Chloroplastic SaNADP-ME4 of C3–C4 Woody Desert Species Salsola laricifolia Confers Drought and Salt Stress Resistance to Arabidopsis
Abstract
:1. Introduction
2. Results
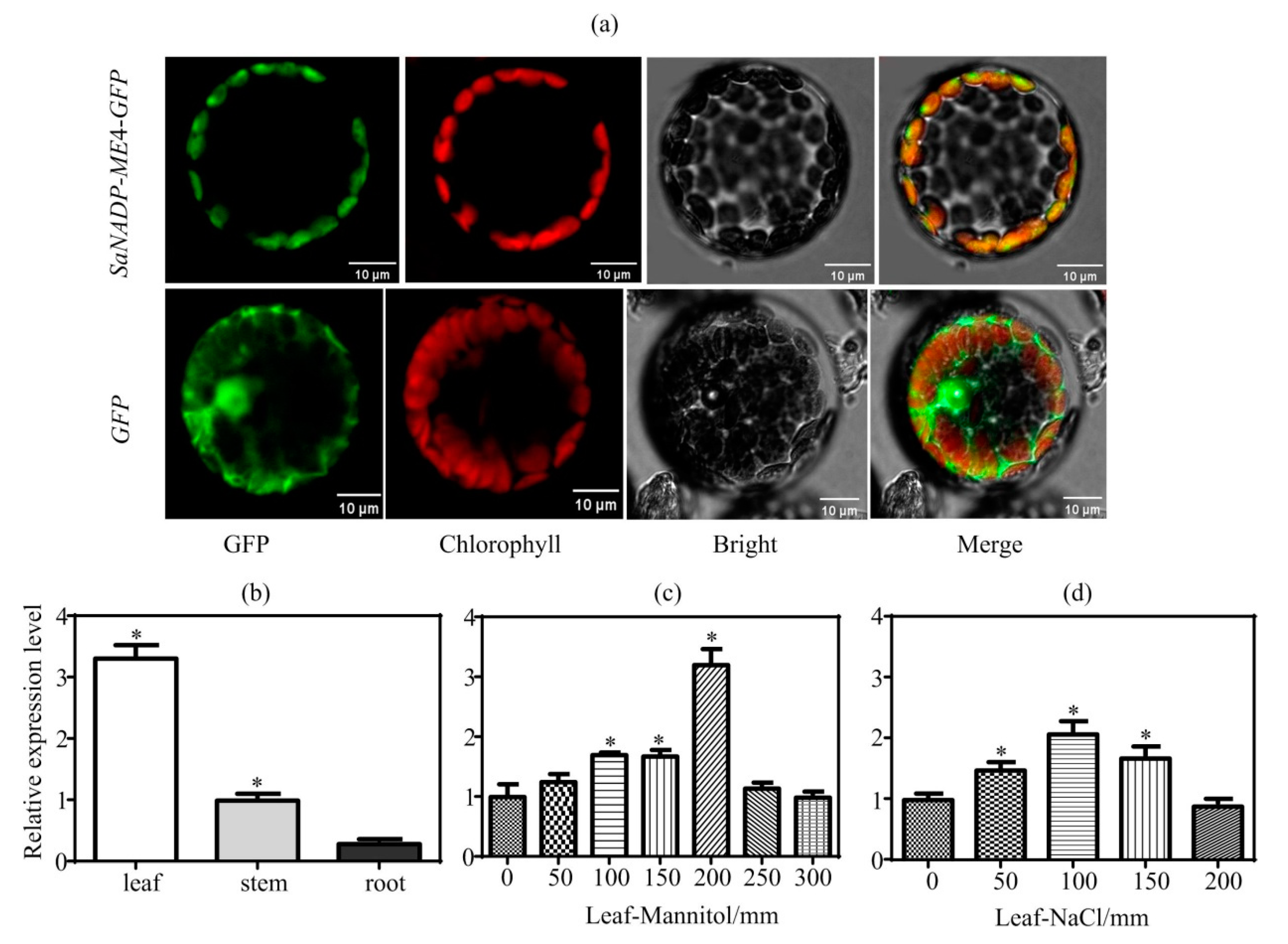
2.1. The Protein SaNADP-ME4 Is Chloroplast-Localized
2.2. Analysis of the Expression of SaNADP-ME4 in S. laricifolia
2.3. Overexpression of SaNADP-ME4 Enhances Osmotic and Salt Tolerances at the Seedling Stage
2.4. Overexpression of SaNADP-ME4 Alleviates the Decreased in Chlorophyll Contents and PSII Photochemical Efficiency under Drought and Salt Stresses
2.5. Overexpression of SaNADP-ME4 Decreases Oxidative Damages under Drought and Salt Stresses
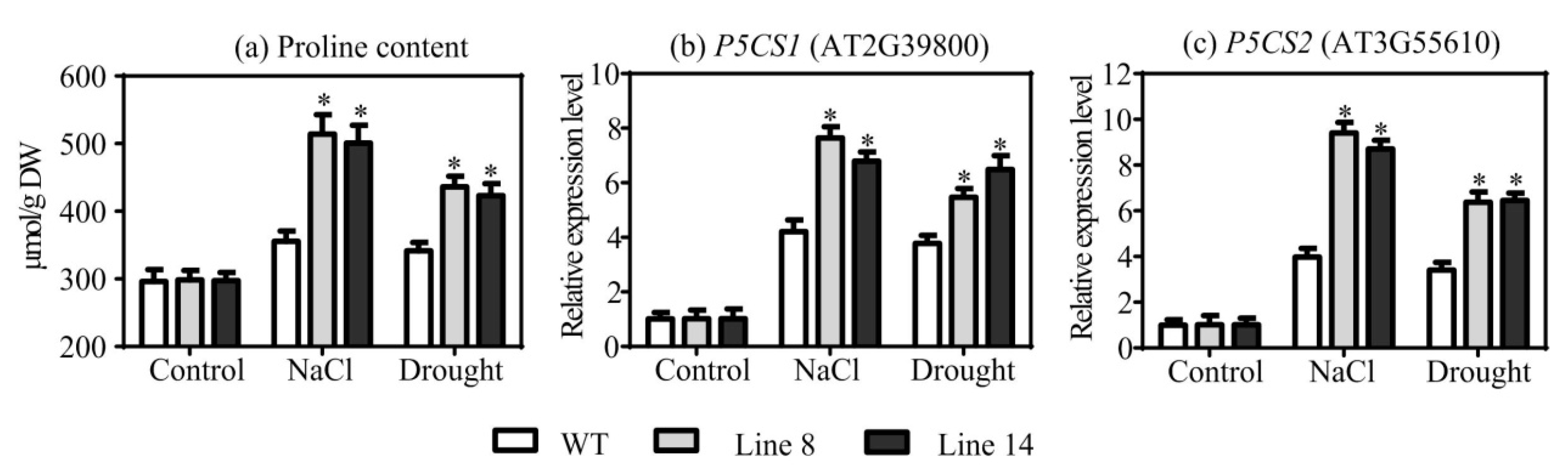
2.6. Overexpression of SaNADP-ME4 Enhances Proline Biosynthesis under Drought and Salt Stresses
2.7. Overexpression of SaNADP-ME4 Regulates of ROS Scavenging Capability under Drought and Salt Stresses
2.8. Overexpression of SaNADP-ME4 Increases of the Total NADP-ME Activity under Drought and Salt Stresses
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
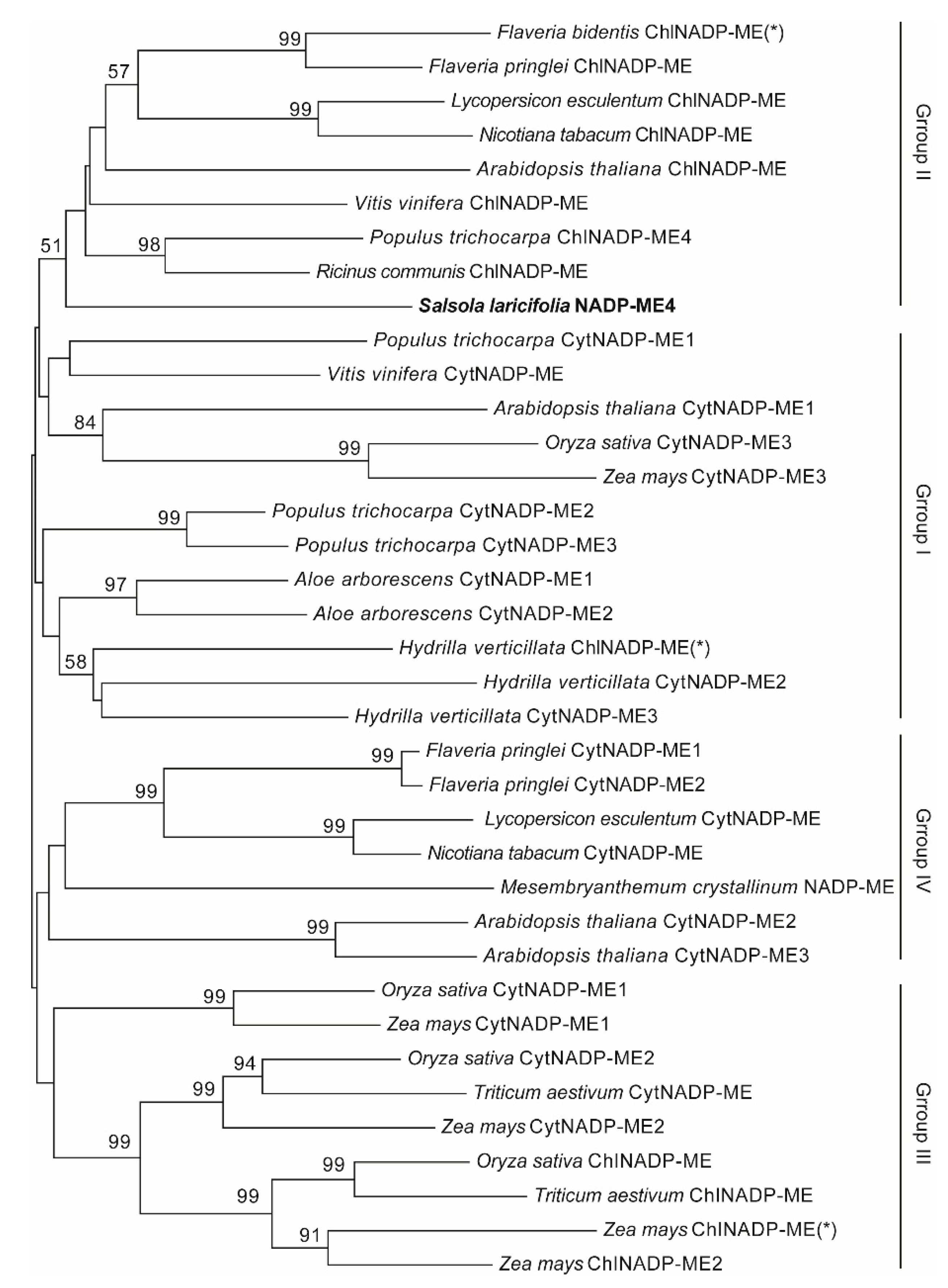
4.2. Sequence and Phylogenetic Analysis
4.3. Subcellular Localization Analysis
4.4. Expression Analysis of SaNADP-ME4 in S. laricifolia under Different Tissues, Mannitol and NaCl Stresses
4.5. Generation of SaNADP-ME4 Overexpressing Arabidopsis
4.6. Assessment of the Mannitol and NaCl Stress Tolerance of Transgenic Arabidopsis at Germination Stage
4.7. Assessment of Mannitol and NaCl Stress Tolerance of Transgenic Arabidopsis at Seedling Stage
4.8. Assessment of Drought and NaCl Stress Tolerance of Transgenic Arabidopsis at Adult Stage
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Edwards, G.E.; Andreo, C.S. NADP-malic enzyme from plants. Phytochemistry 1992, 31, 1845–1857. [Google Scholar]
- Drincovich, M.F.; Casati, P.; Andreo, C.S. NADP-malic enzyme from plants: A ubiquitous enzyme involved in different metabolic pathways. FEBS Lett. 2001, 490, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Maier, A.; Zell, M.B.; Maurino, V.G. Malate decarboxylases: Evolution and roles of NAD(P)-ME isoforms in species performing C4 and C3 photosynthesis. J. Exp. Bot. 2011, 62, 3061–3069. