Agrobacterium-Mediated Genetic Transformation of Taiwanese Isolates of Lemna aequinoctialis
Abstract
:1. Introduction
2. Results
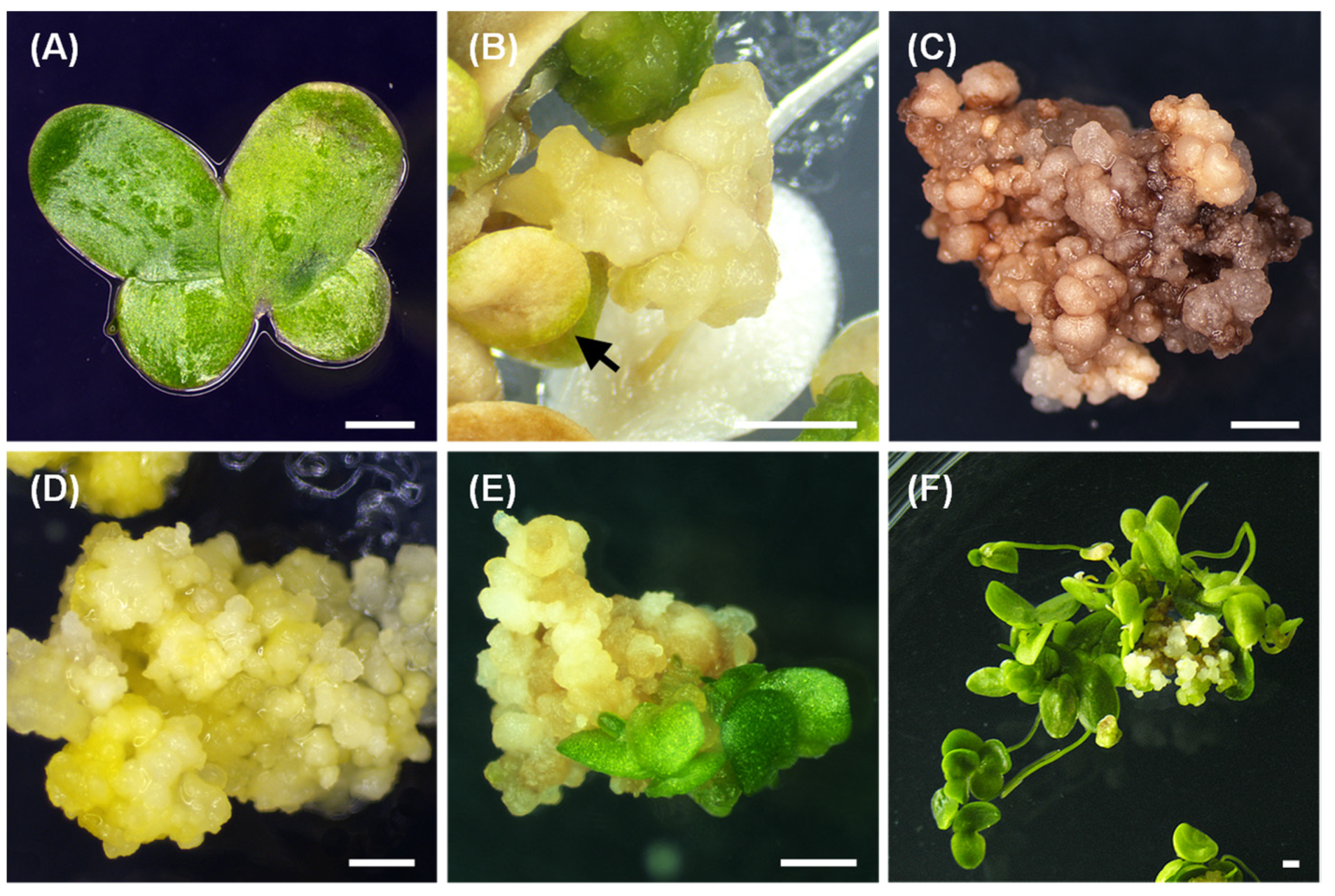
2.1. Callus Induction
2.2. Nodular Callus Proliferation
2.3. Frond Regeneration
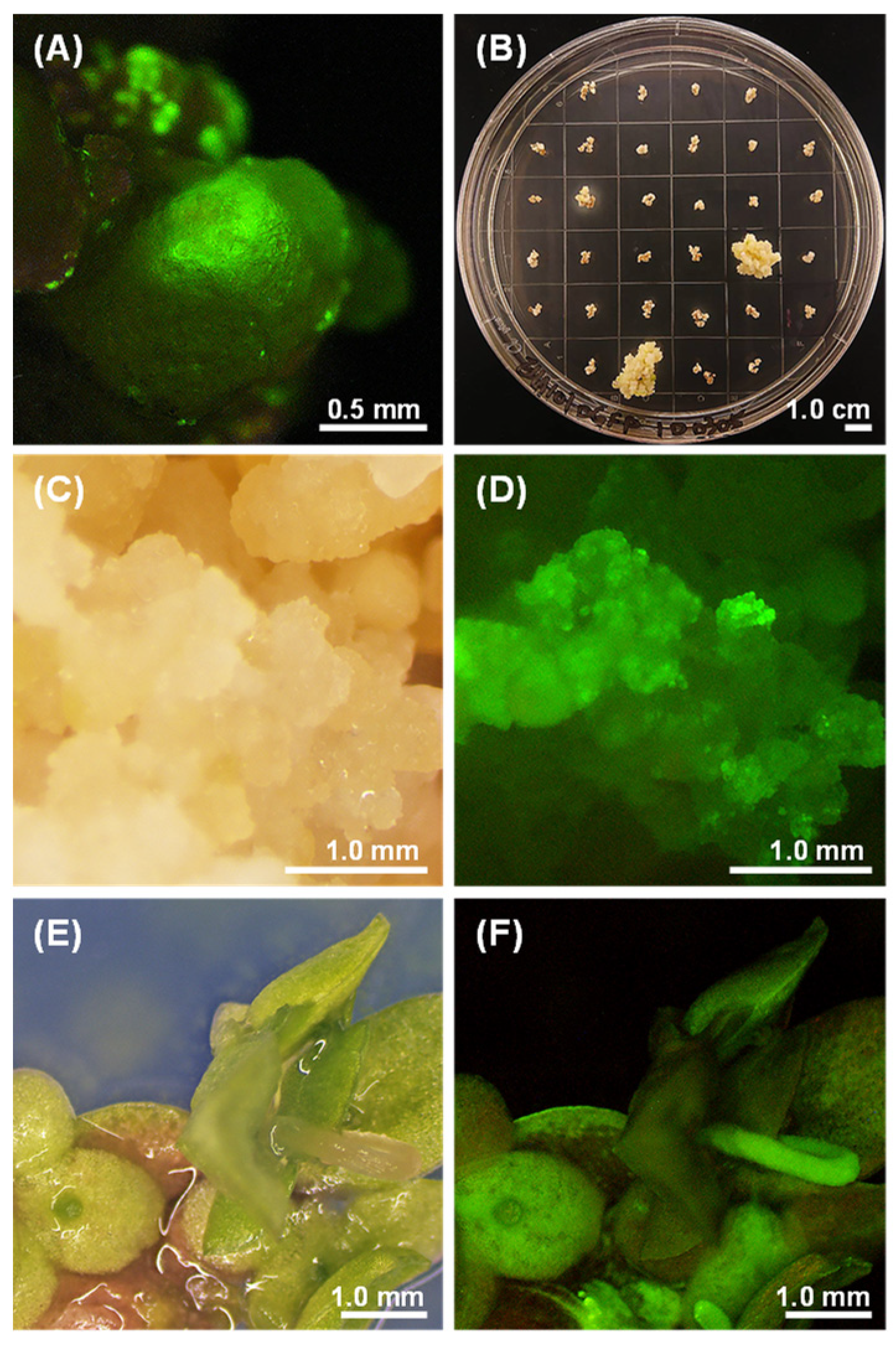
2.4. Optimization of the Transformation Protocol
2.5. Molecular Analysis of Putative Transformants
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Callus Induction
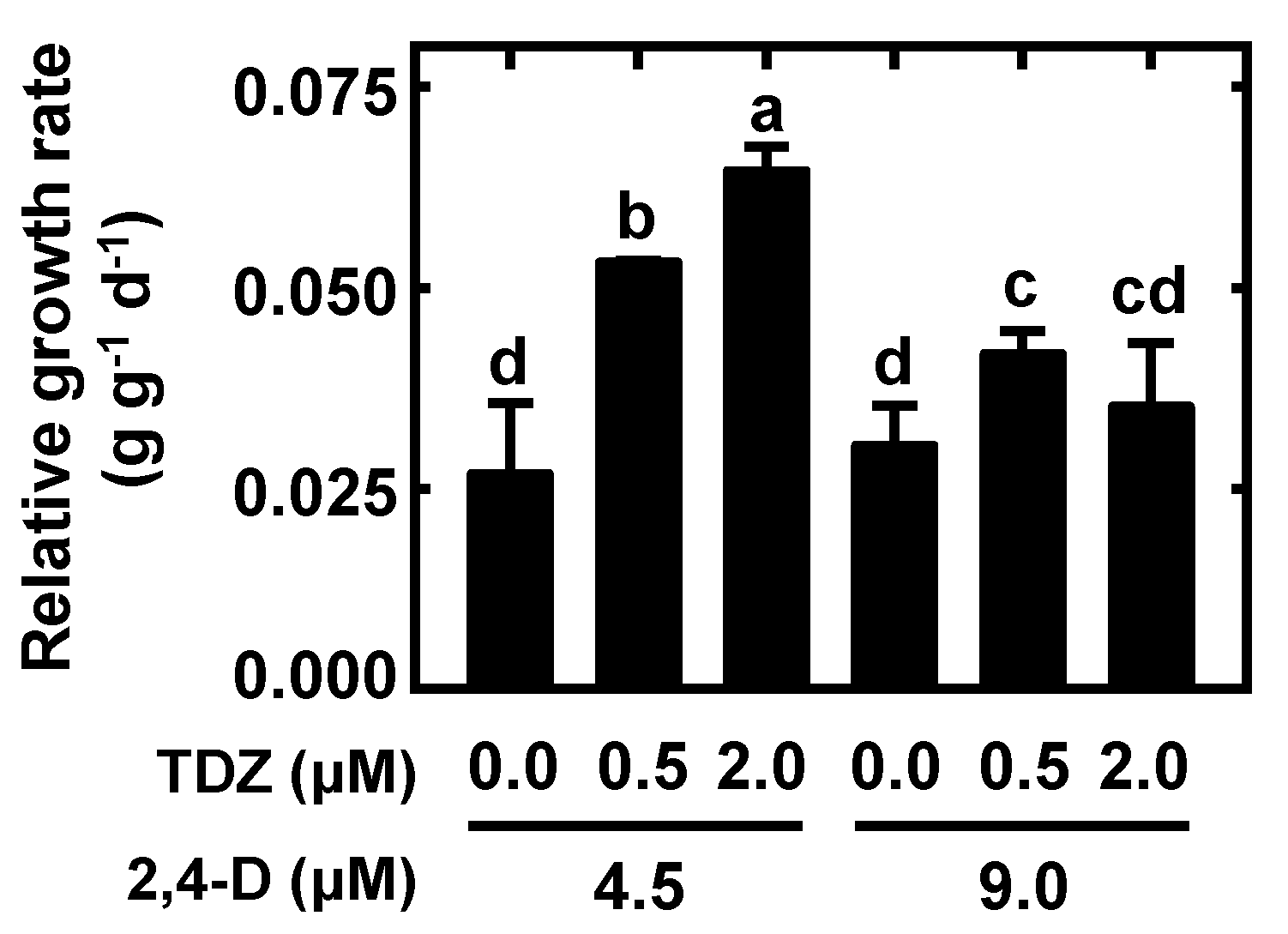
4.3. Nodular Callus Proliferation
4.4. Frond Regeneration
4.5. Agrobacterium Strain and Binary Vector
4.6. Optimization of the Transformation Protocol
4.7. Genomic PCR Analysis of Transgene Integration
4.8. RT- PCR Analysis of Transgenic Duckweed
4.9. Western Blot Analysis of Transgenic Duckweed
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Appenroth, K.; Borisjuk, N.; Lam, E. Telling duckweed apart: Genotyping technologies for the Lemnaceae. Chin. J. Appl. Environ. Biol. 2013, 19, 1–10. [Google Scholar]
- Landolt, E. The family of Lemnaceae-a monographic study. In Biosystematic Investigations in the Family of Duckweeds (Lemnaceae); Veröffentlichungen des Geobotanischen Institutes der ETH; Stiftung Rübel: Zurich, Switzerland, 1986; Volume 1. [Google Scholar]
- Sree, K.S.; Bog, M.; Appenroth, K.J. Taxonomy of duckweeds (Lemnaceae), potential new crop plants. Emir. J. Food Agric. 2016, 28, 291–302. [Google Scholar] [CrossRef] [Green Version]
- Stomp, A.M. The duckweeds: A valuable plant for biomanufacturing. Biotechnol. Annu. Rev. 2005, 11, 69–99. [Google Scholar]
- Yang, J.; Li, G.; Hu, S.; Bishopp, A.; Heenatigala, P.; Kumar, S.; Duan, P.; Yao, L.; Hou, H. A protocol for efficient callus induction and stable transformation of Spirodela polyrhiza (L.) Schleiden using Agrobacterium tumefaciens. Aquat. Bot. 2018, 151, 80–86. [Google Scholar] [CrossRef]
