Genome-Wide Association Studies of Maize Seedling Root Traits under Different Nitrogen Levels
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Experimental Design
2.3. Root and Shoot Phenotype Measurement
2.4. Phenotypic Data Analysis
2.5. Genome-Wide Association Studies
2.6. Candidate Gene Analysis
2.7. Candidate Gene Expression Analysis
- RNA extraction and cDNA preparation
- 2.
- Real-time fluorescent quantitative PCR
3. Results
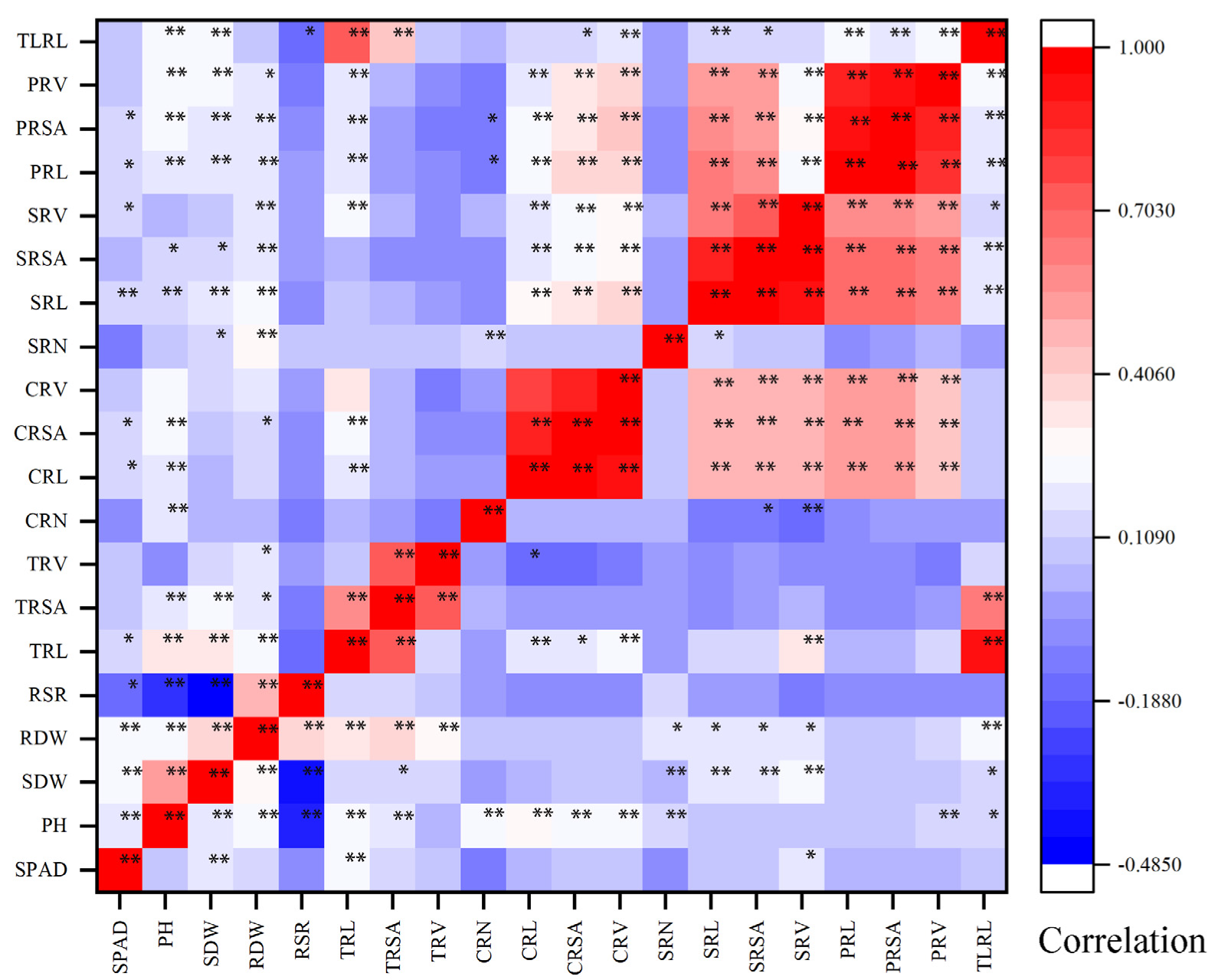
3.1. Phenotypic Analysis of Shoot and Root Traits
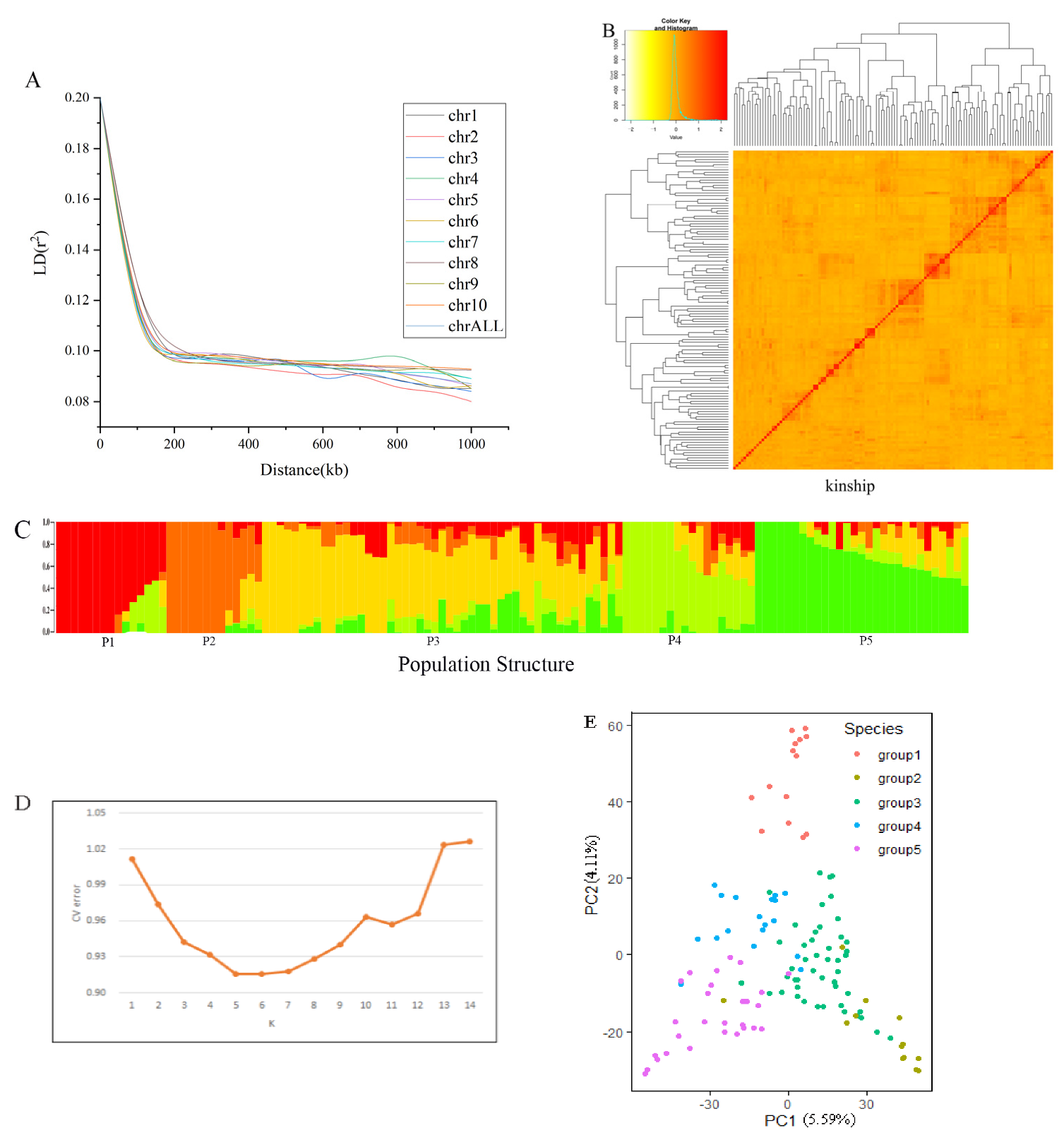
3.2. Population Structure and Linkage Disequilibrium Decay
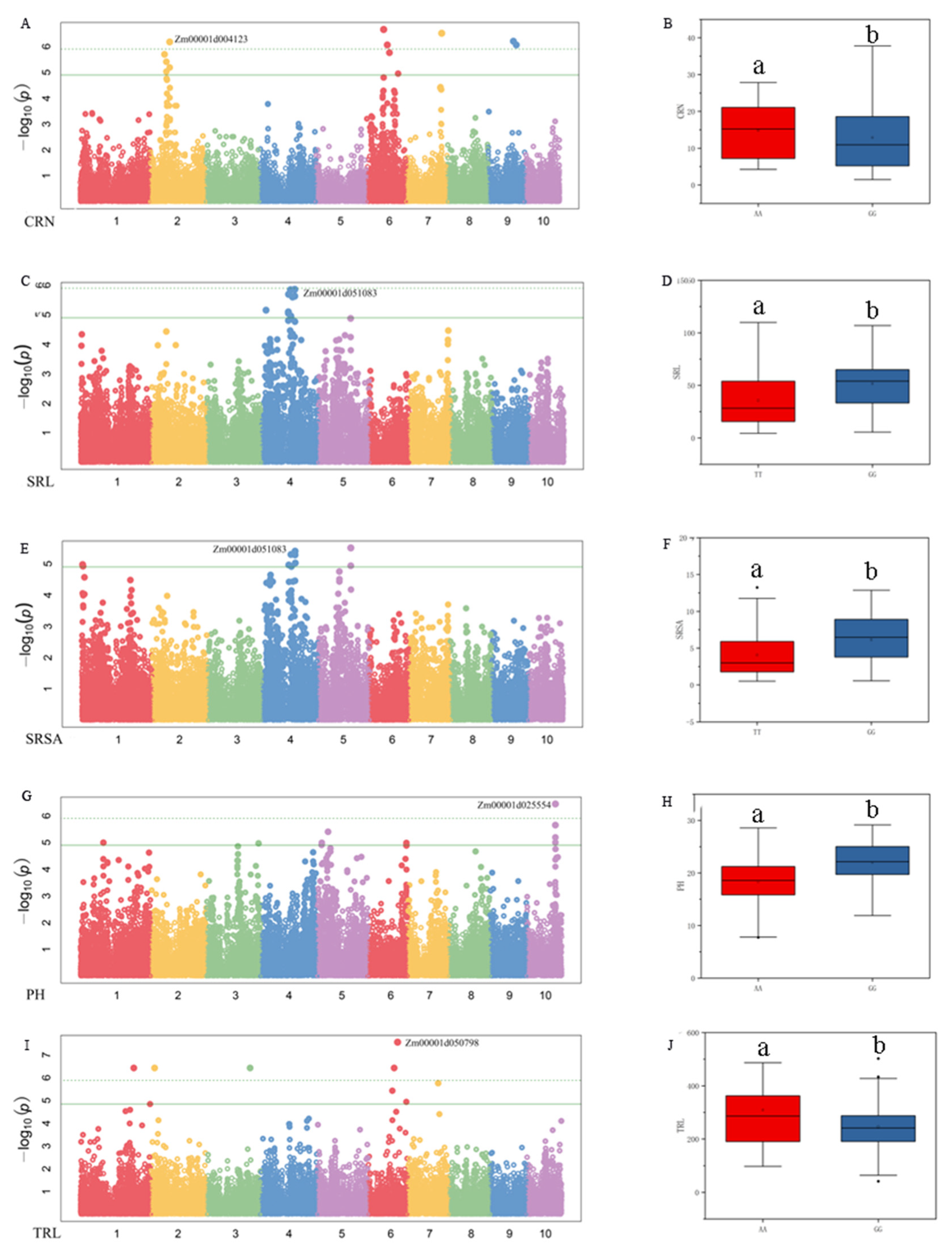
3.3. Genome-Wide Association Studies
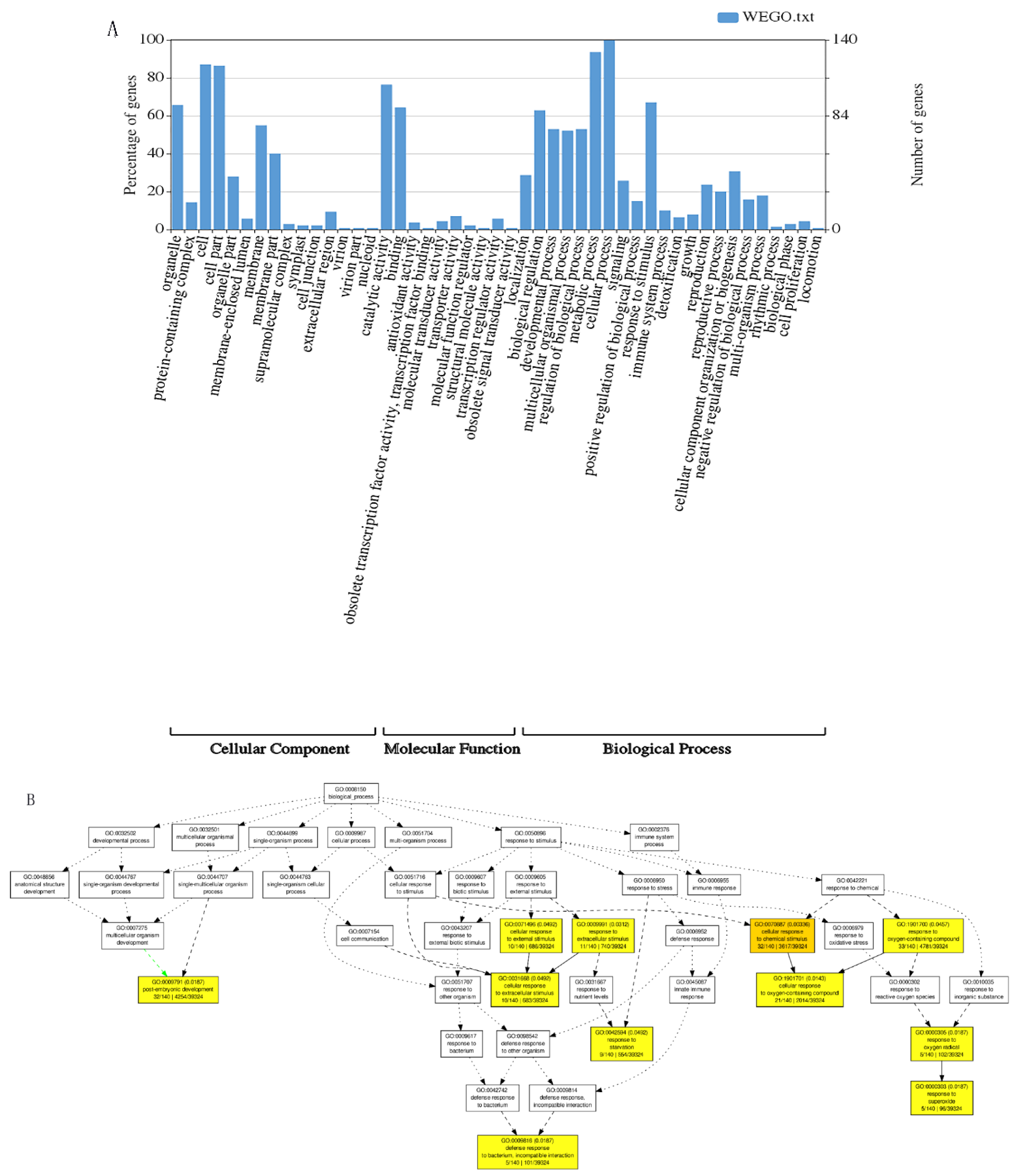
3.4. Tentative Candidate Genes for the Identified SNPs
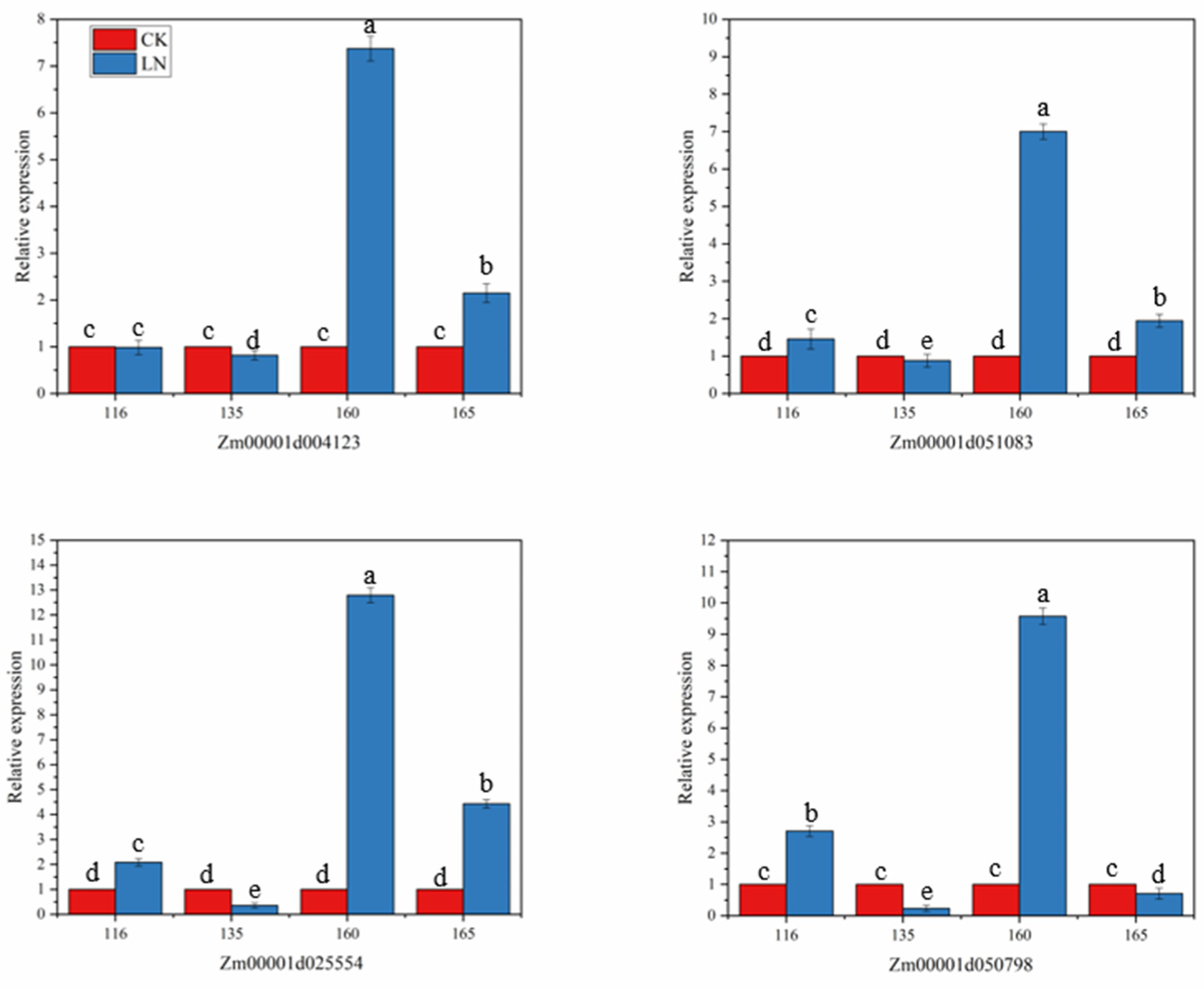
3.5. Candidate Gene Expression Results
4. Discussion
4.1. Phenotypic Evaluation of Traits Related to N Efficiency in Maize under Two N Levels
4.2. The Influence of Population Structure and Kinship on Association Analysis
4.3. Genome-Wide Association Studies for Different Traits under Two N Levels
4.4. Complex Molecular Mechanisms of Root and Shoot Growth under Different N Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, Y.L.; Wang, M.F.; Yang, B.; He, S.Y.; Duan, J.J.; Yang, L.Z.; Xue, L.H. Effects of Film Materials on Ammonia Volatilization Emissions from a Paddy System After Reducing N Fertilizer Application. Environ. Sci. 2021, 42, 477–484. [Google Scholar] [CrossRef]
- Liu, J.G.; You, L.Z.; Amini, M.; Obersteiner, M.; Herrero, M.; Zehnder, A.J.B.; Yang, H. A high-resolution assessment on global N flows in cropland. Proc. Natl. Acad. Sci. USA 2010, 107, 8035–8040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, K.H. Mining of Genetic Loci for N Use Efficiency in Maize Based on Genome-Wide Association Study. Master’s Thesis, Northwest A&F University, Xianyang, China, 2018. [Google Scholar]
- Yu, P.; Li, X.X.; White, P.J.; Li, C.J. A large and deep root system underlies high N-use efficiency in maize production. PLoS ONE 2015, 10, e0126293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.Y. Coupling Effect of Water and N on Maize Grown, N Use Effi-Ciency and Root Spatial Distribution of Spring Maize During the Seeding Stage. Master’s Thesis, Jilin Agricultural University, Changchun, China, 2017. [Google Scholar]