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, C.E.; Saigo, M.; Margarit, E.; Andreo, C.S.; Drincovich, M.F. Kinetics and functional diversity among the five members of the NADP-malic enzyme family from Zea mays, a C4 species. Photosynth. Res. 2013, 115, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Badia, M.B.; Arias, C.L.; Tronconi, M.A.; Maurino, V.G.; Andreo, C.S.; Drincovich, M.F.; Gerrard Wheeler, M.C. Enhanced cytosolic NADP-ME2 activity in A. thaliana affects plant development, stress tolerance and specific diurnal and nocturnal cellular processes. Plant Sci. 2015, 240, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.Q.; Wang, B.P.; Ding, H.Y.; Zhang, J.; Li, S.C. Review: The role of NADP-malic enzyme in plants under stress. Plant Sci. 2019, 281, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Doubnerová Hýsková, V.; Ryšlavá, H. What can enzymes of C4 photosynthesis do for C3 plants under stress? Plant Sci. 2011, 180, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Y.; Song, Y.S.; Zheng, X.X.; Zhang, Y.; Guo, J.R.; Sui, N. NADP-malate dehydrogenase of sweet sorghum improves salt tolerance of Arabidopsis thaliana. J. Agric. Food Chem. 2018, 66, 5992–6002. [Google Scholar] [CrossRef] [PubMed]
- Maurino, V.G.; Gerrard Wheeler, M.C.; Andreo, C.S.; Drincovich, M.F. Redundancy is sometimes seen only by the uncritical: Does Arabidopsis need six malic enzyme isoforms? Plant Sci. 2009, 176, 715–721. [Google Scholar] [CrossRef]
- Chi, W.; Yang, J.; Wu, N.; Zhang, F. Four rice genes encoding NADP malic enzyme exhibit distinct expression profiles. Biosci. Biotech. Biochem. 2004, 68, 1865–1874. [Google Scholar] [CrossRef] [PubMed]
- Gerrard Wheeler, M.C.; Tronconi, M.A.; Drincovich, M.F.; Andreo, C.S.; Flügge, U.I.; Maurino, V.G. A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis thaliana. Plant Physiol. 2005, 139, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Honda, H.; Akagi, H.; Shimada, H. An isozyme of the NADP-malic enzyme of a CAM plant, Aloe arborescens, with variation on conservative amino acid residues. Gene 2000, 243, 85–92. [Google Scholar] [CrossRef]
- Lai, L.B.; Wang, L.; Nelson, T.M. Distinct but conserved functions for two chloroplastic NADP-malic enzyme isoforms in C3 and C4 Flaveria Species. Plant Physiol. 2002, 128, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.S.; Stubbs, J.D.; Taylor, W.C. Two genes encode highly similar chloroplastic NADP-malic enzymes in Flaveria. Plant Physiol. 1996, 111, 1251–1261. [Google Scholar] [CrossRef] [Green Version]
