QTL Mapping for Resistance to Bacterial Wilt Caused by Two Isolates of Ralstonia solanacearum in Chili Pepper (Capsicum annuum L.)
Abstract
:1. Introduction
2. Results
2.1. Bioassay of Bacterial Wilt Resistance Using Three Different Isolates of Ralstonia solanacearum
2.2. GBS Analysis of Two F2 Populations Derived from the Cultivar ‘Konesian Hot’ with Strong Resistance to Bacterial Wilt
2.3. Selection of SNPs Identified by GBS Analysis in Two F2 Populations
2.4. Genetic Linkage Mapping of Pepper in Two F2 Populations
2.5. Identification of QTLs for Bacterial Wilt Resistance in Two F2 Populations
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Pathogen Materials and Inoculum Preparation
4.3. Evaluation of Bacterial Wilt Resistance
4.4. DNA Extraction
4.5. GBS Analysis
4.6. Genetic Linkage Map Construction
4.7. QTL Analysis
4.8. Primer Design for HRM Analysis
4.9. PCR and HRM Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AFLP | amplified fragment length polymorphism |
| CIM | composite interval mapping |
| DI | disease index |
| GBS | genotyping-by-sequencing |
| HRM | high-resolution melting |
| LOD | logarithm of the odds |
| MAS | marker-assisted selection |
| NGS | next-generation sequencing |
| QTL | quantitative trait loci |
| QTL | quantitative trait locus |
| RSSC | Ralstonia solanacearum species complex |
| SNP | single-nucleotide polymorphism |
| SSR | simple sequence repeat |
References
- Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/statistics/en/ (accessed on 8 April 2022).
- Korean Statistical Information Service. Available online: https://kosis.kr/index/index.do/ (accessed on 8 April 2022).
- Hwang, S.M.; Jang, K.S.; Choi, Y.H.; Kim, H.; Choi, G.J. Development of an efficient bioassay method to evaluate resistance of chili pepper cultivars to Ralstonia solanacearum. Res. Plant Dis. 2017, 23, 334–347. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J.; Oh, D.G. Resistance of pepper cultivars to Ralstonia solanacearum isolates from major cultivated areas of chili peppers in Korea. Hortic. Sci. Technol. 2018, 36, 569–576. [Google Scholar]
- Pilet-Nayel, M.L.; Moury, B.; Caffier, V.; Montarry, J.; Kerlan, M.C.; Fournet, S.; Durel, C.E.; Delourme, R. Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Front. Plant Sci. 2017, 8, 1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The List of Plant Diseases in Korea. Available online: http://genebank.rda.go.kr/english/kplantdisease.do/ (accessed on 8 April 2022).
- Kim, J.T.; Kim, S.D. Suppression of bacterial wilt with Bacillus subtilis SKU48-2 strain. Korean J. Microbiol. Biotechnol. 2008, 36, 115–120. [Google Scholar]
- Tran, N.H.; Kim, B.S. Influence of temperature, pathogen strain, inoculum density, seedling age, inoculation method and varietal resistance on infection of pepper seedlings by Ralstonia solanacearum. Hortic. Environ. Biotechnol. 2010, 51, 95–100. [Google Scholar]
- Hayward, A.C. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 1991, 29, 65–87. [Google Scholar] [CrossRef]
- Smith, E.F. A bacterial disease of the tomato, eggplant and Irish potato (Bacillus solanacearum n. sp.). Z. Pflanzenkrankh. 1897, 7, 230–234. [Google Scholar]