- Wang, Y.; Mi, G.H.; Chen, F.J.; Zhang, F.S. Genotypic difference in N efficiency of five maize inbred lines as affected by nitrate levels. Chin. J. Appl. Environ. Biol. 2002, 8, 361–365. [Google Scholar]
- Wang, H.M.; Wei, J.; Li, P.C.; Wang, Y.Y.; Ge, Z.Z.; Qian, J.Y.; Fan, Y.Y.; Ni, J.R.; Xu, Y.; Yang, Z.F.; et al. Integrating GWAS and gene expression analysis identifies candidate genes for root morphology traits in maize at the seedling stage. Genes 2019, 10, 773. [Google Scholar] [CrossRef] [Green Version]
- He, K.H.; Xu, S.T.; Zhang, X.H.; Li, Y.N.; Chang, L.G.; Wang, Y.H.; Shi, Y.Q.; Cui, T.T.; Dong, Y.; Lan, T.R.; et al. Mining of candidate genes for nitrogen use efficiency in maize based on genome-wide association study. Mol. Breed. 2020, 40, 83. [Google Scholar] [CrossRef]
- Sun, X.C.; Ren, W.; Wang, P.; Chen, F.J.; Yuan, L.X.; Pan, Q.H.; Mi, G.H. Evaluation of maize root growth and genome-wide association studies of root traits in response to low N supply at seedling emergence. Crop J. 2020, 9, 794–804. [Google Scholar] [CrossRef]
- Morosini, J.S.; Mendonca, L.D.; Lyra, D.H.; Galli, G.; Vidotti, M.S.; Fritsche-Neto, R. Association mapping for traits related to nitrogen use efficiency in tropical maize lines under field conditions. Plant Soil 2017, 421, 453–463. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Wang, H.R.; Jiang, Z.M.; Wang, W.; Xu, R.N.; Wang, Q.H.; Zhang, Z.H.; Li, A.F.; Liang, Y.; Ou, S.J.; et al. Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 2021, 590, 600–605. [Google Scholar] [CrossRef]
- Lou, G.M.; Chen, P.L.; Hou, H.Z.; Li, P.B.; Xiong, J.W.; Wan, S.S.; Zheng, Y.Y.; Alam, M.; Liu, R.J.; Zhou, H.Y.; et al. FLOURY ENDOSPERM19 encoding a class I glutamine amidotransferase affects grain quality in rice. Mol. Breed. 2021, 41, 36. [Google Scholar] [CrossRef]
- Li, T.; Qu, J.Z.; Wang, Y.H.; Chang, L.G.; He, K.H.; Guo, D.W.; Zhang, X.H.; Xu, S.T.; Xue, J.Q. Genetic characterization of inbred lines from Shaan A and B groups for identifying loci associated with maize grain yield. BMC Genet. 2018, 19, 63. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.X.; He, K.H.; Feng, Z.Q.; Li, Y.N.; Chang, L.G.; Zhang, X.H.; Xu, S.T.; Liu, J.C.; Xue, J.Q. Evaluation of Yield-Based Low Nitrogen Tolerance Indices for Screening Maize (Zea mays L.) Inbred Lines. Agronomy 2019, 9, 240. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, X.; Wang, J.; Li, M.; Wang, Q.; Tian, F.; Su, Z.; Pan, Y.; Liu, D.; Lipka, A.E.; et al. GAPIT Version 2: An Enhanced Integrated Tool for Genomic Association and Prediction. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Peng, Z.Y.; Yang, X.H.; Wang, W.D.; Fu, J.J.; Wang, J.H.; Han, Y.J.; Chai, Y.C.; Guo, T.T.; Yang, N.; et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef]
- Li, Q.; Luo, Y.H.; Long, W.J.; Kong, F.L.; Yang, S.M.; Yuan, J.C. Effect of low nitrogen stress on different low nitrogen tolerance maize cultivars seedling stage growth and physiological characteristics. Acta Pratacult. Sin. 2014, 23, 204–212. [Google Scholar]
- Yang, G.M.; Yang, E.Q.; Qian, X.G.; Tian, R.W. Discussion on the critical Period of Screening Genotypes with Tolerant to Low Nitrogen level in maize. Guizhou Agric. Sci. 2008, 27–30. [Google Scholar]
- Chen, F.J.; Mi, G.H.; Liu, X.S.; Liu, J.A.; Wang, Y.; Zhang, F.S. Combination ability analysis of traits related to nitrogen use efficiency in maize. Sci. Agric. Sin. 2003, 134–139. [Google Scholar]
- Kant, S.; Bi, Y.M.; Rothstein, S.J. Understanding plant response to N limitation for the improvement of crop N use efficiency. J. Exp. Bot. 2011, 62, 1499–1509. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.S.; Wang, X.R.; Wang, J.G. Biological approaches to improve the utilization efficiency of crop nutrient resources. J. China Agric. Univ. 1995, 104–110. [Google Scholar]
- Li, P.C.; Chen, F.J.; Cai, H.G.; Liu, J.C.; Pan, Q.C.; Liu, Z.G.; Gu, R.L.; Mi, G.H.; Zhang, F.S.; Yuan, L.X. A genetic relationship between N use efficiency and seedling root traits in maize as revealed by QTL analysis. J. Exp. Bot. 2015, 66, 3175–3188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postma, J.A.; Dathe, A.; Lynch, J.P. The Optimal Lateral Root Branching Density for Maize Depends on N and Phosphorus Availability. Plant Physiol. 2014, 166, 590–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trachsel, S.; Kaeppler, S.M.; Brown, K.M.; Lynch, J.P. Maize root growth angles become steeper under low N conditions. Field Crop. Res. 2013, 140, 18–31. [Google Scholar] [CrossRef]