- Ziegler, P.; Adelmann, K.; Zimmer, S.; Schmidt, C.; Appenroth, K.J. Relative in vitro growth rates of duckweeds (Lemnaceae)—The most rapidly growing higher plants. Plant Biol. 2015, 17, 33–41. [Google Scholar] [CrossRef]
- Xu, J.; Cheng, J.J.; Stomp, A.M. Growing Spirodela polyrrhiza in swine wastewater for the production of animal feed and fuel ethanol: A pilot study. CLEAN–Soil Air Water 2012, 40, 760–765. [Google Scholar] [CrossRef]
- Xu, J.; Cui, W.; Cheng, J.J.; Stomp, A.M. Production of high-starch duckweed and its conversion to bioethanol. Biosyst. Eng. 2011, 110, 67–72. [Google Scholar] [CrossRef]
- Khellaf, N.; Zerdaoui, M. Growth response of the duckweed Lemna minor to heavy metal pollution. J. Environ. Health Sci. Eng. 2009, 6, 161–166. [Google Scholar]
- Yang, L.; Han, Y.; Wu, D.; Yong, W.; Liu, M.; Wang, S.; Liu, W.; Lu, M.; Wei, Y.; Sun, J. Salt and cadmium stress tolerance caused by overexpression of the Glycine max Na+/H+ Antiporter (GmNHX1) gene in duckweed (Lemna turionifera 5511). Aquat. Toxicol. 2017, 192, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Appenroth, K.; Landesman, L.; Salmeán, A.A.; Lam, E. Duckweed rising at Chengdu: Summary of the 1st International conference on duckweed application and research. Plant Mol. Biol. 2012, 78, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Makhzoum, A.; Benyammi, R.; Moustafa, K.; Trémouillaux-Guiller, J. Recent advances on host plants and expression cassettes’ structure and function in plant molecular pharming. BioDrugs 2014, 28, 145–159. [Google Scholar] [CrossRef]
- Cantó-Pastor, A.; Mollá-Morales, A.; Ernst, E.; Dahl, W.; Zhai, J.; Yan, Y.; Meyers, B.; Shanklin, J.; Martienssen, R. Efficient transformation and artificial miRNA gene silencing in Lemna minor. Plant Biol. 2015, 17, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, Y.T.; Rajbhandari, N.; Lin, X.; Bergmann, B.A.; Nishimura, Y.; Stomp, A.M. Genetic transformation of duckweed Lemna gibba and Lemna minor. Vitr. Cell. Dev. Biol.-Plant 2001, 37, 349–353. [Google Scholar] [CrossRef]