- Yabuuchi, E.; Kosako, Y.; Yano, I.; Hotta, H.; Nishiuchi, Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia Gen. Nov.: Proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) Comb. Nov., Ralstonia solanacearum (Smith 1896) Comb. Nov. and Ralstonia eutropha (Davis 1969) Comb. Nov. Microbiol. Immunol. 1995, 39, 897–904. [Google Scholar] [CrossRef]
- Graham, J.; Lloyd, A.B. An improved indicator plant method for the detection of Pseudomonas solanacearum race 3 in soil [Potatoes, bacterial wilt]. Plant Dis. Rep. 1978, 62, 35–37. [Google Scholar]
- Han, Y.K.; Min, J.S.; Park, J.H.; Han, K.S.; Kin, D.H.; Lee, J.S.; Kin, H.H. Screening of tomato cultivars resistant to bacterial wilts. Res. Plant Dis. 2009, 15, 198–201. [Google Scholar] [CrossRef] [Green Version]
- Wallis, F.M.; Truter, S.J. Histopathology of tomato plants infected with Pseudomonas solanacearum, with emphasis on ultrastructure. Physiol. Plant Pathol. 1978, 13, 307–317. [Google Scholar] [CrossRef]
- Saile, E.; Mc Garvey, J.A.; Schell, M.A.; Denny, T.P. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 1997, 87, 1264–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araud-Razou, I.; Vasse, J.; Montrozier, H.; Etchebar, C.; Trigalet, A. Detection and visualization of the major acidic exopolysaccharide of Ralstonia solanacearum and its role in tomato root infection and vascular colonization. Eur. J. Plant Pathol. 1998, 104, 795–809. [Google Scholar] [CrossRef]
- Genin, S. Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 2010, 187, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.J.; Joo, H.J.; Choi, S.Y.; Lee, S.Y.; Jung, Y.H.; Lee, M.H.; Kong, H.G.; Lee, S.W. Resistance evaluation of tomato germplasm against bacterial wilt by Ralstonia solanacearum. Res. Plant Dis. 2014, 20, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Meng, F.H. The virulence factors of the bacterial wilt pathogen Ralstonia solanacearum. J. Plant Pathol. Microbiol. 2013, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Shen, F.; Yin, W.; Song, S.; Zhang, Z.; Ye, P.; Zhang, Y.; Zhou, J.; He, F.; Li, P.; Deng, Y. Ralstonia solanacearum promotes pathogenicity by utilizing L-glutamic acid from host plants. Mol. Plant Pathol. 2020, 21, 1099–1110. [Google Scholar] [CrossRef]
- Genin, S.; Boucher, C. Lessons learned from the genome analysis of Ralstonia solanacearum. Annu. Rev. Phytopathol. 2004, 42, 107–134. [Google Scholar] [CrossRef]
- Angot, A.; Peeters, N.; Lechner, E.; Vailleau, F.; Baud, C.; Gentzbittel, L.; Sartorel, E.; Genschik, P.; Boucher, C.; Genin, S. Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. USA 2006, 103, 14620–14625. [Google Scholar] [CrossRef] [Green Version]
- Valls, M.; Genin, S.; Boucher, C. Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLoS Pathog. 2006, 2, e82. [Google Scholar] [CrossRef] [Green Version]
- Remigi, P.; Anisimova, M.; Guidot, A.; Genin, S.; Peeters, N. Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 2011, 192, 976–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillings, M.R.; Fahy, P. Genomic Fingerprinting: Towards a Unified View of the Pseudomonas solanacearum Species Complex. In Bacterial Wilt: The Disease and Its Causative Agent, Pseudomonas solanacearum; Hayward, A.C., Hartman, G.L., Eds.; Wallingford CABI: Egham, UK, 1994; pp. 95–112. [Google Scholar]