- Li, Q.; Ren, Y.; Zou, Y.; Liu, J.; Yuan, J.C. Differences in Nitrogen Metabolism and Dry Matter Production between Maize Varieties and Different Nitrogen Efficiencies under Low Nitrogen Stress. Acta Agric. Boreali-Occident. Sin. 2021, 30, 1–9. Available online: http://kns.cnki.net/kcms/detail/detail.aspx?FileName=XBNX202105007&DbName=CJFQ2021 (accessed on 1 September 2021).
- Song, H.; Zhou, W.X.; Yuan, L.Z.; Jin, Y.J.; Li, H.P.; Yang, Y.; You, D.L.; Li, C.H. Effects of Light, Nitrogen and Their Interactions on Nitrogen Absorption, Utilization and Matter Production of Maize. Acta Agron. Sin. 2016, 42, 1844–1852. Available online: http://kns.cnki.net/kcms/detail/detail.aspx?FileName=XBZW201612014&DbName=CJFQ2016 (accessed on 1 September 2021). [CrossRef]
- Thornsberry, J.M.; Goodman, M.M.; Doebley, J.; Kresovich, S.; Nielsen, D.; Buckler, E.S. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 2001, 28, 286–289. [Google Scholar] [CrossRef]
- Liu, L.H.; Wang, L.X.; Zhao, C.P.; Yao, J.; Zhang, F.T.; Zhang, H.; Ye, Z.J.; Qin, Z.L.; Zheng, Y.L. Genetic diversity and alterations of population structure in restorers of dual cross-line hybrid wheat with thermo-photoperiod sensitive male sterile. Chin. J. Biochem. Mol. Biol. 2009, 25, 867–875. [Google Scholar] [CrossRef]
- Wu, Y.S. Association Analysis of Glutamine Synthetase Genes, Glu1-3 and Glu1-4, with N Use Efficiency in Maize. Ph.D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2009. [Google Scholar]
- Gaut, B.S.; Long, A.D. The lowdown on linkage disequilibrium. Plant Cell 2003, 15, 1502–1506. [Google Scholar] [CrossRef] [Green Version]
- Stich, B.; Melchinger, A.E.; Frisch, M.; Maurer, H.P.; Heckenberger, M.; Reif, J.C. Linkage disequilibrium in european elite maize germplasm investigated with SSRs. Theor. Appl. Genet. 2005, 111, 723–730. [Google Scholar] [CrossRef]
- Yang, X.H.; Gao, S.B.; Xu, S.T.; Zhang, Z.X.; Prasanna, B.M.; Li, L.; Li, J.S.; Yan, J.B. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol. Breed. 2011, 28, 511–526. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, H.S.; Fu, Z.J.; Chen, H.; Lin, Y.N.; Yan, P.S.; Li, W.H.; Xie, H.L.; Guo, Z.Y.; Zhang, X.H.; et al. Genetic-based dissection of arsenic accumulation in maize using a genome-wide association analysis method. Plant Biotechnol. J. 2018, 16, 1085–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Tang, Z.; Wei, J.; Qu, H.; Xie, Y.; Xu, G. The OsAMT1.1 gene functions in ammonium uptake and ammoniumepotassium homeostasis over low and high ammonium concentration ranges. J. Genet. Genom. 