- Müller, G.L.; Drincovich, M.F.; Andreo, C.S.; Lara, M.V. Nicotiana tabacum NADP-Malic enzyme: Cloning, characterization and analysis of biological role. Plant Cell Physiol. 2008, 49, 469–480. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.G.; Liu, J.W.; Wang, Z.F.; Nai, J.F.; Lü, M.Y.; Zhou, X.Y.; Chen, Y.X. Characterization of the NADP-malic enzymes in the woody plant Populus trichocarpa. Mol. Biol. Rep. 2013, 40, 1385–1396. [Google Scholar] [CrossRef]
- Estavillo, G.M.; Rao, S.K.; Reiskind, J.B.; Bowes, G. Characterization of the NADP malic enzyme gene family in the facultative, single-cell C4 monocot Hydrilla verticillata. Photosynth. Res. 2007, 94, 43–57. [Google Scholar] [CrossRef]
- Boyer, J.S. Plant productivity and environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef]
- Umezawa, T.; Fujita, M.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Engineering drought tolerance in plants: Discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 2006, 17, 113–122. [Google Scholar] [CrossRef]
- Cheng, Y.; Long, M. A cytosolic NADP-malic enzyme gene from rice (Oryza sativa L.) confers salt tolerance in transgenic Arabidopsis. Biotechnol. Lett. 2007, 29, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Tsugama, D.; Takano, S.; Liu, S.Q. Rice (Oryza sativa L.) OsNADP-ME4 gene responds to adversity stresses. Cell Biochem. Biophys. 2015, 4, 1–7. [Google Scholar]
- Liu, S.; Cheng, Y.; Zhang, X.; Guan, Q.; Nishiuchi, S.; Hase, K.; Takano, T. Expression of an NADP-malic enzyme gene in rice (Oryza sativa L.) is induced by environmental stresses; over-expression of the gene in Arabidopsis confers salt and osmotic stresses tolerance. Plant Mol. Biol. 2007, 64, 49–58. [Google Scholar] [CrossRef]
- Doubnerová Hýsková, V.; Miedzińska, L.; Dobra, J.; Vankova, R.; Ryšlavá, H. Phosphoenolpyruvate carboxylase, NADP-malic enzyme, and pyruvate, phosphate dikinase are involved in the acclimation of Nicotiana tabacum L. to drought stress. J. Plant Physiol. 2014, 171, 19–25. [Google Scholar] [CrossRef]
- Laporte, M.; Shen, B.; Tarczynski, M. Engineering for drought avoidance expression of maize NADP-ME in tobacco results in altered stomatal function. J. Exp. Bot. 2002, 53, 699–705. [Google Scholar] [CrossRef] [Green Version]
