Primer Choice and Xylem-Microbiome-Extraction Method Are Important Determinants in Assessing Xylem Bacterial Community in Olive Trees
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Sampling
2.2. DNA Extraction and Library Sequencing
2.3. Bioinformatics and Statistical Approximation
3. Results
3.1. Illumina Output and Taxa Diversity of Plastids and Bacterial Community
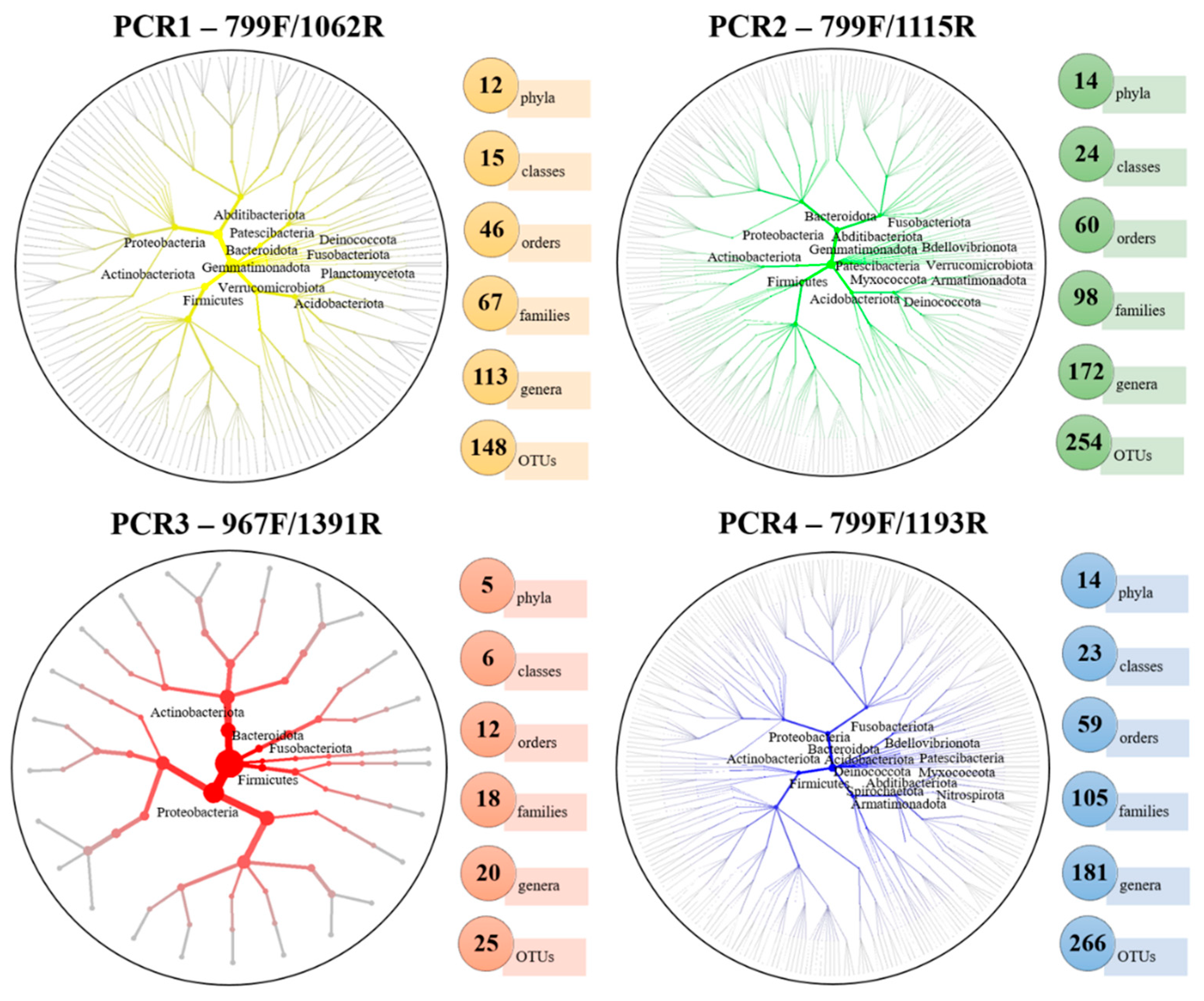
3.2. Xylem-Bacterial-Community Distribution
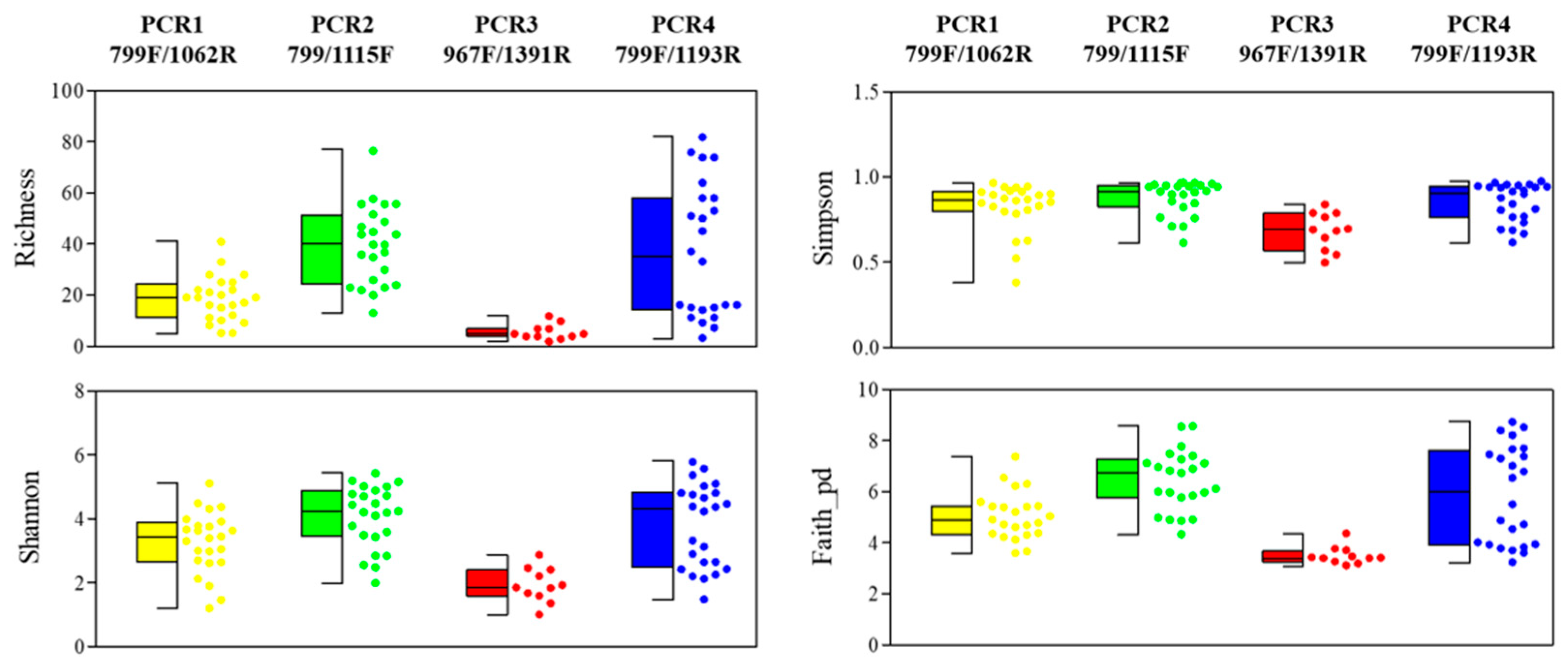
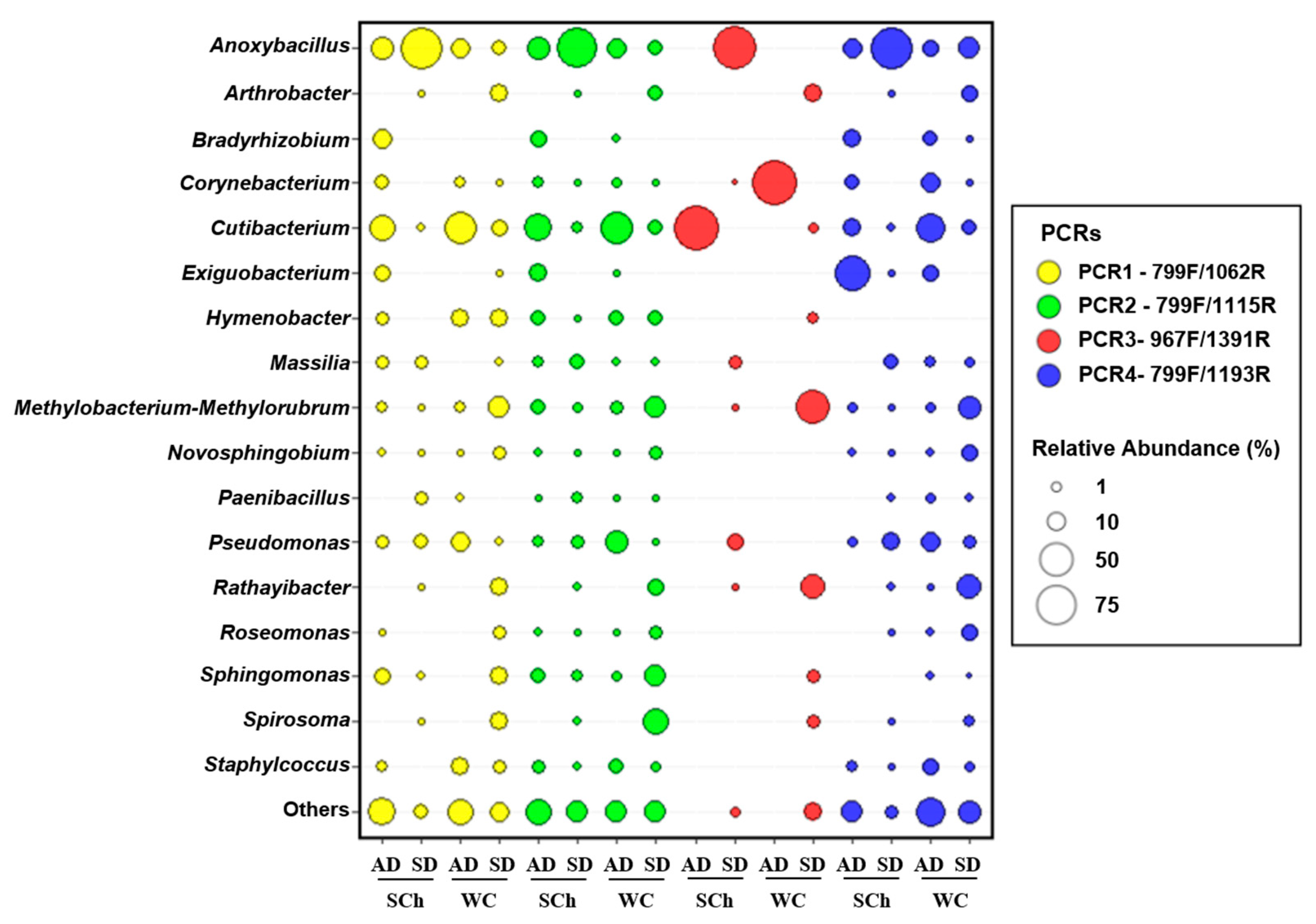
3.3. Bacterial-Abundance Analysis
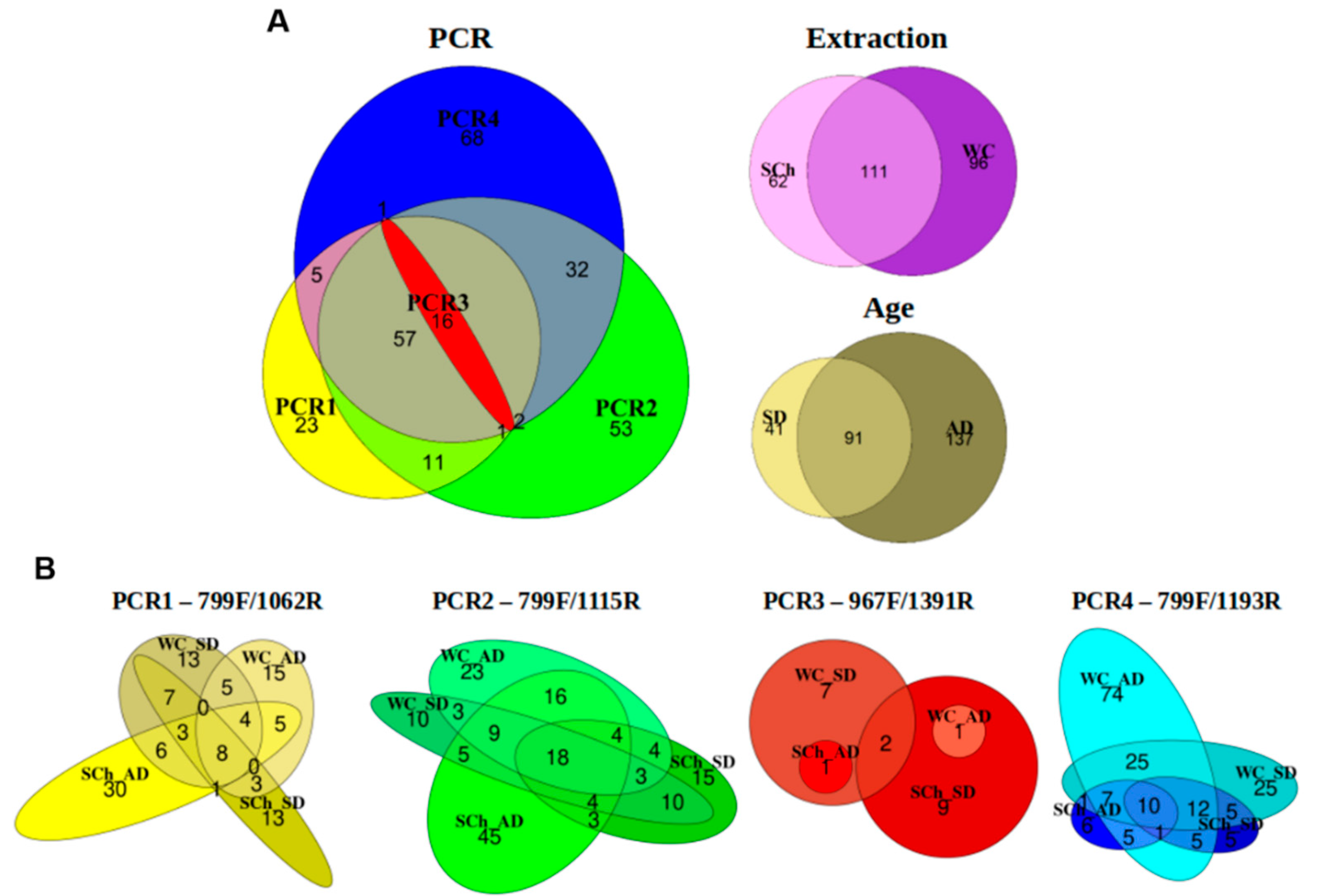
3.4. Effects of PCR-Primer Pairs, Xylem-Extraction Method and Plant Age on Bacterial-Community Structure
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Besnard, G.; Khadari, B.; Navascués, M.; Fernández-Mazuecos, M.; El Bakkali, A.; Arrigo, N.; Baali-Cherif, D.; Brunini-Bronzini de Caraffa, V.; Santoni, S.; Vargas, P.; et al. The complex history of the olive tree: From Late Quaternary diversification of Mediterranean lineages to primary domestication in the northern Levant. Proc. R. Soc. B Biol. Sci. 2013, 280, 20122833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alarcon de la Lastra, C.; Barranco, M.D.; Motilva, V.; Herrerias, J.M. Mediterranean diet and health: Biological importance of olive oil. Curr. Pharm. Des. 2001, 7, 933–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loumou, A.; Giourga, C. Olive groves: “The life and identity of the Mediterranean”. Agric. Hum. Values 2003, 20, 87–95. [Google Scholar] [CrossRef]
- Jiménez-Díaz, R.M.; Cirulli, M.; Bubici, G.; del Mar Jiménez-Gasco, M.; Antoniou, P.P.; Tjamos, E.C. Verticillium Wilt, a major threat to olive production: Current status and future prospects for its management. Plant Dis. 2011, 96, 304–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saponari, M.; Giampetruzzi, A.; Loconsole, G.; Boscia, D.; Saldarelli, P. Xylella fastidiosa in olive in Apulia: Where we stand. Phytopathology 2018, 109, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Almeida, R.P.P.; De La Fuente, L.; Koebnik, R.; Lopes, J.R.S.; Parnell, S.; Scherm, H. Addressing the New Global Threat of Xylella fastidiosa. Phytopathology 2019, 109, 172–174. [Google Scholar] [CrossRef] [Green Version]