2016, 43, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.C.; Cai, H.G.; Chu, Q.; Chen, X.H.; Chen, F.J.; Yuan, L.X.; Mi, G.H.; Zhang, F.S. Genetic analysis of vertical root pulling resistance (VRPR) in maize using two genetic populations. Mol. Breed. 2011, 28, 463–474. [Google Scholar] [CrossRef]
- Luo, B.W.; Tang, H.T.; Liu, H.L.; Su, S.Z.; Zhang, S.Z.; Wu, L.; Liu, D.; Gao, S.B. Mining for low-N tolerance genes by integrating meta-analysis and large-scale gene expression data from maize. Euphytica 2015, 206, 117–131. [Google Scholar] [CrossRef]
- Gallais, A.; Hirel, B. An approach to the genetics of N use efficiency in maize. J. Exp. Bot. 2004, 55, 295–306. [Google Scholar] [CrossRef] [Green Version]
- Tuberosa, R.; Sanguineti, M.C.; Landi, P.; Giuliani, M.M.; Salvi, S.; Conti, S. Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the field at two water regimes. Plant Mol. Biol. 2002, 48, 697–712. [Google Scholar] [CrossRef]
- Zhu, J.M.; Mickelson, S.M.; Kaeppler, S.M.; Lynch, J.P. Detection of quantitative trait loci for seminal root traits in maize (Zea mays L.) seedlings grown under differential phosphorus levels. Theor. Appl. Genet. 2006, 113, 1–10. [Google Scholar] [CrossRef]
- Cai, H.G.; Chen, F.J.; Mi, G.H.; Zhang, F.S.; Maurer, H.P.; Liu, W.X.; Reif, J.C.; Yuan, L.X. Mapping QTLs for root system architecture of maize (Zea mays L.) in the field at different developmental stages. Theor. Appl. Genet. 2012, 125, 1313–1324. [Google Scholar] [CrossRef]
- Liu, Z.G.; Gao, K.; Shan, S.; Gu, R.C.; Wang, Z.K.; Craft, E.J.; Mi, G.H.; Yuan, L.X.; Chen, F.J. Comparative Analysis of Root Traits and the Associated QTLs for Maize Seedlings Grown in Paper Roll, Hydroponics and Vermiculite Culture System. Front. Plant Sci. 2017, 8, 436. [Google Scholar] [CrossRef] [Green Version]
- Bertin, P.; Gallais, A. Genetic variation for nitrogen use efficiency in a set of recombinant inbred lines: II. QTL detection and coincidences. Maydica 2001, 46, 53–68. [Google Scholar]