- Lundgren, M.R.; Christin, P.A. Despite phylogenetic effects, C3-C4 lineages bridge the ecological gap to C4 photosynthesis. J. Exp. Bot. 2017, 68, 241–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sage, R.F.; Khoshravesh, R.; Sage, T.L. From proto-Kranz to C4 Kranz: Building the bridge to C4 photosynthesis. J. Exp. Bot. 2014, 65, 3341–3356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Angiosperm Phylogeny Group. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Wen, Z.B.; Zhang, M.L. Salsola laricifolia, another C3–C4 intermediate species in tribe Salsoleae s.l. (Chenopodiaceae). Photosynth. Res. 2015, 123, 33–43. [Google Scholar] [CrossRef]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drincovich, M.F.; Lara, M.; Maurino, V.G.; Andreo, C. C4 decrboxylases: Different solutions for the same biochemical problem, the provision of CO2 in the bundle sheath cells. In C4 Photosynthesis and Related CO2 Concentrating Mechanisms; Raghavendra, A., Sage, R.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 277–300. [Google Scholar]
- Sui, N.; Han, G. Salt-induced photoinhibition of PSII is alleviated in halophyte Thellungiella halophila by increases of unsaturated fatty acids in membrane lipids. Acta Physiol. Plant 2014, 36, 983–992. [Google Scholar] [CrossRef]
- Kramer, D.M.; Johnson, G.; Kiirats, O.; Edwards, G.E. New Fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth. Res. 2004, 79, 209. [Google Scholar] [CrossRef]
- Wang, X.; Dinler, B.S.; Vignjevic, M.; Jacobsen, S.; Wollenweber, B. Physiological and proteome studies of responses to heat stress during grain filling in contrasting wheat cultivars. Plant Sci. 2015, 230, 33–50. [Google Scholar] [CrossRef]
- Baxter, C.J.; Redestig, H.; Schauer, N.; Repilber, D.; Patil, K.R.; Nielsen, J.; Selbig, J.; Liu, J.; Fernie, A.R.; Sweetlove, L.J. The metabolic response of heterotrophic Arabidopsis cells to oxidative stress. Plant Physiol. 2007, 143, 312–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valderrama, R.; Corpas, F.; Carreras, A.; Gόmez-Rodríguez, M.; Pedrajas, C.J.; Fernández-Ocaña, A.; Del Río, L.; Barroso, J. The dehydrogenase-mediated recycling of NADPH is a key antioxidant system against salt-induced oxidative stress in olive plant. Plant Cell Environ. 2006, 29, 1449–1459. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signally in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.K.; Zhao, Y.Z.; Chu, H.Y.; Wang, A.X.; Zhu, J.H.; Chen, X.J.; Zou, Y.J.; Shi, M.; Liu, R.M.; Su, N.; et al. Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat. Methods 2017, 14, 720–728. [Google Scholar] [CrossRef]
