-specific buckwheat hybrid. Can. J. Plant Sci. 1999, 79, 483–490. [Google Scholar] [CrossRef]
- Matsui, K.; Tetsuka, T.; Nishio, T.; Hara, T. Heteromorphic incompatibility retained in self-compatible plants produced by a cross between common and wild buckwheat. New Phytol. 2003, 159, 701–708. [Google Scholar] [CrossRef]
- Matsui, K.; Mizuno, N.; Ueno, M.; Takeshima, R.; Yasui, Y. Development of co-dominant markers linked to a hemizygous region that is related to the self-compatibility locus (S) in buckwheat (Fagopyrum esculentum). Breed. Sci. 2020, 70, 112–117. [Google Scholar] [CrossRef] [Green Version]
- Matsui, K.; Tetsuka, T.; Hara, T. Two independent gene loci controlling non-brittle pedicels in buckwheat. Euphytica 2003, 134, 203–208. [Google Scholar] [CrossRef]
- Nagano, M.; Aii, J.; Kuroda, M.; Campbell, C.; Adachi, T. Conversion of AFLP Markers Linked to the Sh Allele at the S Locus in Buckwheat to a Simple PCR Based Marker Form. Plant Biotechnol. 2001, 18, 191–196. [Google Scholar] [CrossRef] [Green Version]
- Yasui, Y.; Wang, Y.; Ohnishi, O.; Campbell, C.G. Amplified fragment length polymorphism linkage analysis of common buckwheat (Fagopyrum esculentum) and its wild self-pollinated relative Fagopyrum homotropicum. Genome 2004, 47, 345–351. [Google Scholar] [CrossRef]
- Schoch-Bodmer, H. Zum Heterostylieproblem: Griffelbeschaffenheit und Pollenschlauchwachstum bei Fagopyrum esculentum. Planta 1934, 22, 149–152. [Google Scholar] [CrossRef]
- Miljuš-Đukić, J.; Ninkovic, S.; Nešković, M. Effects of Protein Phosphatase Inhibitors and Calcium Antagonists on Self-Incompatible Reaction in Buckwheat. Biol. Plant. 2003, 46, 475–478. [Google Scholar] [CrossRef]
- Miljuš-Đukić, J.; Ninkovic, S.; Radović, S.; Maksimović, V.; Brkljačić, J.; Nešković, M. Detection of Proteins Possibly Involved in Self-Incompatibility Response in Distylous Buckwheat. Biol. Plant. 2004, 48, 293–296. [Google Scholar] [CrossRef]
- Chen, Q.-F. Hybridization between Fagopyrum (Polygonaceae) species native to China. Bot. J. Linn. Soc. 1999, 131, 177–185. [Google Scholar] [CrossRef]
- Minami, M.; Gomi, M.; Ujihara, A. Pollen tube growth in the interspecific crosses in Fagopyrum. J. Fac. Agric. Shinshu Univ. 1992, 29, 129–135. [Google Scholar]
- Hirose, T.; Ujihara, A.; Kitabayashi, H.; Minami, M. Interspecific Cross-compatibility in Fagopyrum according to Pollen Tube Growth. Jpn. J. Breed. 1994, 44, 307–314. [Google Scholar] [CrossRef] [Green Version]
- Hirose, T.; Ujihara, A.; Kitabayashi, H.; Minami, M. Pollen Tube Behavior Related to Self-incompatibility in Interspecific Crosses of Fagopyrum. Jpn. J. Breed. 1995, 45, 65–70. [Google Scholar] [CrossRef] [Green Version]
- Kachonpadungkitti, Y.; Mangkita, W.; Romchatngoen, S.; Hasegawa, K.; Hisajima, S. Possibility of Cross Breeding of Buckwheat (Fagopyrum esculentum Moench) in vitro. Shokubutsu Kojo Gakkaishi 2003, 15, 98–101. [Google Scholar] [CrossRef] [Green Version]
- Morris, M.R. Cytogenetic studies on buckwheat. J. Hered. 1951, 42, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Ujihara, A.Y.; Nakamura, Y.; Minami, M. Interspecific hybridization in genus Fagopyrum: Properties of hybrids (F. esculentum Moench x F. cymosum Meissner) through ovule culture. Gamma Field Symp. 1990, 29, 33–45. [Google Scholar]
- Samimy, C. Barrier to interspecific crossing of Fagopyrum esculentum with Fagopyrum tataricum: I. Site of pollen-tube arrest. II. Organogenesis from immature embryos of F. tataricum. Euphytica 1991, 54, 215–219. [Google Scholar] [CrossRef]
