Basic β-1,3-Glucanase from Drosera binata Exhibits Antifungal Potential in Transgenic Tobacco Plants
Abstract
:1. Introduction
2. Results
2.1. Antifungal Activity of the Purified rDbGluc1 Protein
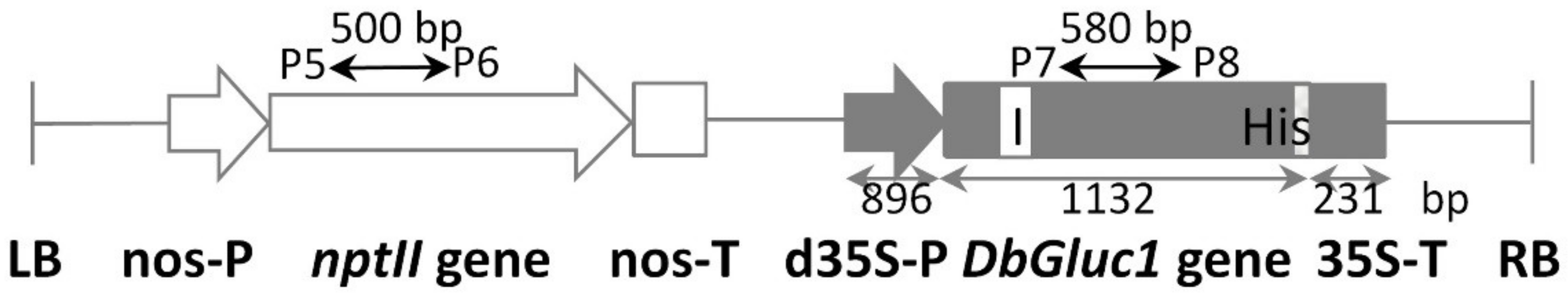
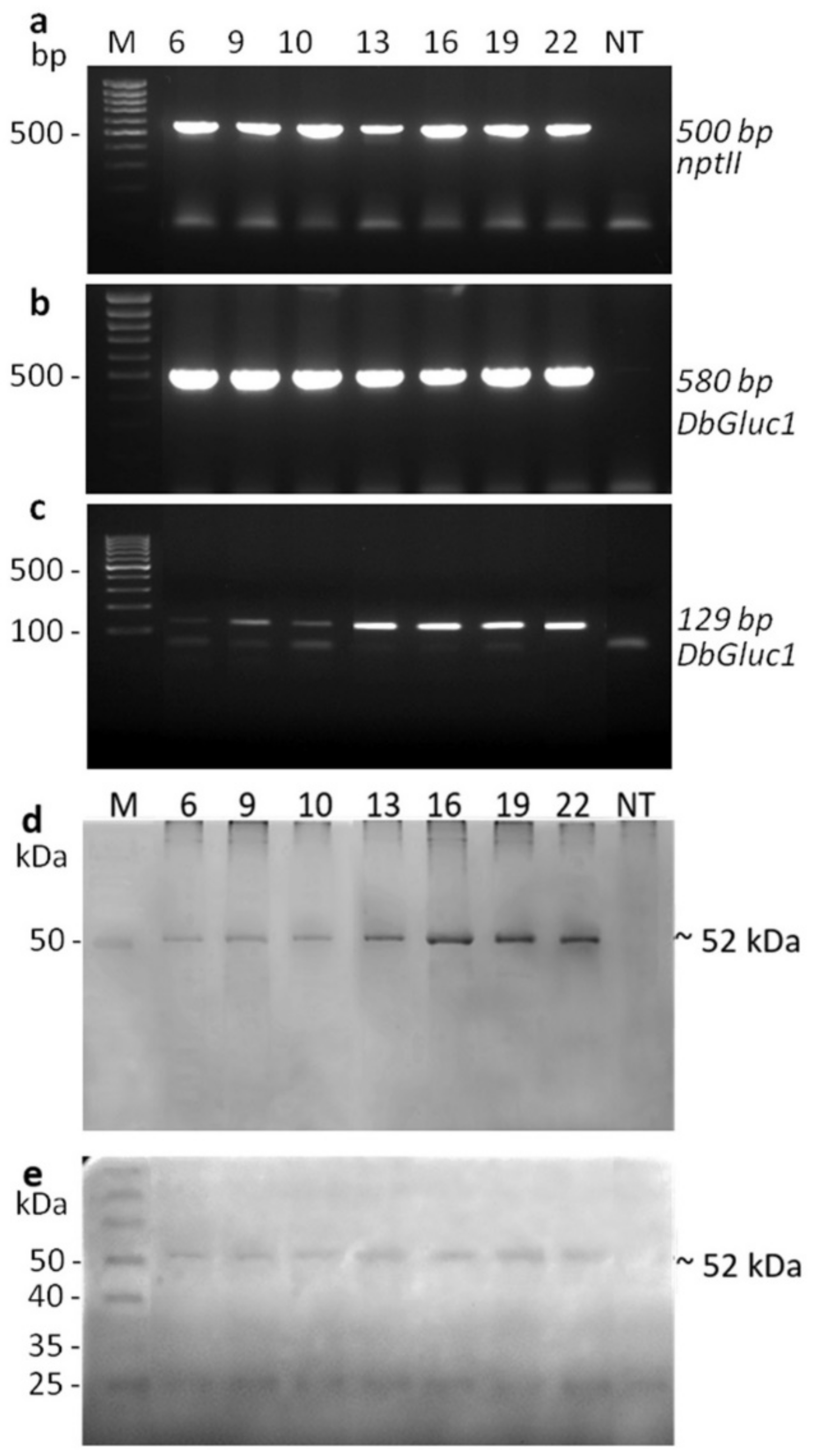
2.2. Production of Transgenic Plants and Molecular Characterisation
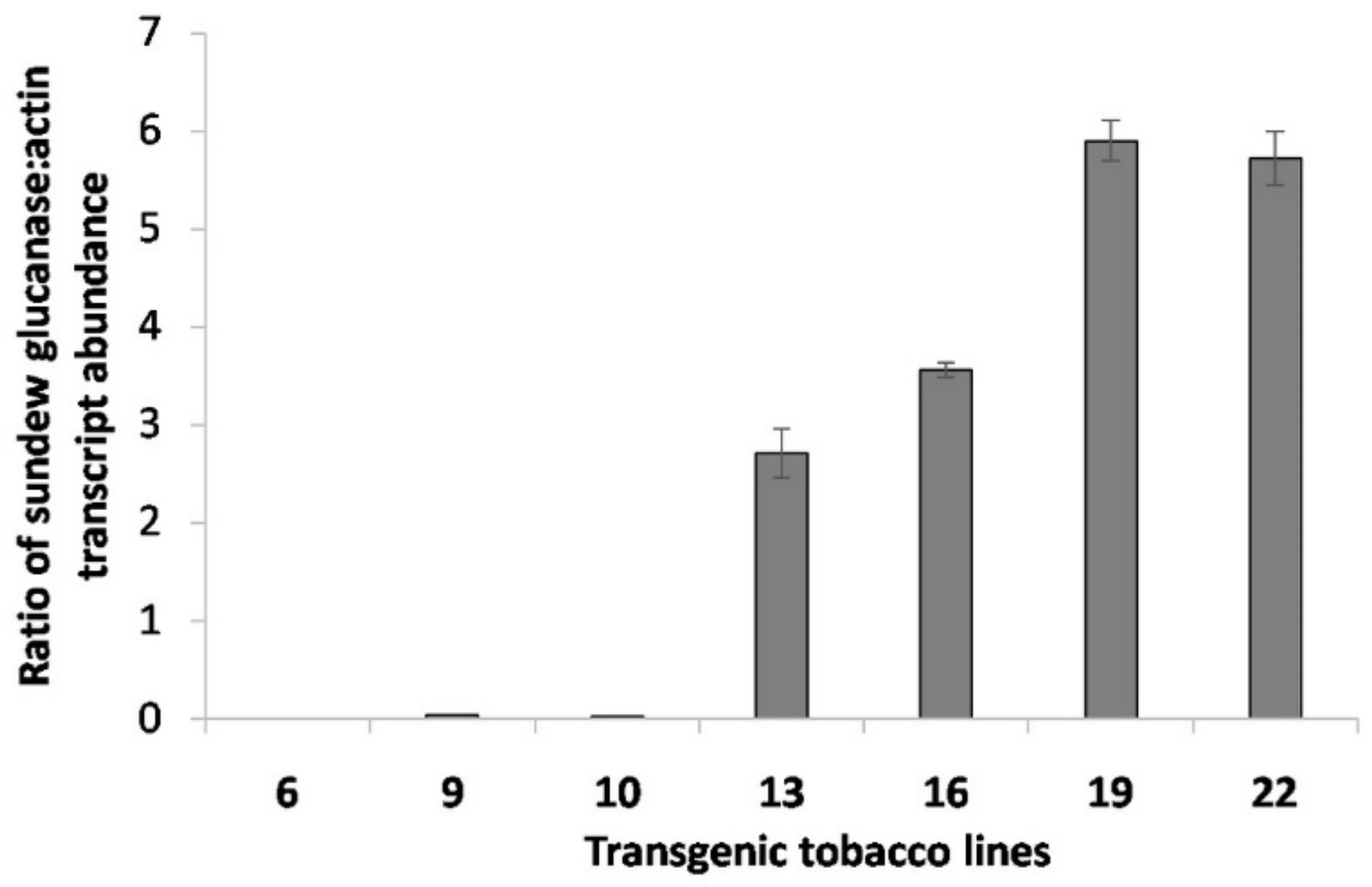
2.3. Quantification of the DbGluc1 Expression Level
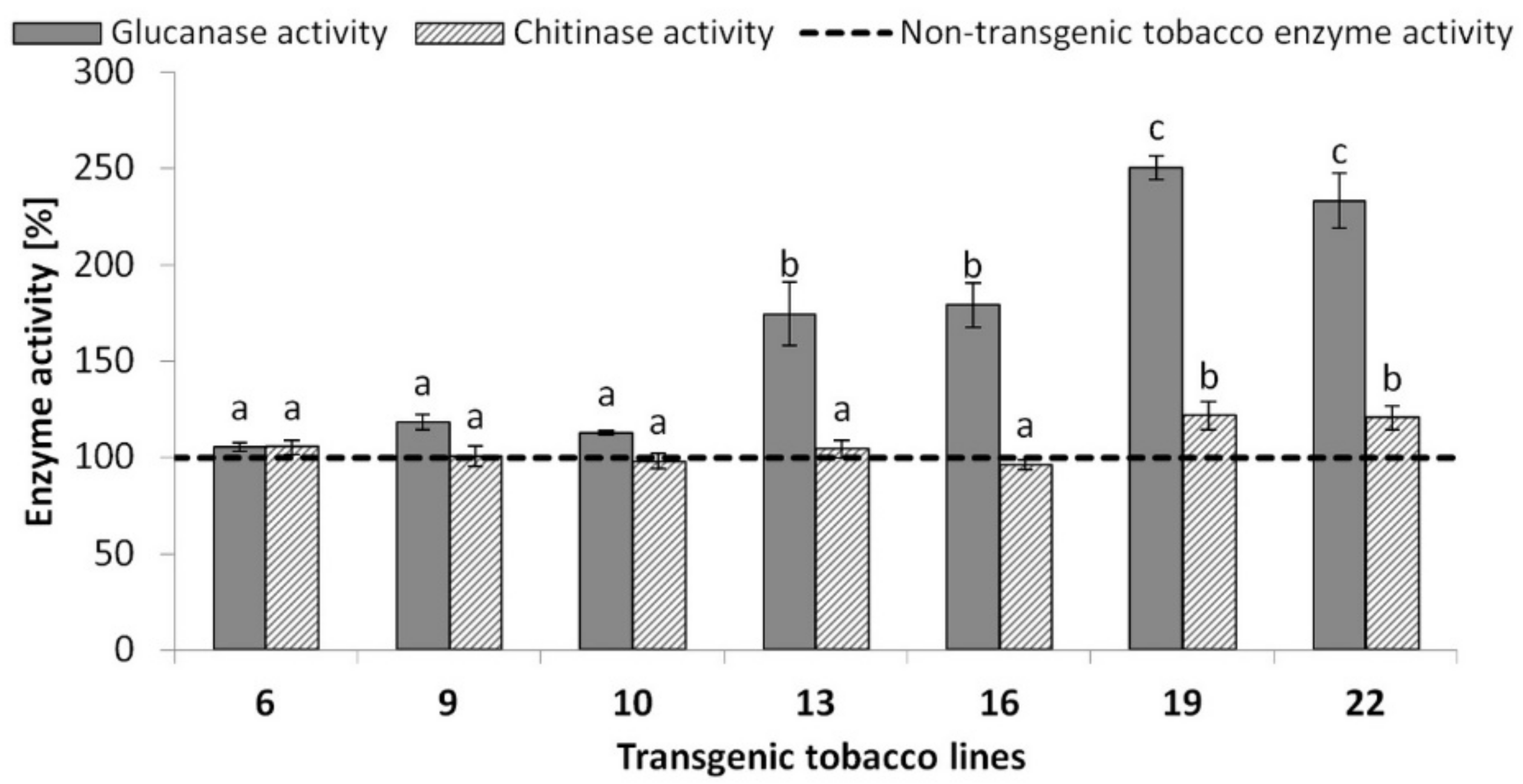
2.4. Quantification of β-1,3-Glucanase Activity
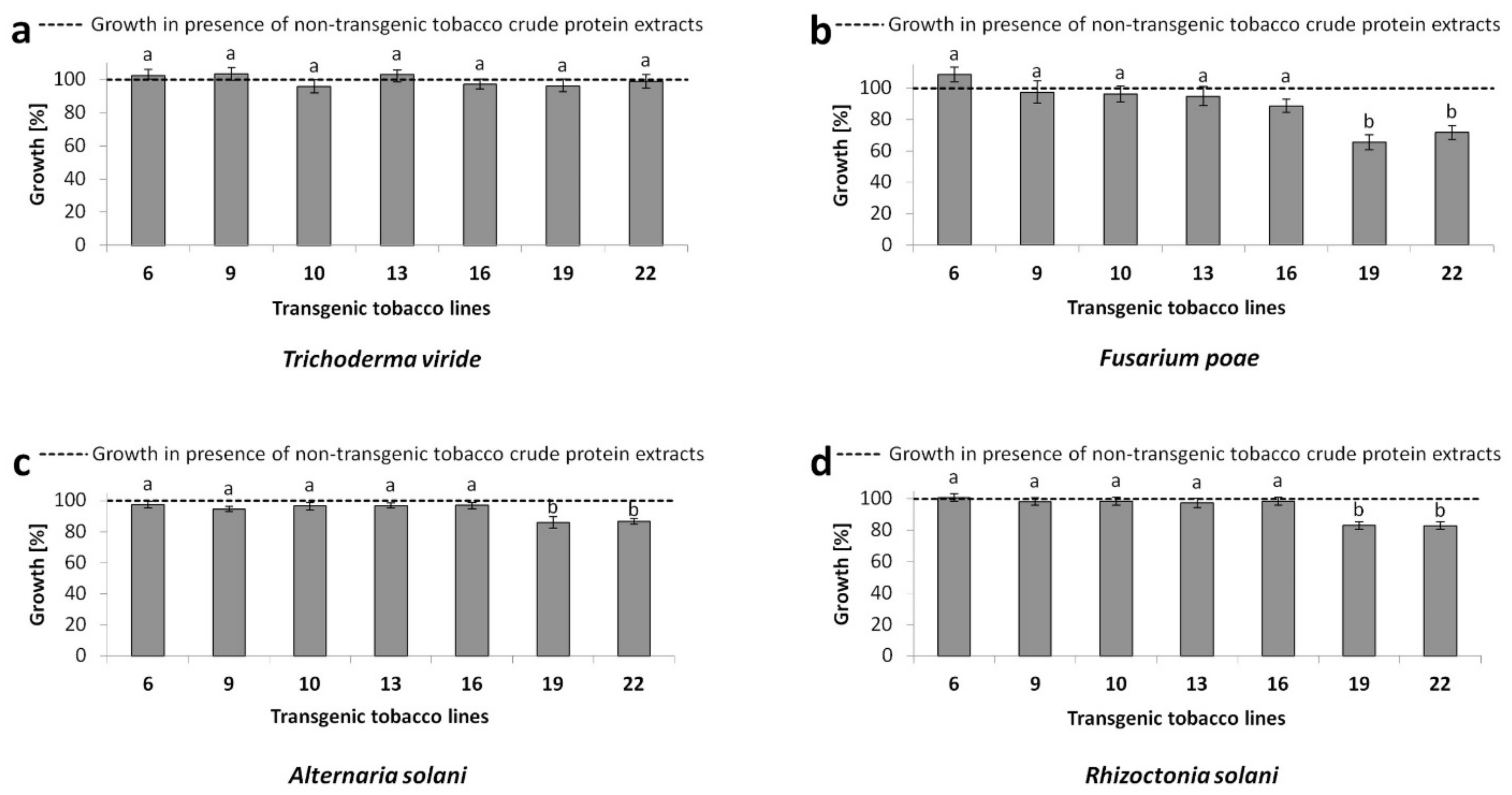
2.5. In-Vitro Antifungal Activity Assay
3. Discussion
4. Materials and Methods
4.1. Plant and Fungal Material
4.2. Expression of rDbGluc1 in Escherichia coli and Its Purification
4.3. Construction of the Plant Expression Vector and Genetic Transformation of Tobacco
4.4. PCR Analysis
4.5. Copy Number Estimation by qPCR
4.6. RT-qPCR
4.7. Fluorescent Detection of the His6-Tag Sequence of DbGluc1 Recombinant Protein
4.8. Enzyme Activity Assay
4.9. In-Vitro Antifungal Activity Assay
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bulcke, M.V.D.; Bauw, G.; Castresana, C.; van Montagu, M.; Vandekerckhove, J. Characterization of vacuolar and extracellular β(1,3)-glucanases of tobacco: Evidence for a strictly compartmentalized plant defense system. Proc. Natl. Acad. Sci. USA 1989, 86, 2673–2677. [Google Scholar] [CrossRef] [Green Version]
- Linthorst, H.J.M.; Melchers, L.S.; Mayer, A.; Vanroekel, J.S.C.; Cornelissen, B.J.C.; Bol, J.F. Analysis of gene families encoding acidic and basic beta-1,3-glucanases of tobacco. Proc. Natl. Acad. Sci. USA 1990, 87, 8756–8760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doxey, A.C.; Yaish, M.W.F.; Moffatt, B.A.; Griffith, M.; McConkey, B.J. Functional divergence in the Arabidopsis beta-1,3-glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol. Biol. Evol 2007, 24, 1045–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, D.H.; Kim, S.T.; Kim, S.G.; Kang, K.Y. Comprehensive analysis of the expression of twenty-seven beta-1, 3-glucanase genes in rice (Oryza sativa L.). Mol. Cells 2007, 23, 207–214. [Google Scholar]