- Cracan, V.; Titov, D.V.; Shen, H.Y.; Grabarek, Z.; Mootha, V.K. A genetically encoded tool for manipulation of NADP+/NADPH in living cells. Nat. Chem. Biol. 2017, 13, 1088–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Z.L.; Wen, X.J.; Wang, Y.C. Betala platuphylla BpHOX2 transcription factor binds to different cis-acting elements and confers osmotic tolerance. J. Integr. Plant Biol. 2020, 62, 1762–1779. [Google Scholar] [CrossRef]
- Zhu, J.J.; Schwörer, S.; Berisa, M.; Kyung, Y.J.; Ryu, K.W.; Yi, J.M.; Jiang, X.J.; Cross, J.R.; Thompson, C.B. Mitochondrial NADP(H) generation is essential for proline biosynthesis. Science 2021, 372, 968–972. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Wen, Z.B.; Wang, Y.L.; Feng, Y. Suitable reference genes for real-time quantitative PCR in Salsola laricifolia under five abiotic stresses. Biol. Plantarum 2019, 63, 380–387. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Ghany, S.E.; Müller-Moulé, P.; Niyogi, K.K.; Pilon, M.; Shikanai, T. Two-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 2005, 17, 1233–1251. [Google Scholar] [CrossRef] [Green Version]
- Candat, A.; Poupart, P.; Andrieu, J.P.; Chevrollier, A.; Reynier, P.; Rogniaux, H.; Avelange-Macherel, M.; Macherel, D. Experimental determination of organelle targeting-peptide cleavage sites using transient expression of green fluorescent protein translational fusions. Anal. Biochem. 2013, 434, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, N.; Pelletier, G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998, 82, 259–266. [Google Scholar] [PubMed]
- Li, X.S.; Liang, Y.Q.; Gao, B.; Mijiti, M.; Bozorov, T.A.; Yang, H.L.; Zhang, D.Y.; Wood, A.J. ScDREB10, an A-5c type of DREB gene of the desert moss Syntrichia caninervis, confers osmotic and salt tolerances to Arabidopsis. Genes 2019, 10, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wintermans, J.F.G.M.; De Mots, A. Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim. Biophys. Acta 1965, 109, 448–453. [Google Scholar] [CrossRef]