- Lee, B.S.; Ujihara, A.; Minami, M.; Hirose, T. Breeding of interspecific hybrids in genus Fagopyrum. (4). Production of interspecific hybrids through ovule culture among F. esculentum, F. tataricum and F. cymosum. Breed. Sci. 1994, 44, 183. [Google Scholar]
- Mendler-Drienyovszki, N.; Cal, A.J.; Dobránszki, J. Progress and prospects for interspecific hybridization in buckwheat and the genus Fagopyrum. Biotechnol. Adv. 2013, 31, 1768–1775. [Google Scholar] [CrossRef] [PubMed]
- Yamane, Y. Induced differentiation of buckwheat plants from subcultured calluses in vitro. Jpn. J. Genet. 1974, 49, 139–146. [Google Scholar] [CrossRef] [Green Version]
- Srejović, V.; Nešković, M. Regeneration of Plants from Cotyledon Fragments of Buckwheat (Fagopyrum esculentum Moench). Z. Pflanzenphysiol. 1981, 104, 37–42. [Google Scholar] [CrossRef]
- Takahata, Y.; Jumonji, E. Plant Regeneration from Hypocotyl Section and Callus in Buckwheat (Fagopyrum esculentum Moench.). Ann. Rep. Fac. Educ. Iwate Univ. 1985, 45, 137–142. [Google Scholar]
- Srejović, V.; Nešković, M. Effect of gibberellic acid on organogenesis in buckwheat tissue culture. Biol. Plant. 1985, 27, 432–437. [Google Scholar] [CrossRef]
- Nešković, M.; Vujičić, R.; Budimir, S. Somatic embryogenesis and bud formation from immature embryos of buckwheat (Fagopyrum esculentum Moench.). Plant Cell Rep. 1987, 6, 423–426. [Google Scholar] [CrossRef]
- Hao, J.; Pei, Y.; Qu, Y.; Zheng, C. Study on callus differentiation conditions of common buckwheat. In Proceedings of the 7th International Symposium on Buckwheat, Winnipeg, MB, Canada, 12–14 August 1998; pp. V-33–V-37. [Google Scholar]
- Lachmann, S.; Adachi, T. Callus regeneration from hypocotyl protoplasts of tartary buckwheat (Fagopyrum tataricum Gartn.). Fagopyrum 1990, 10, 62–64. [Google Scholar]
- Lachmann, S. Plant cell and tissue culture in buckwheat: An approach towards genetic improvements by means of unconventional breeding techniques. In Overcoming Breeding Barriers by Means of Plant Biotechnology; Adachi, T., Ed.; Proceedings of the International College: Miyazaki, Japan, 1991; pp. 145–154. [Google Scholar]
- Miljuš-Djukić, J.; Nesković, M.; Ninković, S.; Crkvenjakov, R. Agrobacterium-mediated transformation and plant regeneration of buckwheat (Fagopyrum esculentum Moench.). Plant Cell Tissue Organ Cult. 1992, 29, 101–108. [Google Scholar] [CrossRef]
- Rajbhandari, B.P.; Dhanbhadel, S.; Gantam, D.M.; Gantam, B.R. Plant regeneration via calli of leaf and stem explants in common buckwheat ecotypes. In Proceedings of the 6th International Symposium on Buckwheat, Shinsu, Japan, 24–29 August 1995; pp. 191–196. [Google Scholar]
- Woo, S.H.; Nair, A.; Adachi, T.; Campbell, C.G. Plant regeneration from cotyledon tissues of common buckwheat (Fagopyrum esculentum Moench). Vitr. Cell. Dev. Biol. Anim. 2000, 36, 358–361. [Google Scholar] [CrossRef]
- Jin, H.; Jia, J.-F.; Hao, J.-G. Efficient plant regeneration in vitro in buckwheat. Plant Cell Tissue Organ Cult. (PCTOC) 2002, 69, 293–295. [Google Scholar] [CrossRef]
- Gumerova, E.A.; Galeeva, E.I.; Chuyenkova, S.A.; Rumyantseva, N.I. Somatic Embryogenesis and Bud Formation on Cultured Fagopyrum esculentum Hypocotyls. Russ. J. Plant Physiol. 2003, 50, 640–645. [Google Scholar] [CrossRef]
- Han, M.H.; Kamal, A.M.; Huh, Y.S.; Jeon, A.Y.; Bae, J.S.; Chung, K.Y.; Lee, M.S.; Park, S.U.; Jeong, H.S.; Woo, S.H. Regeneration of plantlet via somatic embryogenesis from hypocotyls of Tartary Buckwheat (Fagopyrum tataricum). Aust. J. Crop Sci. 2011, 5, 865–869. [Google Scholar]