- Barral, P.; Suarez, C.; Batanero, E.; Alfonso, C.; Alche, J.D.; Rodriguez-Garcia, M.I.; Villalba, M.; Rivas, G.; Rodriguez, R. An olive pollen protein with allergenic activity, Ole e 10, defines a novel family of carbohydrate-binding modules and is potentially implicated in pollen germination. Biochem. J. 2005, 390, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Branco, A.T.; Ferreira, B.D.; Lourenco, G.F.; Marques, V.C.L.; Machado, O.L.T.; Pereira, M.G.; Almeida, J.C.A.; de Souza, G.A. Induction of beta-1,3-glucanase in seeds of maize defective-kernel mutant (827Kpro1). Protein Pept. Lett. 2011, 18, 651–657. [Google Scholar] [CrossRef]
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant beta-1,3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012, 34, 1983–1990. [Google Scholar] [CrossRef]
- Enoki, S.; Fujimori, N.; Yamaguchi, C.; Hattori, T.; Suzuki, S. High Constitutive overexpression of Glycosyl Hydrolase Family 17 delays floral transition by enhancing flc expression in transgenic Arabidopsis. Plants 2017, 6, 31. [Google Scholar] [CrossRef] [Green Version]
- Yaish, M.W.F.; Doxey, A.C.; McConkey, B.J.; Moffatt, B.A.; Griffith, M. Cold-active winter rye glucanases with ice-binding capacity. Plant Physiol. 2006, 141, 1459–1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zur, I.; Golebiowska, G.; Dubas, E.; Golemiec, E.; Matusikova, I.; Libantova, J.; Moravcikova, J. beta-1,3-glucanase and chitinase activities in winter triticales during cold hardening and subsequent infection by Microdochium nivale. Biologia 2013, 68, 241–248. [Google Scholar] [CrossRef]
- Takeda, H.; Yoshikawa, T.; Liu, X.Z.; Nakagawa, N.; Li, Y.Q.; Sakurai, N. Molecular cloning of two exo-beta-glucanases and their in vivo substrates in the cell walls of lily pollen tubes. Plant Cell Physiol. 2004, 45, 436–444. [Google Scholar] [CrossRef] [Green Version]
- Jopcik, M.; Matusikova, I.; Moravcikova, J.; Durechova, D.; Libantova, J. The expression profile of Arabidopsis thaliana beta-1,3-glucanase promoter in tobacco. Mol. Biol. 2015, 49, 543–549. [Google Scholar] [CrossRef]