- Hirel, B.; Bertin, P.; Quillere, I.; Bourdoncle, W.; Attagnant, C.; Dellay, C.; Gouy, A.; Cadiou, S.; Retailliau, C.; Falque, M.; et al. Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol. 2001, 125, 1258–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurino, V.G.; Engqvist, M.K. 2-Hydroxy Acids in Plant Metabolism. Arab. Book 2015, 13, e0182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhowal, B.; Singla-Pareek, S.L.; Sopory, S.K.; Kaur, C. From methylglyoxal to pyruvate: A genome-wide study for the identification of glyoxalases and D-lactate dehydrogenases in Sorghum bicolor. BMC Genom. 2020, 21, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostofa, M.G.; Fujita, M. Salicylic acid alleviates copper toxicity in rice (Oryza sativa L.) seedlings by up-regulating antioxidative and glyoxalase systems. Ecotoxicology 2013, 22, 959–973. [Google Scholar] [CrossRef]
- Alam, P.; Albalawi, T.H.; Altalayan, F.H.; Bakht, M.A.; Ahanger, M.A.; Raja, V.; Ashraf, M.; Ahmad, P. 24-Epibrassinolide (EBR) confers tolerance against NaCl stress in soybean plants by up-regulating antioxidant system, ascorbate-glutathione cycle, and glyoxalase system. Biomolecules 2019, 9, 640. [Google Scholar] [CrossRef] [Green Version]
- Jan, S.; Noman, A.; Kaya, C.; Ashraf, M.; Alyemeni, M.N.; Ahmad, P. 24-Epibrassinolide alleviates the injurious effects of cr(vi) toxicity in tomato plants: Insights into growth, physio-biochemical attributes, antioxidant activity and regulation of ascorbate-glutathione and glyoxalase cycles. J. Plant Growth Regul. 2020, 39, 1587–1604. [Google Scholar] [CrossRef]
- Nishiyama, R.; Le, D.T.; Watanabe, Y.; Matsui, A.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS ONE 2012, 7, e32124. [Google Scholar] [CrossRef] [Green Version]
- Singla-Pareek, S.L.; Reddy, M.K.; Sopory, S.K. Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. USA 2003, 100, 14672–14677. [Google Scholar] [CrossRef] [Green Version]
- Gan, Y.B.; Bernreiter, A.; Filleur, S.; Abram, B.; Forde, B.G. Overexpressing the ANR1 MADS-Box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 2012, 53, 1003–1016. [Google Scholar] [CrossRef] [Green Version]
- El-Shabrawi, H.; Kumar, B.; Kaul, T.; Reddy, M.K.; Singla-Pareek, S.L.; Sopory, S.K. Redox homeostasis, antioxidant defense, and methylglyoxal detoxification as markers for salt tolerance in Pokkali rice. Protoplasma 2010, 245, 85–96. [Google Scholar] [CrossRef]