- Fegan, M.; Prior, P. How Complex is the ‘Ralstonia solanacearum Species Complex’. In Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex; Allen, C., Prior, P., Hayward, A.C., Eds.; APS Press: St. Paul, MN, USA, 2005; pp. 449–462. [Google Scholar]
- Lebeau, A.; Daunay, M.C.; Frary, A.; Palloix, A.; Wang, J.F.; Dintinger, J.; Chirole, F.; Wicker, E.; Prior, P. Bacterial wilt resistance in tomato, pepper, and eggplant: Genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology 2011, 101, 154–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- She, X.; Yu, L.; Lan, G.; Tang, Y.; He, Z. Identification and genetic characterization of Ralstonia solanacearum species complex isolates from Cucurbita maxima in China. Front. Plant Sci. 2017, 8, 1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buddenhagen, I.; Sequeira, L.; Kelman, A. Designation of races in Pseudomonas solanacearum. Phytopathology 1962, 52, 726. [Google Scholar]
- Pegg, K.G.; Moffett, M.L. Host range of the ginger strain of Pseudomonas solanacearum in Queensland. Aust. J. Exp. Agric. Anim. Husb. 1971, 11, 696–698. [Google Scholar] [CrossRef]
- Santana, B.G.; Lopes, C.A.; Alvarez, E.; Barreto, C.C.; Allen, C.; Quirino, B.F. Diversity of Brazilian biovar 2 strains of Ralstonia solanacearum. J. Gen. Plant Pathol. 2012, 78, 190–200. [Google Scholar] [CrossRef]
- Gutarra, L.; Herrera, J.; Fernandez, E.; Kreuze, J.; Lindqvist-Kreuze, H. Diversity, pathogenicity, and current occurrence of bacterial wilt bacterium Ralstonia solanacearum in Peru. Front. Plant Sci. 2017, 8, 1221. [Google Scholar] [CrossRef] [Green Version]
- Wicker, E.; Lefeuvre, P.; de Cambiaire, J.C.; Lemaire, C.; Poussier, S.; Prior, P. Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. ISME J. 2012, 6, 961–974. [Google Scholar] [CrossRef] [Green Version]
- Santiago, T.R.; Lopes, C.A.; Caetano-Anollés, G.; Mizubuti, E.S.G. Phylotype and sequevar variability of Ralstonia solanacearum in Brazil, an ancient center of diversity of the pathogen. Plant Pathol. 2017, 66, 383–392. [Google Scholar] [CrossRef]
- Lee, Y.K.; Kang, H.W. Physiological, biochemical and genetic characteristics of Ralstonia solanacearum strains isolated from pepper plants in Korea. Res. Plant Dis. 2013, 19, 265–272. [Google Scholar] [CrossRef] [Green Version]
- Matos, F.S.A.; Lopes, C.A.; Takatsu, A. Identification of sources of resistance to Pseudomonas solanacearum in Capsicum spp. Hortic. Bras. 1990, 8, 22–23. [Google Scholar]
- Kim, B.S.; Cheung, J.D.; Cha, Y.S.; Hwang, H.S. Resistance to bacterial wilt of introduced peppers. Korean J. Plant Pathol. 1998, 14, 217–219. [Google Scholar]
- Fang, S.; Gu, G.; Chen, Y.; Huang, C.; Chen, S. Colonization and infection of Ralstonia solanacearum in weed roots. Acta Tab. Sin. 2013, 19, 72–81. [Google Scholar]
- Jiang, G.; Wei, Z.; Xu, J.; Chen, H.; Zhang, Y.; She, X.; Macho, A.P.; Ding, W.; Liao, B. Bacterial wilt in China: History, current status, and future perspectives. Front. Plant Sci. 2017, 8, 1549. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Seo, H.J. Antifungal activity of agro-materials against pear scab (Venturia nashicola) and pear rust (Gymnosporangium asiaticum) fungi. Res. Plant Dis. 2018, 24, 33–40. [Google Scholar] [CrossRef]