- Leubner-Metzger, G. beta-1,3-glucanase gene expression in low-hydrated seeds as a mechanism for dormancy release during tobacco after-ripening. Plant J. 2005, 41, 133–145. [Google Scholar] [CrossRef]
- Kauffmann, S.; Legrand, M.; Geoffroy, P.; Fritig, B. Biological function of pathogenesis-related proteins—Four PR proteins of tobacco have 1,3-beta-glucanase activity. EMBO J. 1987, 6, 3209–3212. [Google Scholar] [CrossRef] [PubMed]
- Sela-Buurlage, M.B.; Ponstein, A.S.; Bres-Vloemans, S.A.; Melchers, L.S.; van den Elzen, P.J.M.; Cornelissen, B.J.C. Only specific tobacco (Nicotiana tabacum) chitinases and β-1,3-glucanases exhibit antifungal activity. Plant Physiol. 1993, 101, 857–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulze, W.X.; Sanggaard, K.W.; Kreuzer, I.; Knudsen, A.D.; Bemm, F.; Thogersen, I.B.; Brautigam, A.; Thomsen, L.R.; Schliesky, S.; Dyrlund, T.F.; et al. The protein composition of the digestive fluid from the venus flytrap sheds light on prey digestion mechanisms. Mol. Cell. Proteom. 2012, 11, 1306–1319. [Google Scholar] [CrossRef] [Green Version]
- Singh, D.; Ambroise, A.; Haicour, R.; Sihachakr, D.; Rajam, M.V. Increased resistance to fungal wilts in transgenic eggplant expressing alfalfa glucanase gene. Physiol. Mol. Biol. Plants 2014, 20, 143–150. [Google Scholar] [CrossRef]
- Nayyar, S.; Sharma, B.K.; Kaur, A.; Kalia, A.; Sanghera, G.S.; Thind, K.S.; Yadav, I.S.; Sandhu, J.S. Red rot resistant transgenic sugarcane developed through expression of beta-1,3-glucanase gene. PloS ONE 2017, 12, e0179723. [Google Scholar] [CrossRef]
- Kheiri, H.-R.; Motallebi, M.; Zamani, M.R.; Deljo, A. Beta glucanase (Bgn13.1) expressed in transgenic Brassica napus confers antifungal activity against Sclerotinia sclerotiorum. J. Crop Prot. 2014, 3, 31–42. [Google Scholar]
- Liu, M.; Sun, Z.X.; Zhu, J.; Xu, T.; Harman, G.E.; Lorito, M. Enhancing rice resistance to fungal pathogens by transformation with cell wall degrading enzyme genes from Trichoderma atroviride. J. Zhejiang Uni. Sci. 2004, 5, 133–136. [Google Scholar] [CrossRef]