- Landa, B.B.; Pérez, A.G.; Luaces, P.; Montes-Borrego, M.; Navas-Cortés, J.A.; Sanz, C. Insights into the effect of Verticillium dahliae defoliating-pathotype infection on the content of phenolic and volatile compounds related to the sensory properties of virgin olive oil. Front. Plant Sci. 2019, 10, 232. [Google Scholar] [CrossRef] [Green Version]
- Schneider, K.; van der Werf, W.; Cendoya, M.; Mourits, M.; Navas-Cortés, J.A.; Vicent, A.; Oude Lansink, A. Impact of Xylella fastidiosa subspecies pauca in European olives. Natl. Acad. Sci. 2020, 117, 9250–9259. [Google Scholar] [CrossRef] [Green Version]
- Reinhold-Hurek, B.; Hurek, T. Living inside plants: Bacterial endophytes. Curr. Opin. Plant Biol. 2011, 14, 435–443. [Google Scholar] [CrossRef]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. Unraveling the plant microbiome: Looking back and future perspectives. Front. Microbiol. 2014, 5, 148. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Weller, D.M.; Raaijmakers, J.M.; Gardener, B.B.M.; Thomashow, L.S. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 2002, 40, 309–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anguita-Maeso, M.; Trapero-Casas, J.L.; Olivares-García, C.; Ruano-Rosa, D.; Palomo-Ríos, E.; Jiménez-Díaz, R.M.; Navas-Cortés, J.A.; Landa, B.B. Verticillium dahliae inoculation and in vitro propagation modify the xylem microbiome and disease reaction to Verticillium wilt in a wild olive genotype. Front. Plant Sci. 2021, 12, 250. [Google Scholar] [CrossRef]
- Martín, J.A.; Witzell, J.; Blumenstein, K.; Rozpedowska, E.; Helander, M.; Sieber, T.N.; Gil, L. Resistance to Dutch Elm Disease Reduces Presence of Xylem Endophytic Fungi in Elms (Ulmus spp.). PLoS ONE 2013, 8, e56987. [Google Scholar] [CrossRef] [Green Version]
- Deyett, E.; Roper, M.C.; Ruegger, P.; Yang, J.-I.; Borneman, J.; Rolshausen, P.E. Microbial Landscape of the Grapevine Endosphere in the Context of Pierce’s Disease. Phytobiomes J. 2017, 1, 138–149. [Google Scholar] [CrossRef]
- Fausto, C.; Mininni, A.N.; Sofo, A.; Crecchio, C.; Scagliola, M.; Dichio, B.; Xiloyannis, C. Olive orchard microbiome: Characterisation of bacterial communities in soil-plant compartments and their comparison between sustainable and conventional soil management systems. Plant Ecol. Divers. 2018, 11, 597–610. [Google Scholar] [CrossRef]
- Deyett, E.; Rolshausen, P.E. Temporal dynamics of the sap microbiome of grapevine under high Pierce’s Disease pressure. Front. Plant Sci. 2019, 10, 1246. [Google Scholar] [CrossRef]