- Takahata, Y. Plant regeneration from cultured immature inflorescence of common buckwheat (Fagopyrum esculentum Moench) and perennial buckwheat (F. cymosum Meisn.). Jpn. J. Breed. 1988, 38, 409–413. [Google Scholar] [CrossRef] [Green Version]
- Adachi, T.; Yamaguchi, A.; Miike, Y.; Hoffmann, F. Plant regeneration from protoplasts of common buckwheat (fagopyrum esculentum). Plant Cell Rep. 1989, 8, 247–250. [Google Scholar] [CrossRef]
- Bohanec, B.; Nešković, M.; Vujičić, R. Anther culture and androgenetic plant regeneration in buckwheat (Fagopyrum esculentum Moench). Plant Cell, Tissue Organ Cult. 1993, 35, 259–266. [Google Scholar] [CrossRef]
- Luthar, Z.; Marchetti, S. Plant regeneration from mature cotyledons in a buckwheat (Fagopyrum esculentum Moench.) germplasm collection. Fagopyrum 1994, 14, 65–69. [Google Scholar]
- Berbec, A.; Doroszewska, T. Regeneration in vitro of three cultivars of buckwheat (Fagopyrum esculentum Moench.) as affected by medium compositron. Fagopyrum 1999, 16, 49–52. [Google Scholar]
- Gumerova, E.; Rumyantseva, N.; Gatina, E. PECC formation in hypocotyl tissues of common buckwheat (Fagopyrum esculentum Moench). In Proceedings of the 7th International Symposium on Buckwheat, Winnipeg, MB, Canada, 12–14 August 1998; pp. V-53–V-57. [Google Scholar]
- Gumerova, E.; Gatina, E.; Chuenkova, S.; Rumyantseva, N. Somatic embryogenesis in common buckwheat Fagopyrum esculentum Moench. In Proceedings of the 8th International Symposium on Buckwheat, Chunchon, Korea, 30 August–2 September 2001; pp. 377–381. [Google Scholar]
- Rumyantseva, N.; Sergeeva, N.; Hakimova, L.; Gumerova, E.; Lozovaya, V. Morphogenesis and plant regeneration in buckwheat tissue culture. In Proceedings of the 4th International Symposium on Buckwheat, Orel, Russia, 11–15 July 1989; pp. 322–328. [Google Scholar]
- Rumyantseva, N.I.; Sal’nikov, V.V.; Fedoseeva, N.V.; Lozovaya, V.V. Peculiarities of Morphogenesis in Long-Term Cultivated Buckwheat Calluses. Fiziol. Rast. 1992, 39, 143–151. [Google Scholar]
- Kostyukova, Y.; Rumyantseva, N. Histological study of embryoidogenic callus induction in immature embryos of Tartary buckwheat Fagopyrum tataricum (L.) Gaertn. In Proceedings of the 11th International Symposium on Buckwheat, Orel, Russia, 19–23 July 2010; pp. 167–174. [Google Scholar]
- Kwon, S.-J.; Han, M.-H.; Huh, Y.-S.; Roy, S.K.; Lee, C.-W.; Woo, S.H. Plantlet Regeneration via Somatic Embryogenesis from Hypocotyls of Common Buckwheat (Fagopyrum esculentum Moench.). Korean J. Crop. Sci. 2013, 58, 331–335. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Park, C. Multiple shoot organogenesis and plant regeneration from cotyledons of buckwheat (Fagopyrum esculentum Moench). In Proceedings of the 8th International Symposium on Buckwheat, Cunchon, Korea, 30 August–2 September 2001; pp. 427–430. [Google Scholar]
- Adachi, T.; Suputitada, S.; Miike, Y. Plant regeneration from anther culture in common buckwheat (F. esculentum). Fagopyrum 1988, 8, 5–9. [Google Scholar]
- Skof, S.; Bohanec, B.; Kastelec, D.; Luthar, Z. Spontaneous induction of tetraploidy in hop using adventitious shoot regeneration method. Plant Breed. 2007, 126, 416–421. [Google Scholar] [CrossRef]
- Kong, F.; Shong, Y.; Wang, Z.; Yang, L. Study on plant regeneration from anther culture in common buckwheat (Fagopyrum esculentum). In Proceedings of the 5th International Symposium on Buckwheat, Taiyuan, China, 20–26 August 1992; pp. 309–314. [Google Scholar]