- Mondal, K.K.; Bhattacharya, R.C.; Koundal, K.R.; Chatterjee, S.C. Transgenic Indian mustard (Brassica juncea) expressing tomato glucanase leads to arrested growth of Alternaria brassicae. Plant Cell Rep. 2007, 26, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Maziah, M.; Sariah, M.; Sreeramanan, S. Transgenic banana Rastali (AAB) with β-1, 3-glucanase gene for tolerance to fusarium wilt race 1 disease via Agrobacterium-mediated transformation system. Plant Pathol. J. 2007, 6, 271–282. [Google Scholar] [CrossRef]
- Sridevi, G.; Parameswari, C.; Sabapathi, N.; Raghupathy, V.; Veluthambi, K. Combined expression of chitinase and beta-1,3-glucanase genes in indica rice (Oryza sativa L.) enhances resistance against Rhizoctonia solani. Plant Sci. 2008, 175, 283–290. [Google Scholar] [CrossRef]
- Borkowska, M.; Krzymowska, M.; Talarczyk, A.; Awan, M.F.M.; Yakovleva, L.; Kleczkowski, K.; Wielgat, B. Transgenic potato plants expressing soybean beta-1,3-endoglucanase gene exhibit an increased resistance to Phytophthora infestans. Zeitschrift für Naturforschung C 1998, 53, 1012–1016. [Google Scholar] [CrossRef]
- Moravcikova, J.; Libantova, J.; Heldak, J.; Salaj, J.; Bauer, M.; Matusikova, I.; Galova, Z.; Mlynarova, L. Stress-induced expression of cucumber chitinase and Nicotiana plumbaginifolia beta-1,3-glucanase genes in transgenic potato plants. Acta Physiol. Plant. 2007, 29, 133–141. [Google Scholar] [CrossRef]
- Sundaresha, S.; Kumar, A.M.; Rohini, S.; Math, S.A.; Keshamma, E.; Chandrashekar, S.C.; Udayakumar, M. Enhanced protection against two major fungal pathogens of groundnut, Cercospora arachidicola and Aspergillus flavus in transgenic groundnut over-expressing a tobacco beta 1-3 glucanase. Eur. J. Plant Pathol. 2010, 126, 497–508. [Google Scholar] [CrossRef]
- Liu, D.; He, X.; Li, W.; Chen, C.; Ge, F. A β-1,3-glucanase gene expressed in fruit of Pyrus pyrifolia enhances resistance to several pathogenic fungi in transgenic tobacco. Eur. J. Plant Pathol. 2013, 135, 265–277. [Google Scholar] [CrossRef]
- Fujimori, N.; Enoki, S.; Suzuki, A.; Naznin, H.A.; Shimizu, M.; Suzuki, S. Grape apoplasmic beta-1,3-glucanase confers fungal disease resistance in Arabidopsis. Sci. Hortic. 2016, 200, 105–110. [Google Scholar] [CrossRef]