- Chhabra, G.; Chaudhary, D.; Sainger, M.; Jaiwal, P.K. Genetic transformation of Indian isolate of Lemna minor mediated by Agrobacterium tumefaciens and recovery of transgenic plants. Physiol. Mol. Biol. Plants 2011, 17, 129–136. [Google Scholar] [CrossRef] [Green Version]
- Firsov, A.; Tarasenko, I.; Mitiouchkina, T.; Ismailova, N.; Shaloiko, L.; Vainstein, A.; Dolgov, S. High-yield expression of M2e peptide of avian influenza virus H5N1 in transgenic duckweed plants. Mol. Biotechnol. 2015, 57, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Firsov, A.; Tarasenko, I.; Mitiouchkina, T.; Shaloiko, L.; Kozlov, O.; Vinokurov, L.; Rasskazova, E.; Murashev, A.; Vainstein, A.; Dolgov, S. Expression and immunogenicity of M2e peptide of avian influenza virus H5N1 fused to ricin toxin b chain produced in duckweed plants. Front. Chem. 2018, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.M.; Sun, H.J.; Oh, M.J.; Song, I.J.; Kim, M.J.; Sin, H.S.; Goh, C.H.; Kim, Y.W.; Lim, P.O.; Lee, H.Y. Expression of the protective antigen for PEDV in transgenic duckweed, Lemna minor. Hortic. Environ. Biotechnol. 2011, 52, 511–515. [Google Scholar] [CrossRef]
- Sun, Y.; Cheng, J.J.; Himmel, M.E.; Skory, C.D.; Adney, W.S.; Thomas, S.R.; Tisserat, B.; Nishimura, Y.; Yamamoto, Y.T. Expression and characterization of Acidothermus cellulolyticus E1 endoglucanase in transgenic duckweed Lemna minor 8627. Bioresour. Technol. 2007, 98, 2866–2872. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.L.; Fang, Y.; Xu, Y.L.; Tan, L.; Li, Q.; Liu, Y.; Lai, F.; Jin, Y.-L.; Du, A.-P.; He, K.