- Berbec, A.; Doroszewska, T. Callus formation and plant regeneration in anther culture of three buckwheat (Fagopyrum esculentum Moench) cultivars. In Proceedings of the 7th International Symposium on Buckwheat, Winnipeg, MB, Canada, 12–14 August 1998; pp. V-48–V-52. [Google Scholar]
- Yui, M.; Yoshida, T. Callus induction and plant regeneration in anther culture of Japanese buckwheat cultivars (Fagopyrum esculentum Moench). Fagopyrum 2001, 18, 27–35. [Google Scholar]
- Wang, Y.; Campbell, C. Effect of genotypes, pretreatments and media in anther culture of common (Fagopyrum esculentum) and self-pollinated buckwheat. Fagopyrum 2006, 23, 29–35. [Google Scholar]
- Bohanec, B. Progress of buckwheat in vitro culture techniques with special aspect on induction of haploid plants. Curr. Adv. Buckwheat Res. 1995, 1, 205–209. [Google Scholar]
- Lachmann, S.; Kishima, Y.; Adachi, T. Protoplast fusion on buckwheat: Preliminary results of somatic hybridization. Fagopyrum 1994, 14, 7–12. [Google Scholar]
- Tanaka, N.; Yoshimatsu, K.; Shimomura, K.; Ishimaru, K. Rutin and other polyphenols in Fagopyrum esculentum hairy roots. Nat. Med. 1996, 50, 269–272. [Google Scholar]
- Lee, S.; Cho, S.; Park, M.; Kim, Y.; Choi, J.; Park, S. Growth and Rutin Production in Hairy Root Cultures of Buckwheat (Fagopyrum esculentum M.). Prep. Biochem. Biotechnol. 2007, 37, 239–246. [Google Scholar] [CrossRef]
- Kim, Y.K.; Xu, H.; Park, W.T.; Park, N.I.; Lee, S.Y.; Park, S.U. Genetic transformation of buckwheat (Fagopyrum esculentum M.) with Agrobacterium rhizogenes and production of rutin in transformed root cultures. Aust. J. Crop Sci. 2010, 4, 485–490. [Google Scholar]
- Park, N.I.; Li, X.; Thwe, A.A.; Lee, S.Y.; Kim, S.G.; Wu, Q.; Park, S.U. Enhancement of rutin in Fagopyrum esculentum hairy root cultures by the Arabidopsis transcription factor AtMYB12. Biotechnol. Lett. 2011, 34, 577–583. [Google Scholar] [CrossRef]
- Kim, Y.K.; Li, X.; Xu, H.; Park, N.I.; Uddin, M.R.; Pyon, J.Y.; Park, S.U. Production of phenolic compounds in hairy root culture of tartary buckwheat (Fagopyrum tataricum Gaertn). J. Crop Sci. Biotechnol. 2009, 12, 53–57. [Google Scholar] [CrossRef]
- Park, N.; Xiaohua, L.; Uddin, R.; Park, S. Phenolic compound production by different morphological phenotypes in hairy root cultures of Fagopyrum tataricum Gaertn. Arch. Biol. Sci. 2011, 63, 193–198. [Google Scholar] [CrossRef]
- Thwe, A.A.; Kim, J.K.; Li, X.; Kim, Y.B.; Uddin, R.; Kim, S.-J.; Suzuki, T.; Park, N.I.; Park, S.U. Metabolomic Analysis and Phenylpropanoid Biosynthesis in Hairy Root Culture of Tartary Buckwheat Cultivars. PLoS ONE 2013, 8, e65349. [Google Scholar] [CrossRef]
- Thwe, A.A.; Kim, Y.; Li, X.; Kim, Y.B.; Park, N.-I.; Kim, H.H.; Kim, S.-J.; Park, S.U. Accumulation of Phenylpropanoids and Correlated Gene Expression in Hairy Roots of Tartary Buckwheat under Light and Dark Conditions. Appl. Biochem. Biotechnol. 2014, 174, 2537–2547. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; AyeThwe, A.; Kim, S.-J.; Park, J.S.; Arasu, M.; Al-Dhabi, N.A.; Park, N.I.; Park, S.U. Effect of Auxins on Anthocyanin Accumulation in Hairy Root Cultures of Tartary Buckwheat Cultivar Hokkai T10. Nat. Prod. Commun. 2016, 11, 1283–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolff, K.; Schoen, E.D.; Rijn, J.P.-V. Optimizing the generation of random amplified polymorphic DNAs in chrysanthemum. Theor. Appl. Genet. 1993, 86, 1033–1037. [Google Scholar] [CrossRef]