- Huet, G. Breeding for resistances to Ralstonia solanacearum. Front. Plant Sci. 2014, 5, 715. [Google Scholar] [CrossRef] [Green Version]
- Collard, B.C.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. Ser. B 2008, 363, 557–572. [Google Scholar] [CrossRef] [Green Version]
- Ganal, M.W.; Altmann, T.; Röder, M.S. SNP identification in crop plants. Curr. Opin. Plant Biol. 2009, 12, 211–217. [Google Scholar] [CrossRef]
- Mimura, Y.; Kageyama, T.; Minamiyama, Y.; Hirai, M. QTL analysis for resistance to Ralstonia solanacearum in Capsicum accession ‘LS2341’. J. Jpn. Soc. Hortic. Sci. 2009, 78, 307–313. [Google Scholar] [CrossRef] [Green Version]
- Tran, N.H.; Kim, B.S. Inheritance of resistance to bacterial wilt (Ralstonia solanacearum) in pepper (Capsicum annuum L.). Hortic. Environ. Biotechnol. 2010, 51, 431–439. [Google Scholar]
- Du, H.; Wen, C.; Zhang, X.; Xu, X.; Yang, J.; Chen, B.; Geng, S. Identification of a major QTL (qRRs-10.1) that confers resistance to Ralstonia solanacearum in pepper (Capsicum annuum L.) using SLAF-BSA and QTL mapping. Int. J. Mol. Sci. 2019, 20, 5887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, S.Y.; Lee, K.; Do, J.W.; Hong, S.C.; Lee, K.H.; Cho, M.C.; Yang, E.Y.; Yoon, J.B. QTL Mapping of resistance to bacterial wilt in pepper plants (Capsicum annuum) using genotyping-by-sequencing (GBS). Horticulturae 2022, 8, 115. [Google Scholar] [CrossRef]
- Kumar, S.; Banks, T.W.; Cloutier, S. SNP discovery through next-generation sequencing and its applications. Int. J. Plant Genom. 2012, 2012, 831460. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [Green Version]
- Beissinger, T.M.; Hirsch, C.N.; Sekhon, R.S.; Foerster, J.M.; Johnson, J.M.; Muttoni, G.; Vaillancourt, B.; Buell, C.R.; Kaeppler, S.M.; de Leon, N. Marker density and read depth for genotyping populations using genotyping-by-sequencing. Genetics 2013, 193, 1073–1081. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.X.; Liu, H.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef] [Green Version]
- Barka, G.D.; Lee, J. Molecular marker development and gene cloning for diverse disease resistance in pepper (Capsicum annuum L.): Current status and prospects. Plant Breed. Biotechnol. 2020, 8, 89–113. [Google Scholar] [CrossRef]
- Lee, Y.R.; Lee, J. A genetic linkage map of allo-octoploid strawberry (Fragaria × ananassa Duch.) using SNP markers. Korean J. Breed. Sci. 2017, 49, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.E.; Oh, S.K.; Lee, J.H.; Lee, B.M.; Jo, S.H. Genome wide SNP calling using next generation sequencing data in tomato. Mol. Cells 2014, 37, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Kosambi, D.D. The estimation of map distance from recombination value. Ann. Eugen. 1943, 12, 172–175. [Google Scholar] [CrossRef]
- Voorrips, R.E. MAPCHART: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [Green Version]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Longman Group: London, UK, 1996; p. 463. [Google Scholar]
- Kearsey, M.J. The principles of QTL analysis (a minimal mathematics approach). J. Exp. Bot. 1998, 49, 1619–1623. [Google Scholar] [CrossRef]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits; Sinauer Associates, Inc.: Sunderland, MA, USA, 1998. [Google Scholar]