- Heubl, G.; Bringmann, G.; Meimberg, H. Molecular phylogeny and character evolution of carnivorous plant families in caryophyllales—Revisited. Plant Biol. 2006, 8, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Arai, N.; Ohno, Y.; Jumyo, S.; Hamaji, Y.; Ohyama, T. Organ-specific expression and epigenetic traits of genes encoding digestive enzymes in the lance-leaf sundew (Drosera adelae). J. Exp. Bot. 2020, 72, 1946–1961. [Google Scholar] [CrossRef]
- Fukushima, K.; Fang, X.D.; Alvarez-Ponce, D.; Cai, H.M.; Carretero-Paulet, L.; Chen, C.; Chang, T.H.; Farr, K.M.; Fujita, T.; Hiwatashi, Y.; et al. Genome of the pitcher plant Cephalotus reveals genetic changes associated with carnivory. Nat. Ecol. Evol. 2017, 1, 0059. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2020, 10, 2993. [Google Scholar] [CrossRef]
- Schlumbaum, A.; Mauch, F.; Vogeli, U.; Boller, T. Plant chitinases are potent inhibitors of fungal growth. Nature 1986, 324, 365–367. [Google Scholar] [CrossRef]
- Harries, E.; Berruezo, L.A.; Galvan, M.Z.; Rajal, V.B.; Cardenas, G.E.M. Soil properties related to suppression of Rhizoctonia solani on tobacco fields from northwest Argentina. Plant Pathol. 2020, 69, 77–86. [Google Scholar] [CrossRef]
- Petrunak, D.M.; Christ, B.J. Isozyme variability in Alternaria solani and Alternaria alternata. Phytopathology 1992, 82, 1343–1347. [Google Scholar] [CrossRef]
- Stenglein, S.; Barreto, D.; Nicholson, P.; Chandler, E.; Brambilla, V.; Piris, E.M.; Saliva, V.; Mitidieri, M.; Salerno, G. First report of Fusarium poae on tomato in Argentina. Plant Pathol. 2009, 58, 401. [Google Scholar] [CrossRef]
- Rajninec, M.; Jopcik, M.; Danchenko, M.; Libantova, J. Biochemical and antifungal characteristics of recombinant class I chitinase from Drosera rotundifolia. Int. J. Biol. Macromol. 2020, 161, 854–863. [Google Scholar] [CrossRef]
- Liu, B.Y.; Lu, Y.; Xin, Z.Y.; Zhang, Z.Y. Identification and antifungal assay of a wheat beta-1,3-glucanase. Biotechnol. Lett. 2009, 31, 1005–1010. [Google Scholar] [CrossRef]