- Sofo, A.; Mininni, A.N.; Fausto, C.; Scagliola, M.; Crecchio, C.; Xiloyannis, C.; Dichio, B. Evaluation of the possible persistence of potential human pathogenic bacteria in olive orchards irrigated with treated urban wastewater. Sci. Total Environ. 2019, 658, 763–767. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; Olivares-García, C.; Haro, C.; Imperial, J.; Navas-Cortés, J.A.; Landa, B.B. Culture-dependent and culture-independent characterization of the olive xylem microbiota: Effect of sap extraction methods. Front. Plant Sci. 2020, 10, 1708. [Google Scholar] [CrossRef] [Green Version]
- Haro, C.; Anguita-Maeso, M.; Metsis, M.; Navas-Cortés, J.A.; Landa, B.B. Evaluation of established methods for DNA extraction and primer pairs targeting 16S rRNA gene for bacterial microbiota profiling of olive xylem sap. Front. Plant Sci. 2021, 12, 296. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anguita-Maeso, M.; Haro, C.; Montes-Borrego, M.; De La Fuente, L.; Navas-Cortés, J.A.; Landa, B.B. Metabolomic, ionomic and microbial characterization of olive xylem sap reveals differences according to plant age and genotype. Agronomy 2021, 11, 1179. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; Baptista, P.; Morelli, M.; Cameirão, C.; Lino Neto, T.; Costa, D.; D’Attoma, G.; Abou Kubaa, R.; Altamura, G.; Saponari, M.; et al. Differences in the endophytic microbiome of olive cultivars infected by Xylella fastidiosa across seasons. Pathogens 2020, 9, 723. [Google Scholar] [CrossRef]
- Dastogeer, K.M.G.; Tumpa, F.H.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant Microbiome–An account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; Ares-Yebra, A.; Haro, C.; Román-Écija, M.; Olivares-García, C.; Costa, J.; Marco-Noales, E.; Ferrer, A.; Navas-Cortés, J.A.; Landa, B.B. Xylella Fastidiosa Infection Reshapes Microbial Composition and Network Associations in the Xylem of Almond Trees. Front. Microbiol. 2022; in press. [Google Scholar]
- Lundberg, D.S.; Yourstone, S.; Mieczkowski, P.; Jones, C.D.; Dangl, J.L. Practical innovations for high-throughput amplicon sequencing. Nat. Methods 2013, 10, 999–1002. [Google Scholar] [CrossRef]
- Hanshew, A.S.; Mason, C.J.; Raffa, K.F.; Currie, C.R. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J. Microbiol. Methods 2013, 95, 149–155. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, J.; Singh, K.; Fern, A.; Kirton, E.; He, S.; Woyke, T.; Lee, J.; Chen, F.; Dangl, J.; Tringe, S. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015, 6, 771. [Google Scholar] [CrossRef] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012, 41, e1. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. Msystems 2016, 1, e00009-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comeau, A.M.; Li, W.K.W.; Tremblay, J.-É.; Carmack, E.C.; Lovejoy, C. Arctic Ocean Microbial Community Structure before and after the 2007 Record Sea Ice Minimum. PLoS ONE 2011, 6, e27492. [Google Scholar] [CrossRef]
- Dyall, S.D.; Brown, M.T.; Johnson, P.J. Ancient invasions: From endosymbionts to organelles. Science 2004, 304, 253–257. [Google Scholar] [CrossRef] [Green Version]
- Beckers, B.; Op De Beeck, M.; Thijs, S.; Truyens, S.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Performance of 16s rDNA primer pairs in the study of rhizosphere and endosphere bacterial microbiomes in metabarcoding studies. Front. Microbiol. 2016, 7, 650. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos, M.; Kavamura, V.; Reynaldo, E.; Souza, D.; Da Silva, E.; May, A. Bacterial structure of agricultural soils with high and low yields. J. Plant Pathol. Microbiol. 2017, 8, 405. [Google Scholar] [CrossRef]
- Ahumada-Orellana, L.; Ortega-Farías, S.; Poblete-Echeverría, C.; Searles, P.S. Estimation of stomatal conductance and stem water potential threshold values for water stress in olive trees (cv. Arbequina). Irrig. Sci. 2019, 37, 461–467. [Google Scholar] [CrossRef]
- García-Tejera, O.; López-Bernal, Á.; Orgaz, F.; Testi, L.; Villalobos, F.J. The pitfalls of water potential for irrigation scheduling. Agric. Water Manag. 2021, 243, 106522. [Google Scholar] [CrossRef]
- Iniesta, F.; Testi, L.; Orgaz, F.; Villalobos, F.J. The effects of regulated and continuous deficit irrigation on the water use, growth and yield of olive trees. Eur. J. Agron. 2009, 30, 258–265. [Google Scholar] [CrossRef]
- Chelius, M.K.; Triplett, E.W. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 2001, 41, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Diskin, S.; Feygenberg, O.; Maurer, D.; Droby, S.; Prusky, D.; Alkan, N. Microbiome Alterations Are Correlated with Occurrence of Postharvest Stem-End Rot in Mango Fruit. Phytobiomes J. 2017, 1, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Laforest-Lapointe, I.; Paquette, A.; Messier, C.; Kembel, S.W. Leaf bacterial diversity mediates plant diversity and ecosystem function relationships. Nature 2017, 546, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Sogin, M.L.; Morrison, H.G.; Huber, J.A.; Welch, D.M.; Huse, S.M.; Neal, P.R.; Arrieta, J.M.; Herndl, G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef] [Green Version]