- Weigel, R.R.; Bäuscher, C.; Pfitzner, A.J.; Pfitzner, U.M. NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol. Biol. 2001, 46, 143–160. [Google Scholar] [CrossRef]
- Hu, L.X.; Li, H.Y.; Pang, H.C.; Fu, J.M. Responses of antioxidant gene, protein and enzymes to salinity stress in two genotypes of perennial ryegrass (Lolium perenne) differing in salt tolerance. J. Plant Physiol. 2012, 169, 146–156. [Google Scholar] [CrossRef]
- Li, H.X.; Xiao, Y.; Cao, L.L.; Yan, X.; Li, C.; Shi, H.Y.; Wang, J.W.; Ye, Y.H. Cerebroside C increases tolerance to chilling injury and alters lipid composition in wheat roots. PLoS ONE 2013, 8, e73380. [Google Scholar] [CrossRef] [Green Version]
- Jana, S.; Choudhuri, M.A. Glycolate metabolism of three submerged aquatic angiosperms during aging. Aquat. Bot. 1981, 12, 345–354. [Google Scholar] [CrossRef]
- Draper, H.H.; Hadley, M. Malondialdehyde determination as index of lipid peroxidation. Methods Enzymol. 1990, 186, 421–431. [Google Scholar]
- Ries, S.K.; Giannopolitis, C.N. Superoxide dismutases: II. Purification and quantitative relationship with water-soluble protein in seedlings. Plant Physiol. 1977, 59, 315–318. [Google Scholar]
- Chance, M.; Maehly, A.C. Assay of catalases and peroxidases. Method Enzy. 1955, 2, 764–817. [Google Scholar]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, Z.; Wang, Y.; Xia, C.; Zhang, Y.; Zhang, H. Chloroplastic SaNADP-ME4 of C3–C4 Woody Desert Species Salsola laricifolia Confers Drought and Salt Stress Resistance to Arabidopsis. Plants 2021, 10, 1827. https://doi.org/10.3390/plants10091827
Wen Z, Wang Y, Xia C, Zhang Y, Zhang H. Chloroplastic SaNADP-ME4 of C3–C4 Woody Desert Species Salsola laricifolia Confers Drought and Salt Stress Resistance to Arabidopsis. Plants. 2021; 10(9):1827. https://doi.org/10.3390/plants10091827
Chicago/Turabian StyleWen, Zhibin, Yulan Wang, Chunlan Xia, Yuhui Zhang, and Hongxiang Zhang. 2021. "Chloroplastic SaNADP-ME4 of C3–C4 Woody Desert Species Salsola laricifolia Confers Drought and Salt Stress Resistance to Arabidopsis" Plants 10, no. 9: 1827. https://doi.org/10.3390/plants10091827
APA StyleWen, Z., Wang, Y., Xia, C., Zhang, Y., & Zhang, H. (2021). Chloroplastic SaNADP-ME4 of C3–C4 Woody Desert Species Salsola laricifolia Confers Drought and Salt Stress Resistance to Arabidopsis. Plants, 10(9), 1827. https://doi.org/10.3390/plants10091827