- Zhang, S.B.; Zhang, W.J.; Zhai, H.C.; Lv, Y.Y.; Cai, J.P.; Jia, F.; Wang, J.S.; Hu, Y.S. Expression of a wheat beta-1,3-glucanase in Pichia pastoris and its inhibitory effect on fungi commonly associated with wheat kernel. Protein Expres. Purif. 2019, 154, 134–139. [Google Scholar] [CrossRef]
- Mauch, F.; Hadwiger, L.A.; Boller, T. Antifungal hydrolases in pea tissue. 1. Purification and characterization of two chitinases and two beta-1,3-glucanases differentially regulated during development and in response to fungal infection. Plant Physiol. 1988, 87, 325–333. [Google Scholar] [CrossRef] [Green Version]
- Arlorio, M.; Ludwig, A.; Boller, T.; Bonfante, P. Inhibition of fungal growth by plant chitinases and beta-1,3-glucanases. Protoplasma 1992, 171, 34–43. [Google Scholar] [CrossRef]
- Free, S.J. Chapter two—Fungal cell wall organization and biosynthesis. In Advances in Genetics; Friedmann, T., Dunlap, J.C., Goodwin, S.F., Eds.; Academic Press: Cambridge, MA, USA, 2013; Volume 81, pp. 33–82. [Google Scholar] [CrossRef]
- Beerhues, L.; Kombrink, E. Primary structure and expression of messenger-rnas encoding basic chitinase and 1,3-beta-glucanase in potato. Plant Mol. Biol. 1994, 24, 353–367. [Google Scholar] [CrossRef]
- Kim, Y.J.; Hwang, B.K. Isolation of a basic 34 kiloDalton beta-1,3-glucanase with inhibitory activity against Phytophthora capsici from pepper stems. Physiol. Mol. Plant Pathol. 1997, 50, 103–115. [Google Scholar] [CrossRef]
- Wrobel-Kwiatkowska, M.; Lorenc-Kukula, K.; Starzycki, M.; Oszmianski, J.; Kepczynska, E.; Szopa, J. Expression of beta-1,3-glucanase in flax causes increased resistance to fungi. Physiol. Mol. Plant Pathol. 2004, 65, 245–256. [Google Scholar] [CrossRef]
- Taif, S.; Zhao, Q.; Pu, L.M.; Li, X.; Liu, D.Q.; Cui, X.M. A beta-1,3-glucanase gene from Panax notoginseng confers resistance in tobacco to Fusarium solani. Ind. Crops Prod. 2020, 143. [Google Scholar] [CrossRef]
- Broekaert, W.F.; Marien, W.; Terras, F.R.G.; Debolle, M.F.C.; Proost, P.; Vandamme, J.; Dillen, L.; Claeys, M.; Rees, S.B.; Vanderleyden, J.; et al. Antimicrobial peptides from Amaranthus caudatus seeds with sequence homology to the cysteine glycine-rich domain of chitin-binding proteins. Biochemistry 1992, 31, 4308–4314. [Google Scholar] [CrossRef] [PubMed]