- Walker, J.J.; Pace, N.R. Phylogenetic Composition of Rocky Mountain Endolithic Microbial Ecosystems. Appl. Environ. Microbiol. 2007, 73, 3497–3504. [Google Scholar] [CrossRef] [Green Version]
- Bodenhausen, N.; Horton, M.W.; Bergelson, J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 2013, 8, e56329. [Google Scholar] [CrossRef]
- Kibbe, W.A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007, 35, W43–W46. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Prepr. 2018, 6, e27295v2. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, T.C.; Ma, K.H.; Chao, A. iNEXT: An R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Larsson, J. Eulerr: Area-Proportional Euler and Venn Diagrams with Ellipses. Bachelor’s Thesis, Lund University, Lund, Sweden, 2018. [Google Scholar]
- Foster, Z.S.L.; Sharpton, T.J.; Grünwald, N.J. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 2017, 13, e1005404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peres-Neto, P.R.; Legendre, P.; Dray, S.; Borcard, D. Variation Partitioning of Species Data Matrices: Estimation and Comparison of Fractions. Ecology 2006, 87, 2614–2625. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [Green Version]
- Lucaciu, R.; Pelikan, C.; Gerner, S.M.; Zioutis, C.; Köstlbacher, S.; Marx, H.; Herbold, C.W.; Schmidt, H.; Rattei, T. A Bioinformatics Guide to Plant Microbiome Analysis. Front. Plant Sci. 2019, 10, 1313. [Google Scholar] [CrossRef] [Green Version]
- Thijs, S.; Op De Beeck, M.; Beckers, B.; Truyens, S.; Stevens, V.; Van Hamme, J.D.; Weyens, N.; Vangronsveld, J. Comparative Evaluation of Four Bacteria-Specific Primer Pairs for 16S rRNA Gene Surveys. Front. Microbiol. 2017, 8, 494. [Google Scholar] [CrossRef]
- Dong, Y.; Lang, Z.; Kong, X.; Lu, D.; Liu, Z. Kinetic and multidimensional profiling of accelerated degradation of oil sludge by biostimulation. Environ. Sci. Process. Impacts 2015, 17, 763–774. [Google Scholar] [CrossRef]