- Paran, I.; Michelmore, R.W. Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor. Appl. Genet. 1993, 85, 985–993. [Google Scholar] [CrossRef]
- Aii, J.; Nagano, M.; Woo, S.H.; Campbell, C.; Adachi, T. Development of the SCAR markers linked to the Sh gene in buckwheat. Fagopyrum 1999, 16, 19–22. [Google Scholar]
- Tsuji, K.; Ohnishi, O. Origin of cultivated Tatary buckwheat (Fagopyrum tataricum Gaertn.) revealed by RAPD analyses. Genet. Resour. Crop. Evol. 2000, 47, 431–438. [Google Scholar] [CrossRef]
- Sharma, T.; Jana, S. Random amplified polymorphic DNA (RAPD) variation in Fagopyrum tataricum Gaertn. accessions from China and the Himalayan region. Euphytica 2002, 127, 327–333. [Google Scholar] [CrossRef]
- Kump, B.; Javornik, B. Evaluation of genetic variability among common buckwheat (Fagopyrum esculentum Moench) populations by RAPD markers. Plant Sci. 1996, 114, 149–158. [Google Scholar] [CrossRef]
- Kump, B.; Javornik, B. Genetic diversity and relationships among cultivated and wild accessions of tartary buckwheat (Fagopyrum tataricum Gaertn.) as revealed by RAPD markers. Genet. Resour. Crop. Evol. 2002, 49, 565–572. [Google Scholar] [CrossRef]
- Rout, A.; Chrungoo, N.K. Genetic variation and species relationships in Himalayan buckwheats as revealed by SDS PAGE of endosperm proteins extracted from single seeds and RAPD based DNA fingerprints. Genet. Resour. Crop. Evol. 2007, 54, 767–777. [Google Scholar] [CrossRef]
- Pan, S.-J.; Chen, Q.-F. Genetic mapping of common buckwheat using DNA, protein and morphological markers. Hereditas 2010, 147, 27–33. [Google Scholar] [CrossRef]
- Asaduzzaman, M.; Minami, M.; Matsushima, K.; Nemoto, K. Characterization of Interspecific Hybrid Between F. tataricum and F. esculentum. J. Biol. Sci. 2009, 9, 137–144. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Scarth, R.; Campbell, C. Interspecific hybridization between diploid Fagopyrum esculentum and tetraploid F. homotropicum. Can. J. Plant Sci. 2005, 85, 41–48. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Van De Lee, T.; Hornes, M.; Friters, A.; Pot, J.; Paleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [Green Version]
- Acquaah, G. Principles of Plant Genetics and Breeding, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2012. [Google Scholar]
- Matsui, K.; Kiryu, Y.; Komatsuda, T.; Kurauchi, N.; Ohtani, T.; Tetsuka, T. Identification of AFLP makers linked to non-seed shattering locus (sht1) in buckwheat and conversion to STS markers for marker-assisted selection. Genome 2004, 47, 469–474. [Google Scholar] [CrossRef]
- Yasui, Y.; Mori, M.; Matsumoto, D.; Ohnishi, O.; Campbell, C.G.; Ota, T. Construction of a BAC library for buckwheat genome research -An application to positional cloning of agriculturally valuable traits-. Genes Genet. Syst. 2008, 83, 393–401. [Google Scholar] [CrossRef] [Green Version]
- Konishi, T.; Yasui, Y.; Ohnishi, O. Original birthplace of cultivated common buckwheat inferred from genetic relationships among cultivated populations and natural populations of wild common buckwheat revealed by AFLP analysis. Genes Genet. Syst. 2005, 80, 113–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.J.; Zang, Z.W.; Wu, B.; Li, Y.Q. Genetic Diversity in Tartary Buckwheat Revealed by AFLP Analysis. Sci. Agric. Sin. 2009, 42, 4166–4174. [Google Scholar] [CrossRef]
- Gupta, N.; Sharma, S.K.; Rana, J.C.; Chauhan, R.S. AFLP fingerprinting of tartary buckwheat accessions (Fagopyrum tataricum) displaying rutin content variation. Fitoterapia 2012, 83, 1131–1137. [Google Scholar] [CrossRef]