-Z. Frond transformation system mediated by Agrobacterium tumefaciens for Lemna minor. Plant Mol. Biol. 2018, 98, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Han, H.; Liu, M.; Zuo, Z.; Zhou, K.; Lü, J.; Zhu, Y.; Bai, Y.; Wang, Y. Overexpression of the Arabidopsis photorespiratory pathway gene, serine: Glyoxylate aminotransferase (AtAGT1), leads to salt stress tolerance in transgenic duckweed (Lemna minor). Plant Cell Tissue Organ Cult. (PCTOC) 2013, 113, 407–416. [Google Scholar] [CrossRef]
- Rival, S.; Wisniewski, J.P.; Langlais, A.; Kaplan, H.; Freyssinet, G.; Vancanneyt, G.; Vunsh, R.; Perl, A.; Edelman, M. Spirodela (duckweed) as an alternative production system for pharmaceuticals: A case study, aprotinin. Transgenic Res. 2008, 17, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Vunsh, R.; Li, J.; Hanania, U.; Edelman, M.; Flaishman, M.; Perl, A.; Wisniewski, J.P.; Freyssinet, G. High expression of transgene protein in Spirodela. Plant Cell Rep. 2007, 26, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Khvatkov, P.; Chernobrovkina, M.; Okuneva, A.; Pushin, A.; Dolgov, S. Transformation of Wolffia arrhiza (L.) Horkel ex Wimm. Plant Cell Tissue Organ Cult. (PCTOC) 2015, 123, 299–307. [Google Scholar] [CrossRef]
- Khvatkov, P.; Chernobrovkina, M.; Okuneva, A.; Shvedova, A.; Chaban, I.; Dolgov, S. Callus induction and regeneration in Wolffia arrhiza (L.) Horkel ex Wimm. Plant Cell Tissue Organ Cult. (PCTOC) 2015, 120, 263–273. [Google Scholar] [CrossRef]
- Heenatigala, P.P.M.; Yang, J.; Bishopp, A.; Sun, Z.; Li, G.; Kumar, S.; Hu, S.; Wu, Z.; Lin, W.; Yao, L.; et al. Development of efficient protocols for stable and transient gene transformation for Wolffia globosa using Agrobacterium. Front. Chem. 2018, 6, 227. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wang, Y.; Xu, S.; Tang, X.; Zhao, J.; Yu, C.; He, G.; Xu, H.; Wang, S.; Tang, Y. Efficient genetic transformation and CRISPR/Cas9-mediated genome editing in Lemna aequinoctialis. Plant Biotechnol. J. 2019, 17, 2143–2152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Li, Y.; Ma, J.; Cheng, J. Survey of duckweed diversity in Lake Chao and total fatty acid, triacylglycerol, profiles of representative strains. Plant Biol. 2015, 17, 1066–1072. [Google Scholar] [CrossRef]