- Kharbush, J.J.; Kejriwal, K.; Aluwihare, L.I. Distribution and Abundance of Hopanoid Producers in Low-Oxygen Environments of the Eastern Pacific Ocean. Microb. Ecol. 2016, 71, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Serebryakova, A.; Aires, T.; Viard, F.; Serrão, E.A.; Engelen, A.H. Summer shifts of bacterial communities associated with the invasive brown seaweed Sargassum muticum are location and tissue dependent. PLoS ONE 2018, 13, e0206734. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.-Y.; Zhang, C.-S. Illumina-Based Analysis of Endophytic and Rhizosphere Bacterial Diversity of the Coastal Halophyte Messerschmidia sibirica. Front. Microbiol. 2017, 8, 2288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlaeppi, K.; Dombrowski, N.; Oter, R.G.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. USA 2014, 111, 585–592. [Google Scholar] [CrossRef] [Green Version]
- Campisano, A.; Ometto, L.; Compant, S.; Pancher, M.; Antonielli, L.; Yousaf, S.; Varotto, C.; Anfora, G.; Pertot, I.; Sessitsch, A.; et al. Interkingdom Transfer of the Acne-Causing Agent, Propionibacterium acnes, from Human to Grapevine. Mol. Biol. Evol. 2014, 31, 1059–1065. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Abdelfattah, A.; Norelli, J.; Burchard, E.; Schena, L.; Droby, S.; Wisniewski, M. Apple endophytic microbiota of different rootstock/scion combinations suggests a genotype-specific influence. Microbiome 2018, 6, 18. [Google Scholar] [CrossRef]
- Beckers, B.; Op De Beeck, M.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 2017, 5, 25. [Google Scholar] [CrossRef] [Green Version]
- Pham, N.T.; Meier-Dinkel, A.; Höltken, A.M.; Quambusch, M.; Mahnkopp, F.; Winkelmann, T. Endophytic bacterial communities in in vitro shoot cultures derived from embryonic tissue of hybrid walnut (Juglans × intermedia). Plant Cell Tissue Organ Cult. 2017, 130, 153–165. [Google Scholar] [CrossRef]
- Bruez, E.; Haidar, R.; Alou, M.T.; Vallance, J.; Bertsch, C.; Mazet, F.; Fermaud, M.; Deschamps, A.; Guerin-Dubrana, L.; Compant, S.; et al. Bacteria in a wood fungal disease: Characterization of bacterial communities in wood tissues of esca-foliar symptomatic and asymptomatic grapevines. Front. Microbiol. 2015, 6, 1137. [Google Scholar] [CrossRef] [Green Version]
- Bao, L.; Gu, L.; Sun, B.; Cai, W.; Zhang, S.; Zhuang, G.; Bai, Z.; Zhuang, X. Seasonal variation of epiphytic bacteria in the phyllosphere of Gingko biloba, Pinus bungeana and Sabina chinensis. FEMS Microbiol. Ecol. 2020, 96, fiaa017. [Google Scholar] [CrossRef]
- Espenshade, J.; Thijs, S.; Gawronski, S.; Bové, H.; Weyens, N.; Vangronsveld, J. Influence of Urbanization on Epiphytic Bacterial Communities of the Platanus × hispanica Tree Leaves in a Biennial Study. Front. Microbiol. 2019, 10, 675. [Google Scholar] [CrossRef] [Green Version]