- Ohnishi, O.; Ohta, T. Construction of a linkage map in common buckwheat, Fagopyrum esculentum Moench. Jpn. J. Genet. 1987, 62, 397–414. [Google Scholar] [CrossRef] [Green Version]
- Konishi, T.; Iwata, H.; Yashiro, K.; Tsumura, Y.; Ohsawa, R.; Yasui, Y.; Ohnishi, O. Development and Characterization of Microsatellite Markers for Common Buckwheat. Breed. Sci. 2006, 56, 277–285. [Google Scholar] [CrossRef] [Green Version]
- Konishi, T.; Ohnishi, O. A linkage map for common buckwheat based on microsatellite and AFLP markers. Fagopyrum 2006, 23, 1–6. [Google Scholar]
- Yabe, S.; Hara, T.; Ueno, M.; Enoki, H.; Kimura, T.; Nishimura, S.; Yasui, Y.; Ohsawa, R.; Iwata, H. Rapid genotyping with DNA micro-arrays for high-density linkage mapping and QTL mapping in common buckwheat (Fagopyrum esculentum Moench). Breed. Sci. 2014, 64, 291–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enoki, H.; Nishimura, S.; Murakami, A. Method for Designing Probe in Dna Microarray, and DNA Microarray Provided with Probe Designed Thereby. U.S. Patent Application No. US20120190582A1, 26 July 2012. [Google Scholar]
- Iehisa, J.C.M.; Ohno, R.; Kimura, T.; Enoki, H.; Nishimura, S.; Okamoto, Y.; Nasuda, S.; Takumi, S. A High-Density Genetic Map with Array-Based Markers Facilitates Structural and Quantitative Trait Locus Analyses of the Common Wheat Genome. DNA Res. 2014, 21, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Yabe, S.; Hara, T.; Ueno, M.; Enoki, H.; Kimura, T.; Nishimura, S.; Yasui, Y.; Ohsawa, R.; Iwata, H. Potential of Genomic Selection in Mass Selection Breeding of an Allogamous Crop: An Empirical Study to Increase Yield of Common Buckwheat. Front. Plant Sci. 2018, 9, 276. [Google Scholar] [CrossRef] [Green Version]
- Meuwissen, T.H.E.; Hayes, B.; Goddard, M. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Penin, A.A.; Kasianov, A.S.; Klepikova, A.V.; Kirov, I.V.; Gerasimov, E.S.; Fesenko, A.N.; Logacheva, M.D. High-Resolution Transcriptome Atlas and Improved Genome Assembly of Common Buckwheat, Fagopyrum esculentum. Front. Plant Sci. 2021, 12, 264. [Google Scholar] [CrossRef]
- Logacheva, M.D.; Schelkunov, M.I.; Fesenko, A.N.; Kasianov, A.S.; Penin, A.A. Mitochondrial Genome of Fagopyrum esculentum and the Genetic Diversity of Extranuclear Genomes in Buckwheat. Plants 2020, 9, 618. [Google Scholar] [CrossRef]
- Cho, K.-S.; Yun, B.-K.; Yoon, Y.-H.; Hong, S.-Y.; Mekapogu, M.; Kim, K.-H.; Yang, T.-J. Complete Chloroplast Genome Sequence of Tartary Buckwheat (Fagopyrum tataricum) and Comparative Analysis with Common Buckwheat (F. esculentum). PLoS ONE 2015, 10, e0125332. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Ma, B.; Gao, Q.; Du, H.; Han, Y.; Li, Y.; Cao, Y.; Qi, M.; Zhu, Y.; et al. The Tartary Buckwheat Genome Provides Insights into Rutin Biosynthesis and Abiotic Stress Tolerance. Mol. Plant 2017, 10, 1224–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.-L.; Ding, M.-Q.; Zou, C.-Y.; Zhu, X.-M.; Tang, Y.; Zhou, M.-L.; Shao, J.-R. Comparative Analysis of Four Buckwheat Species Based on Morphology and Complete Chloroplast Genome Sequences. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Z.; Wang, A.; Zheng, T.; Huang, L.; Sun, W.; Zhang, Y.; Jin, W.; Zhan, J.; Cai, Y.; et al. Genome-Wide Investigation of the Auxin Response Factor Gene Family in Tartary Buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 2018, 19, 3526. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Huang, L.; Ma, Z.; Sun, W.