- Taira, J.; Kida, Y.; Yamaguchi, H.; Kuwano, K.; Higashimoto, Y.; Kodama, H. Modifications on amphiphilicity and cationicity of unnatural amino acid containing peptides for the improvement of antimicrobial activity against pathogenic bacteria. J. Pep. Sci. 2010, 16, 607–612. [Google Scholar] [CrossRef]
- Durechova, D.; Jopcik, M.; Rajninec, M.; Moravcikova, J.; Libantova, J. Expression of Drosera rotundifolia chitinase in transgenic tobacco plants enhanced their antifungal potential. Mol. Biotechnol. 2019, 61, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Stintzi, A.; Heitz, T.; Prasad, V.; Wiedemannmerdinoglu, S.; Kauffmann, S.; Geoffroy, P.; Legrand, M.; Fritig, B. Plant pathogenesis-related proteins and their role in defense against pathogens. Biochimie 1993, 75, 687–706. [Google Scholar] [CrossRef]
- Maglovski, M.; Gregorova, Z.; Rybansky, L.; Meszaros, P.; Moravcikova, J.; Hauptvogel, P.; Adamec, L.; Matusikova, I. Nutrition supply affects the activity of pathogenesis-related β-1,3-glucanases and chitinases in wheat. Plant Growth Regul. 2017, 81, 443–453. [Google Scholar] [CrossRef]
- Tull, D.; Phillipson, B.A.; Kramhoft, B.; Knudsen, S.; Olsen, O.; Svensson, B. Enhanced amylolytic activity in germinating barley through synthesis of a bacterial alpha-amylase. J.Cereal Sci. 2003, 37, 71–80. [Google Scholar] [CrossRef]
- Libantova, J.; Jopcik, M.; Fratrikova, M.; Rajninec, M. In gel detection of a his-tagged transgene following the separation of crude plant protein extracts with SDS PAGE. J. Microbiol. Biotechnol. Food Sci. 2019, 9, 127–131. [Google Scholar] [CrossRef]
- Sharma, V.; Salwan, R.; Shanmugam, V. Unraveling the multilevel aspects of least explored plant beneficial Trichodermasaturnisporum isolate GITX-Panog (C). Eur. J. Plant Pathol. 2018, 152, 169–183. [Google Scholar] [CrossRef]
- Dana, M.D.; Pintor-Toro, J.A.; Cubero, B. Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol. 2006, 142, 722–730. [Google Scholar] [CrossRef] [Green Version]