- Moyes, A.B.; Kueppers, L.M.; Pett-Ridge, J.; Carper, D.L.; Vandehey, N.; O’Neil, J.; Frank, A.C. Evidence for foliar endophytic nitrogen fixation in a widely distributed subalpine conifer. New Phytol. 2016, 210, 657–668. [Google Scholar] [CrossRef] [Green Version]
- Gardner, J.M.; Feldman, A.W.; Zablotowicz, R.M. Identity and behavior of xylem-residing bacteria in rough lemon roots of Florida citrus trees. Appl. Environ. Microbiol. 1982, 43, 1335–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, H.M.; Chew, T.H.; Hudson, A.O.; Savka, M.A. Genome Sequence of Methylobacterium sp. Strain GXF4, a Xylem-Associated Bacterium Isolated from Vitis vinifera L. Grapevine. J. Bacteriol. 2012, 194, 5157–5158. [Google Scholar] [CrossRef] [PubMed]
- Podolich, O.; Laschevskyy, V.; Ovcharenko, L.; Kozyrovska, N.; Pirttilä, A.M. Methylobacterium sp. resides in unculturable state in potato tissues in vitro and becomes culturable after induction by Pseudomonas fluorescens IMGB163. J. Appl. Microbiol. 2009, 106, 728–737. [Google Scholar] [CrossRef] [PubMed]
- Bacilio-Jiménez, M.; Aguilar-Flores, S.; del Valle, M.V.; Pérez, A.; Zepeda, A.; Zenteno, E. Endophytic bacteria in rice seeds inhibit early colonization of roots by Azospirillum brasilense. Soil Biol. Biochem. 2001, 33, 167–172. [Google Scholar] [CrossRef]
- Poonguzhali, S.; Madhaiyan, M.; Yim, W.-J.; Kim, K.-A.; Sa, T.-M. Colonization pattern of plant root and leaf surfaces visualized by use of green-fluorescent-marked strain of Methylobacterium suomiense and its persistence in rhizosphere. Appl. Microbiol. Biotechnol. 2008, 78, 1033–1043. [Google Scholar] [CrossRef]
- Macey, M.C.; Pratscher, J.; Crombie, A.T.; Murrell, J.C. Impact of plants on the diversity and activity of methylotrophs in soil. Microbiome 2020, 8, 31. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef] [Green Version]
- Walters, W.A.; Jin, Z.; Youngblut, N.; Wallace, J.G.; Sutter, J.; Zhang, W.; González-Peña, A.; Peiffer, J.; Koren, O.; Shi, Q.; et al. Large-Scale replicated field study of maize rhizosphere identifies heritable microbes. Proc. Natl. Acad. Sci. USA 2018, 115, 7368–7373. [Google Scholar] [CrossRef] [Green Version]