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, expression analysis and functional study of the GRAS gene family in Tartary buckwheat (Fagopyrum tataricum). BMC Plant Biol. 2019, 19, 342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Sun, W.; Ma, Z.; Zheng, T.; Huang, L.; Wu, Q.; Zhao, G.; Tang, Z.; Bu, T.; Li, C.; et al. Genome-wide investigation of the AP2/ERF gene family in tartary buckwheat (Fagopyum tataricum). BMC Plant Biol. 2019, 19, 84. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Ma, Z.; Liu, M. Cytochrome P450 family: Genome-wide identification provides insights into the rutin synthesis pathway in Tartary buckwheat and the improvement of agricultural product quality. Int. J. Biol. Macromol. 2020, 164, 4032–4045. [Google Scholar] [CrossRef]
- Hou, S.; Sun, Z.; Linghu, B.; Xu, D.; Wu, B.; Zhang, B.; Wang, X.; Han, Y.; Zhang, L.; Qiao, Z.; et al. Genetic Diversity of Buckwheat Cultivars (Fagopyrum tartaricum Gaertn.) Assessed with SSR Markers Developed from Genome Survey Sequences. Plant Mol. Biol. Rep. 2015, 34, 233–241. [Google Scholar] [CrossRef]
- Shi, T.; Li, R.; Chen, Q.; Li, Y.; Pan, F.; Chen, Q. De novo sequencing of seed transcriptome and development of genic-SSR markers in common buckwheat (Fagopyrum esculentum). Mol. Breed. 2017, 37, 147. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, M.; Liu, L. Identification of Genetic Locus Underlying Easy Dehulling in Rice-Tartary for Easy Postharvest Processing of Tartary Buckwheat. Genes 2020, 11, 459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuie, Y.; Shimoyama, H.; Morishita, T.; Tsugama, D.; Fujino, K. A putative AGAMOUS ortholog is a candidate for the gene determining ease of dehulling in Tartary buckwheat (Fagopyrum tataricum). Planta 2020, 251, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Zhang, Y.; Zhang, Y.; Huang, K.; Yang, W.; Li, X.; Zhang, Z.; Wu, K.; Xu, X.; Ruan, R.; et al. De novo transcriptome assembly and identification of genes related to seed size in common buckwheat (Fagopyrum esculentum M.). Breed. Sci. 2019, 69, 487–497. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Bai, X.; Zhao, W.; Xiang, D.; Wan, Y.; Yan, J.; Zou, L.; Zhao, G. De Novo Assembly and Analysis of Tartary Buckwheat (Fagopyrum tataricum Garetn.) Transcriptome Discloses Key Regulators Involved in Salt-Stress Response. Genes 2017, 8, 255. [Google Scholar] [CrossRef] [Green Version]
- Ishida, Y.; Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of maize. Nat. Protoc. 2007, 2, 1614–1621. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, A.; Aichi, I.; Matsuoka, M. A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 2006, 1, 2796–2802. [Google Scholar] [CrossRef]
- Richardson, T.; Thistleton, J.; Higgins, T.; Howitt, C.; Ayliffe, M. Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell Tissue Organ Cult. 2014, 119, 647–659. [Google Scholar] [CrossRef]
- Sakamoto, S.; Matsui, K.; Oshima, Y.; Mitsuda, N. Efficient transient gene expression system using buckwheat hypocotyl protoplasts for large-scale experiments. Breed. Sci. 2020, 70, 128–134. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Kang, H.J.; Lee, Y.T.; Lee, S.Y.; Ko, J.A.; Rha, E.S. Direct regeneration of transgenic buckwheat from hypocotyl segment by Agrobacterium-mediated transformation. Korean J. Crop Sci. 2001, 45, 375–379. [Google Scholar]