- Xu, Y.; Ma, S.; Huang, M.; Peng, M.; Bog, M.; Sree, K.S.; Appenroth, K.-J.; Zhang, J. Species distribution, genetic diversity and barcoding in the duckweed family (Lemnaceae). Hydrobiologia 2015, 743, 75–87. [Google Scholar] [CrossRef]
- Huang, M.; Fu, L.; Sun, X.; Di, R.; Zhang, J. Rapid and highly efficient callus induction and plant regeneration in the starch-rich duckweed strains of Landoltia punctata. Acta Physiol. Plant. 2016, 38, 122. [Google Scholar] [CrossRef]
- Wang, Y. Callus induction and frond regeneration in Spirodela polyrhiza. Czech J. Genet. Plant Breed. 2016, 52, 114–119. [Google Scholar] [CrossRef] [Green Version]
- Ball, S.T.; Zhou, H.; Konzak, C.F. Influence of 2,4-D, IAA, and duration of callus induction in anther cultures of spring wheat. Plant Sci. 1993, 90, 195–200. [Google Scholar] [CrossRef]
- Adil, M.; Ren, X.; Kang, D.I.; Jeong, B.R. Effect of explant type and plant growth regulators on callus induction, growth and secondary metabolites production in Cnidium officinale Makino. Mol. Biol. Rep. 2018, 45, 1919–1927. [Google Scholar] [CrossRef]
- Zheng, M.; Konzak, C. Effect of 2,4-dichlorophenoxyacetic acid on callus induction and plant regeneration in anther culture of wheat (Triticum aestivum L.). Plant Cell Rep. 1999, 19, 69–73. [Google Scholar] [CrossRef]
- Moon, H.; Stomp, A. Effects of medium components and light on callus induction, growth, and frond regeneration in Lemna gibba (Duckweed). Vitr. Cell. Dev. Biol.-Plant 1997, 33, 20–25. [Google Scholar] [CrossRef]
- Moon, H.K.; Yang, M.S. Nodular somatic embryogenesis and frond regeneration in duckweed, Lemna gibba G3. J. Plant Biol. 2002, 45, 154–160. [Google Scholar] [CrossRef]