- Jia, H.T.; Li, M.F.; Li, W.Y.; Liu, L.; Jian, Y.A.; Yang, Z.X.; Shen, X.M.; Ning, Q.; Du, Y.F.; Zhao, R.; et al. A serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 regulates maize grain yield. Nat. Commun. 2020, 11, 988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.L.; Yu, Q.Y.; Tang, L.L.; Ji, W.; Bai, X.; Cai, H.; Liu, X.F.; Ding, X.D.; Zhu, Y.M. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J. Plant Physiol. 2013, 170, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Azad, I.; Alemzadeh, A. Bioinformatic and empirical analysis of a gene encoding serine/threonine protein kinase regulated in response to chemical and biological fertilizers in two maize (Zea mays L.) cultivars. Mol. Biol. Res. Commun. 2017, 6, 65–75. [Google Scholar] [PubMed]
- Xia, Y.J.; Suzuki, H.; Borevitz, J.; Blount, J.; Guo, Z.J.; Patel, K.; Dixon, R.A.; Lamb, C. An extracellular aspartic protease functions in arabidopsis disease resistance signaling. EMBO J. 2004, 23, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, G.R.; Huang, F.L.; Chen, Y.S. Gene cloning, expression and purification of Ricinus communis L. aspartic proteases. Chin. J. Biol. 2020, 33, 1122–1127. [Google Scholar] [CrossRef]
- Wang, Y.R.; Wu, Y. The research progress on the functions of plant aspartic proteases. Chin. Bull. Life Sci. 2016, 28, 384–390. [Google Scholar] [CrossRef]
- Mochizuki, S.; Jikumaru, Y.; Nakamura, H.; Koiwai, H.; Sasaki, K.; Kamiya, Y.; Ichikawa, H.; Minami, E.; Nishizawa, Y. Ubiquitin ligase EL5 maintains the viability of root meristems by influencing cytokinin-mediated N effects in rice. J. Exp. Bot. 2014, 65, 2307–2318. [Google Scholar] [CrossRef]
- Luo, X. Functional Verification of Maize Salt-Tolerant Candidate Genes and Establishment of Salt-Tolerant RILs. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2020. [Google Scholar]


| Trait Name | Abbreviations | Trait Measurements and Descriptions |
|---|---|---|
| SPAD value of the 2nd leaf | SPAD | Measured with a chlorophyll meter |
| Plant height (cm) | PH | Measured with a ruler |
| Shoot dry weight (g) | SDW | Dried and weighed on a balance (0.001 g) |
| Root dry weight (g) | RDW | Dried and weighed on a balance (0.001 g) |
| Root-to-shoot ratio | RSR | Root dry weight/shoot dry weight |
| Total root length (cm) | TRL | Length of the whole root system |
| Total root surface area (cm2) | TRSA | Surface area of the whole root system |
| Total root volume (cm3) | TRV | Volume of the whole root system |