- Chen, L.-H.; Zhang, B.; Xu, Z.-Q. Salt tolerance conferred by overexpression of Arabidopsis vacuolar Na+/H+ antiporter gene AtNHX1 in common buckwheat (Fagopyrum esculentum). Transgenic Res. 2008, 17, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Kojima, M.; Arai, Y.; Iwase, N.; Shirotori, K.; Shioiri, H.; Nozue, M. Development of a Simple and Efficient Method for Transformation of Buckwheat Plants (Fagopyrum esculentum) Using Agrobacterium tumefaciens. Biosci. Biotechnol. Biochem. 2000, 64, 845–847. [Google Scholar] [CrossRef] [PubMed]
- Suvorova, G. Buckwheat Tissue Cultures and Genetic Transformation. In Molecular Breeding and Nutritional Aspects of Buckwheat; Elsevier BV: Amsterdam, The Netherlands, 2016; pp. 365–375. [Google Scholar]
- Kojima, M.; Hihara, M.; Koyama, S.-I.; Shioiri, H.; Nozue, M.; Yamamoto, K.; Sasaki, T. Buckwheat Transformed with cDNA of a Rice MADS Box Gene Is Stimulated in Branching. Plant Biotechnol. 2000, 17, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Bratic, A.; Majic, D.; Miljus-Djukic, J.; Jovanovic, Z.; Maksimovic, V. In planta transformation of buckwheat (Fagopyrum esculentum Moench.). Arch. Biol. Sci. 2007, 59, 135–138. [Google Scholar] [CrossRef]
- Chawla, A.; Singh, H.; Kant, A. Development of In planta transformation method for Tartary Buckwheat (Fagopyrum tataricum). Int. Conf. Emerg. Trends Biotechnol. 2012, 45–46. [Google Scholar]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Sandhya, D.; Jogam, P.; Allini, V.R.; Abbagani, S.; Alok, A. The present and potential future methods for delivering CRISPR/Cas9 components in plants. J. Genet. Eng. Biotechnol. 2020, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Alok, A.; Sandhya, D.; Jogam, P.; Rodrigues, V.; Bhati, K.K.; Sharma, H.; Kumar, J. The rise of the CRISPR/Cpf1 system for efficient genome editing in plants. Front. Plant Sci. 2020, 11, 264. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, S.; Kim, J.-S.; Velasco, R.; Kanchiswamy, C.N. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef] [PubMed]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Alok, A.; Shivani; Kaur, N.; Pandey, P.; Awasthi, P.; Tiwari, S. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genom. 2018, 18, 89–99. [Google Scholar] [CrossRef]
- Lee, K.; Zhang, Y.; Kleinstiver, B.; Guo, J.A.; Aryee, M.J.; Miller, J.; Malzahn, A.; Zarecor, S.; Lawrence-Dill, C.J.; Joung, J.K.; et al. Activities and specificities of CRISPR /Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol. J. 2019, 17, 362–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arndell, T.; Sharma, N.; Langridge, P.; Baumann, U.; Watson-Haigh, N.S.; Whitford, R. gRNA validation for wheat genome editing with the CRISPR-Cas9 system. BMC Biotechnol. 2019, 19, 71. [Google Scholar] [CrossRef]
- Joshi, D.C.; Zhang, K.; Wang, C.; Chandora, R.; Khurshid, M.; Li, J.; He, M.; Georgiev, M.; Zhou, M. Strategic enhancement of genetic gain for nutraceutical development in buckwheat: A genomics-driven perspective. Biotechnol. Adv. 2020, 39, 107479. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luthar, Z.; Fabjan, P.; Mlinarič, K. Biotechnological Methods for Buckwheat Breeding. Plants 2021, 10, 1547. https://doi.org/10.3390/plants10081547
Luthar Z, Fabjan P, Mlinarič K. Biotechnological Methods for Buckwheat Breeding. Plants. 2021; 10(8):1547. https://doi.org/10.3390/plants10081547
Chicago/Turabian StyleLuthar, Zlata, Primož Fabjan, and Katja Mlinarič. 2021. "Biotechnological Methods for Buckwheat Breeding" Plants 10, no. 8: 1547. https://doi.org/10.3390/plants10081547
APA StyleLuthar, Z., Fabjan, P., & Mlinarič, K. (2021). Biotechnological Methods for Buckwheat Breeding. Plants, 10(8), 1547. https://doi.org/10.3390/plants10081547