- Li, J.; Jain, M.; Vunsh, R.; Vishnevetsky, J.; Hanania, U.; Flaishman, M.; Perl, A.; Edelman, M. Callus induction and regeneration in Spirodela and Lemna. Plant Cell Rep. 2004, 22, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Khvatkov, P.; Firsov, A.; Shvedova, A.; Shaloiko, L.; Kozlov, O.; Chernobrovkina, M.; Pushin, A.; Tarasenko, I.; Chaban, I.; Dolgov, S. Development of Wolffia arrhiza as a producer for recombinant human granulocyte colony-stimulating factor. Front. Chem. 2018, 6, 304. [Google Scholar] [CrossRef]
- Aileni, M.; Abbagani, S.; Zhang, P. Highly efficient production of transgenic Scoparia dulcis L. mediated by Agrobacterium tumefaciens: Plant regeneration via shoot organogenesis. Plant Biotechnol. Rep. 2011, 5, 147–156. [Google Scholar] [CrossRef]
- Subramanyam, K.; Subramanyam, K.; Sailaja, K.; Srinivasulu, M.; Lakshmidevi, K. Highly efficient Agrobacterium-mediated transformation of banana cv. Rasthali (AAB) via sonication and vacuum infiltration. Plant Cell Rep. 2011, 30, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Indurker, S.; Misra, H.S.; Eapen, S. Agrobacterium-mediated transformation in chickpea (Cicer arietinum L.) with an insecticidal protein gene: Optimisation of different factors. Physiol. Mol. Biol. Plants 2010, 16, 273–284. [Google Scholar] [CrossRef] [Green Version]
- Bakshi, S.; Sadhukhan, A.; Mishra, S.; Sahoo, L. Improved Agrobacterium-mediated transformation of cowpea via sonication and vacuum infiltration. Plant Cell Rep. 2011, 30, 2281–2292. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Y.; Yan, Y.; Ermakova, M.; Kerstetter, R.; Messing, J. DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol. 2010, 10, 205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, Y.; Sawa, Y.; Nagaoka, N.; Kozai, T. Photoautotrophic growth of Pleioblastus pygmaea plantlets in vitro and ex vitro as affected by types of supporting material in vitro. In Transplant Production in the 21st Century; Springer: Dordrecht, The Netherlands, 2000; pp. 226–230. [Google Scholar]