- Roberts, C.S.; Rajagopal, S.; Smith, L.A.; Nguyen, T.A.; Yang, W.; Nugroho, S.; Ravi, K.S.; Vijayachandra, K.; Harcourt, K.L.; Dransfield, L.; et al. A comprehensive set of modular vectors for advanced manipulations and efficient transformation of plants by both Agrobacterium and direct DNA uptake methods; CAMBIA: Canberra, Australia, 2002. [Google Scholar]
- Mlynarova, L.; Loonen, A.; Heldens, J.; Jansen, R.C.; Keizer, P.; Stiekema, W.J.; Nap, J.P. Reduced position effect in mature transgenic plants conferred by the chicken lysozyme matrix-associated region. Plant Cell 1994, 6, 417–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Greenblatt, I.M.; Dellaporta, S.L. Molecular analysis of Ac transposition and DNA replication. Genetics 1992, 130, 665–676. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucl. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Shrestha, K.L.; Liu, S.W.; Huang, C.P.; Wu, H.M.; Wang, W.C.; Li, Y.K. Characterization and identification of essential residues of the glycoside hydrolase family 64 laminaripentaose-producing--1, 3-glucanase. Protein Eng. Des. Sel. 2011, 24, 617–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broekaert, W.F.; Terras, F.R.G.; Cammue, B.P.A.; Vanderleyden, J. An automated quantitative assay for fungal growth inhibition. FEMS Microbiol. Lett. 1990, 69, 55–59. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajninec, M.; Fratrikova, M.; Boszoradova, E.; Jopcik, M.; Bauer, M.; Libantova, J. Basic β-1,3-Glucanase from Drosera binata Exhibits Antifungal Potential in Transgenic Tobacco Plants. Plants 2021, 10, 1747. https://doi.org/10.3390/plants10081747
Rajninec M, Fratrikova M, Boszoradova E, Jopcik M, Bauer M, Libantova J. Basic β-1,3-Glucanase from Drosera binata Exhibits Antifungal Potential in Transgenic Tobacco Plants. Plants. 2021; 10(8):1747. https://doi.org/10.3390/plants10081747
Chicago/Turabian StyleRajninec, Miroslav, Monika Fratrikova, Eva Boszoradova, Martin Jopcik, Miroslav Bauer, and Jana Libantova. 2021. "Basic β-1,3-Glucanase from Drosera binata Exhibits Antifungal Potential in Transgenic Tobacco Plants" Plants 10, no. 8: 1747. https://doi.org/10.3390/plants10081747