- Marques, J.M.; da Silva, T.F.; Vollu, R.E.; Blank, A.F.; Ding, G.-C.; Seldin, L.; Smalla, K. Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS Microbiol. Ecol. 2014, 88, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Micallef, S.A.; Channer, S.; Shiaris, M.P.; Colon-Carmona, A. Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal. Behav. 2009, 4, 777–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Wang, G.; Jin, J.; Liu, J.; Zhang, Q.; Liu, X. Bacterial communities in soybean rhizosphere in response to soil type, soybean genotype, and their growth stage. Soil Biol. Biochem. 2009, 41, 919–925. [Google Scholar] [CrossRef]
- Carper, D.L.; Carrell, A.A.; Kueppers, L.M.; Frank, A.C. Bacterial endophyte communities in Pinus flexilis are structured by host age, tissue type, and environmental factors. Plant Soil 2018, 428, 335–352. [Google Scholar] [CrossRef] [Green Version]
- Meaden, S.; Metcalf, C.J.E.; Koskella, B. The effects of host age and spatial location on bacterial community composition in the English Oak tree (Quercus robur). Environ. Microbiol. Rep. 2016, 8, 649–658. [Google Scholar] [CrossRef] [Green Version]
- Caliz, J.; Montes-Borrego, M.; Triadó-Margarit, X.; Metsis, M.; Landa, B.B.; Casamayor, E.O. Influence of edaphic, climatic, and agronomic factors on the composition and abundance of nitrifying microorganisms in the rhizosphere of commercial olive crops. PLoS ONE 2015, 10, e0125787. [Google Scholar] [CrossRef] [Green Version]
- Müller, H.; Berg, C.; Landa, B.B.; Auerbach, A.; Moissl-Eichinger, C.; Berg, G. Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Front. Microbiol. 2015, 6, 138. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhu, A.; Tan, H.; Cao, L.; Zhang, R. Engineering banana endosphere microbiome to improve Fusarium wilt resistance in banana. Microbiome 2019, 7, 74. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anguita-Maeso, M.; Haro, C.; Navas-Cortés, J.A.; Landa, B.B. Primer Choice and Xylem-Microbiome-Extraction Method Are Important Determinants in Assessing Xylem Bacterial Community in Olive Trees. Plants 2022, 11, 1320. https://doi.org/10.3390/plants11101320
Anguita-Maeso M, Haro C, Navas-Cortés JA, Landa BB. Primer Choice and Xylem-Microbiome-Extraction Method Are Important Determinants in Assessing Xylem Bacterial Community in Olive Trees. Plants. 2022; 11(10):1320. https://doi.org/10.3390/plants11101320
Chicago/Turabian StyleAnguita-Maeso, Manuel, Carmen Haro, Juan A. Navas-Cortés, and Blanca B. Landa. 2022. "Primer Choice and Xylem-Microbiome-Extraction Method Are Important Determinants in Assessing Xylem Bacterial Community in Olive Trees" Plants 11, no. 10: 1320. https://doi.org/10.3390/plants11101320