- Wu, T.M.; Lin, K.C.; Liau, W.S.; Chao, Y.Y.; Yang, L.H.; Chen, S.Y.; Lu, C.A.; Hong, C.Y. A set of GFP-based organelle marker lines combined with DsRed-based gateway vectors for subcellular localization study in rice (Oryza sativa L.). Plant Mol. Biol. 2016, 90, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Benke, D.; Cicin-Sain, A.; Mertens, S.; Mohler, H. Immunochemical identification of the α1-and α3–subuntis of the GabaA-receptor in rat brain. J. Recept. Res. 1991, 11, 407–424. [Google Scholar] [CrossRef] [PubMed]

| 2,4-D 1 (μM) | Cytokinin 2 (μM) | % Fronds Forming Callus 3 | |
|---|---|---|---|
| 5 | 0.0 | 57 ± 6 gf | |
| 10 | 65 ± 5 ef | ||
| 25 | 50 ± 9 g | ||
| 50 | 27 ± 6 h | ||
| 5 | 6-BA | 0.5 | 28 ± 14 h |
| 1.0 | 55 ± 5 gf | ||
| 2.0 | 55 ± 5 gf | ||
| 10 | 0.5 | 65 ± 5 ef | |
| 1.0 | 67 ± 3 ef | ||
| 2.0 | 73 ± 14 e | ||
| 25 | 0.5 | 87 ± 3 bcd | |
| 1.0 | 95 ± 5 abc | ||
| 2.0 | 100 ± 0 a | ||
| 50 | 0.5 | 72 ± 3 e | |
| 1.0 | 93 ± 6 abc | ||
| 2.0 | 87 ± 6 bcd | ||
| 5 | TDZ | 0.5 | 75 ± 5 de |
| 1.0 | 88 ± 3 bc | ||
| 2.0 | 85 ± 5 cd | ||
| 10 | 0.5 | 93 ± 6 abc | |
| 1.0 | 95 ± 5 abc | ||
| 2.0 | 87 ± 12 bcd | ||
| 25 | 0.5 | 100 ± 0 a | |
| 1.0 | 95 ± 5 abc | ||
| 2.0 | 100 ± 0 a | ||
| 50 | 0.5 | 92 ± 10 abc | |
| 1.0 | 98 ± 3 ab | ||
| 2.0 | 97 ± 3 abc |
| Basal Medium 1 | 6-BA (μM) | % Callus Regenerating Fronds 2 | Number of Fronds Regenerated/Callus Piece 2 |
|---|---|---|---|
| 0 | 0 d | 0 D | |
| ½SH | 1 | 0 d | 0 D |
| 2 | 61 ± 5 b | 9 ± 2 C | |
| 0 | 60 ± 5 b | 10 ± 2 C | |
| SH | 1 | 100 ± 0 a | 42 ± 4 A |
| 2 | 47 ± 2 c | 18 ± 3 B | |
| 0 | 0 d | 0 D | |
| ½MS | 1 | 0 d | 0 D |
| 2 | 0 d | 0 D | |
| 0 | 0 d | 0 D | |
| MS | 1 | 0 d | 0 D |
| 2 | 0 d | 0 D |
| AS 1 (µM) | Coculture (Day) | Photoperiod | GFP Expression 2 (%) | |