| Crown root number | CRN | Count of the axial first whorl crown roots |
| Crown root length (cm) | CRL | Measured with a ruler |
| Crown root surface area (cm2) | CRSA | Measured with root scanner |
| Crown root volume (cm3) | CRV | Measured with root scanner |
| Seminal root number | SRN | Count of the axial seminal roots |
| Seminal root length (cm) | SRL | Measured with a ruler |
| Seminal root surface area (cm2) | SRSA | Measured with root scanner |
| Seminal root volume (cm3) | SRV | Measured with root scanner |
| Primary root length (cm) | PRL | Measured with a ruler |
| Primary root surface area (cm2) | PRSA | Surface area of the whole primary root |
| Primary root volume (cm3) | PRV | Volume of the whole primary root |
| Total lateral root length | TLRL | The total length of all of the lateral roots of the entire root system |
| Traits | Env | Total | |
|---|---|---|---|
| CK | LN | ||
| SPAD | 7 | 2 | 9 |
| PH | 9 | 2 | 11 |
| SDW | 6 | 1 | 7 |
| RDW | 2 | 4 | 6 |
| RSR | 0 | 9 | 9 |
| TRL | 1 | 7 | 8 |
| TRSA | 1 | 2 | 3 |
| TRV | 2 | 1 | 3 |
| CRN | 14 | 1 | 15 |
| CRL | 3 | 6 | 9 |
| CRSA | 3 | 2 | 5 |
| CRV | 6 | 3 | 9 |
| SRN | 3 | 1 | 4 |
| SRL | 2 | 4 | 6 |
| SRSA | 4 | 3 | 7 |
| SRV | 1 | 4 | 5 |
| PRL | 10 | 0 | 10 |
| PRSA | 5 | 3 | 8 |
| PRV | 11 | 13 | 24 |
| TLRL | 11 | 16 | 27 |
| Total | 101 | 84 | 185 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Y.; Liu, J.; Xia, Z.; Wang, Q.; Zhang, S.; Zhang, G.; Lu, H. Genome-Wide Association Studies of Maize Seedling Root Traits under Different Nitrogen Levels. Plants 2022, 11, 1417. https://doi.org/10.3390/plants11111417
Fu Y, Liu J, Xia Z, Wang Q, Zhang S, Zhang G, Lu H. Genome-Wide Association Studies of Maize Seedling Root Traits under Different Nitrogen Levels. Plants. 2022; 11(11):1417. https://doi.org/10.3390/plants11111417
Chicago/Turabian StyleFu, Yafang, Jianchao Liu, Zhenqing Xia, Qi Wang, Shibo Zhang, Guixin Zhang, and Haidong Lu. 2022. "Genome-Wide Association Studies of Maize Seedling Root Traits under Different Nitrogen Levels" Plants 11, no. 11: 1417. https://doi.org/10.3390/plants11111417
APA StyleFu, Y., Liu, J., Xia, Z., Wang, Q., Zhang, S., Zhang, G., & Lu, H. (2022). Genome-Wide Association Studies of Maize Seedling Root Traits under Different Nitrogen Levels. Plants, 11(11), 1417. https://doi.org/10.3390/plants11111417