|---|---|---|---|---|
| Day 9 | Week 10 | |||
| 0 | 1 | 12 h light/12 h dark | 0 e | 0 B |
| 24 h dark | 0 e | 0 B | ||
| 2 | 12 h light/12 h dark | 0 e | 0 B | |
| 24 h dark | 0 e | 0 B | ||
| 3 | 12 h light/12 h dark | 0 e | 0 B | |
| 24 h dark | 0 e | 0 B | ||
| 50 | 1 | 12 h light/12 h dark | 0 e | 0 B |
| 24 h dark | 0 e | 0 B | ||
| 2 | 12 h light/12 h dark | 0 e | 0 B | |
| 24 h dark | 0 e | 0 B | ||
| 3 | 12 h light/12 h dark | 0 e | 0 B | |
| 24 h dark | 0 e | 0 B | ||
| 100 | 1 | 12 h light/12 h dark | 0 e | 0 B |
| 24 h dark | 3 ± 3 de | 0 B | ||
| 2 | 12 h light/12 h dark | 0 e | 0 B | |
| 24 h dark | 0 e | 0 B | ||
| 3 | 12 h light/12 h dark | 0 e | 0 B | |
| 24 h dark | 0 e | 0 B | ||
| 200 | 1 | 12 h light/12 h dark | 2 ± 2 de | 0 B |
| 24 h dark | 49 ± 4 a | 3 ± 1 A | ||
| 2 | 12 h light/12 h dark | 2 ± 2 de | 0 B | |
| 24 h dark | 14 ± 2 c | 0 B | ||
| 3 | 12 h light/12 h dark | 4 ± 2 d | 0 B | |
| 24 h dark | 25 ± 5 b | 0 B | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.-T.; Hong, M.-C.; Wu, Y.-S.; Wu, T.-M. Agrobacterium-Mediated Genetic Transformation of Taiwanese Isolates of Lemna aequinoctialis. Plants 2021, 10, 1576. https://doi.org/10.3390/plants10081576
Wang K-T, Hong M-C, Wu Y-S, Wu T-M. Agrobacterium-Mediated Genetic Transformation of Taiwanese Isolates of Lemna aequinoctialis. Plants. 2021; 10(8):1576. https://doi.org/10.3390/plants10081576
Chicago/Turabian StyleWang, Kuang-Teng, Ming-Chang Hong, Yu-Sheng Wu, and Tsung-Meng Wu. 2021. "Agrobacterium-Mediated Genetic Transformation of Taiwanese Isolates of Lemna aequinoctialis" Plants 10, no. 8: 1576. https://doi.org/10.3390/plants10081576
APA StyleWang, K.-T., Hong, M.-C., Wu, Y.-S., & Wu, T.-M. (2021). Agrobacterium-Mediated Genetic Transformation of Taiwanese Isolates of Lemna aequinoctialis. Plants, 10(8), 1576. https://doi.org/10.3390/plants10081576