- Silva, L.D.C.E.; Wang, S.; Zeng, Z.B. Composite Interval Mapping and Multiple Interval Mapping: Procedures and Guidelines for Using Windows QTL Cartographer. In Quantitative Trait Loci (QTL); Humana Press: Totowa, NJ, USA, 2012; pp. 75–119. [Google Scholar]
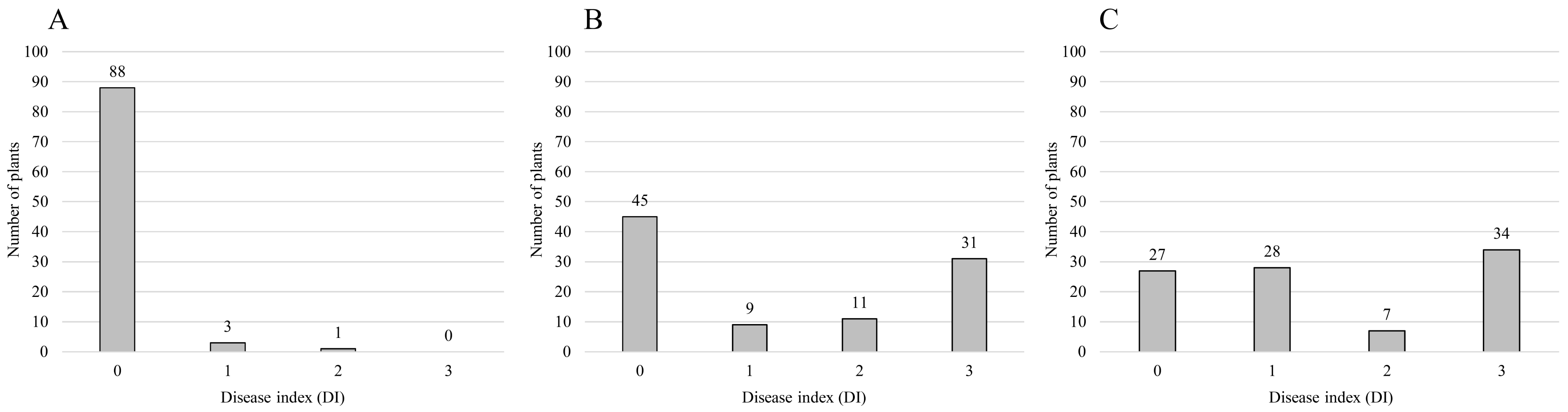

| Population | Isolate of Ralstonia solanacearumz | Number of Plants | Segregating Ratio | Chi-Square Value | Probability Value | ||
|---|---|---|---|---|---|---|---|
| Resistant (DIy = 0 or 1) | Susceptible (DI = 2 or 3) | Total | |||||
| F2 of ‘Konesian Hot’ | ‘IS’ | 91 | 1 | 92 | 15:1 | 4.186 | 0.041 |
| F2 of ‘Konesian Hot’ | ‘HS’ | 54 | 42 | 96 | 9:7 | 0 | 1 |
| F2 of ‘Konesian Hot’ | ‘HWA’ | 55 | 41 | 96 | 9:7 | 0.042 | 0.837 |
| GBS Statistics | ‘HS’-Inoculated Population | ‘HWA’-Inoculated Population |
|---|---|---|
| Number of F2 plants for multiplexing | 96 | 96 |
| Sequencing system | Illumina HiSeq 2500 | Illumina HiSeq X |
| Average length of raw reads (bp) | 101 (100%) | 151 (100%) |
| Total number of sequenced raw reads | 592,251,242 (100%) | 725,489,528 (100%) |
| Total length of sequenced raw data (bp) | 59,817,375,442 (100%) | 109,548,918,728 (100%) |
| Total number of demultiplexed reads | 579,885,814 (97.7%) | 630,875,996 (87.0%) |
| Average number of demultiplexed reads/sample | 6,040,477 | 6,571,625 |
| Total length of demultiplexed reads (bp) | 58,568,467,214 (97.9%) | 95,262,275,396 (87.0%) |
| Number of trimmed reads | 484,089,570 (81.7%) | 574,385,318 (79.2%) |
| Total length of trimmed reads (bp) | 40,076,781,039 (67.0%) | 65,375,963,492 (59.7%) |
| Average length of trimmed reads (bp) | 82.7 (81.9%) | 113.78 (75.4%) |
| Total length of pepper reference genome (Capsicum annuum cv. CM334 ver.1.55) (bp) | 2,753,501,687 | 2,753,501,687 |
| Number of mapped reads | 429,461,067 (72.5%) | 491,518,788 (67.7%) |
| Average number of mapped regions | 108,919 | 112,352 |
| Average depth of mapped region | 17.27 | 14.80 |
| Average length of mapped region (bp) | 144.32 | 266.14 |
| Reference genome coverage (%) | 0.5739 | 1.1020 |
| Chr.z No. | ‘HS’-Inoculated Population | ‘HWA’-Inoculated Population | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of Markers | Length of Linkage Distance (cM) | Average Marker Interval (cM/marker) | Number of Markers | Length of Linkage Distance (cM) | Average Marker Interval (cM/marker) | |||||
| GBSy- Based SNP | HRMx | Total | GBS-Based SNP | HRM | Total | |||||
| 1 | 95 | 0 | 95 | 308.8 | 3.25 | 107 | 0 | 107 | 184.1 | 1.72 |
| 2 | 90 | 2 | 92 | 184.9 | 2.01 | 100 | 1 | 101 | 172.8 | 1.71 |
| 3 | 125 | 4 | 129 | 267.0 | 2.07 | 145 | 2 | 147 | 246.0 | 1.67 |
| 4 | 87 | 0 | 87 | 185.7 | 2.13 | 70 | 0 | 70 | 195.1 | 2.79 |
| 5 | 61 | 4 | 65 | 185.6 | 2.86 | 108 | 6 | 114 | 167.6 | 1.47 |
| 6 | 91 | 0 | 91 | 207.0 | 2.27 | 96 | 0 | 96 | 204.1 | 2.13 |
| 7 | 103 | 14 | 117 | 168.9 | 1.44 | 92 | 9 | 101 | 183.3 | 1.81 |
| 8 | 78 | 7 | 85 | 179.1 | 2.11 | 81 | 3 | 84 | 160.1 | 1.91 |
| 9 | 60 | 15 | 75 | 155.1 | 2.07 | 80 | 4 | 84 | 193.2 | 2.30 |
| 10 | 65 | 3 | 68 | 229.5 | 3.38 | 99 | 2 | 101 | 203.4 | 2.01 |
| 11 | 124 | 0 | 124 | 154.8 | 1.25 | 148 | 0 | 148 | 193.2 | 1.31 |
| 12 | 90 | 2 | 92 | 206.5 | 2.24 | 78 | 4 | 82 | 211.7 | 2.58 |
| Total | 1069 | 51 | 1120 | 2432.8 | 2.17 | 1204 | 31 | 1235 | 2314.6 | 1.87 |
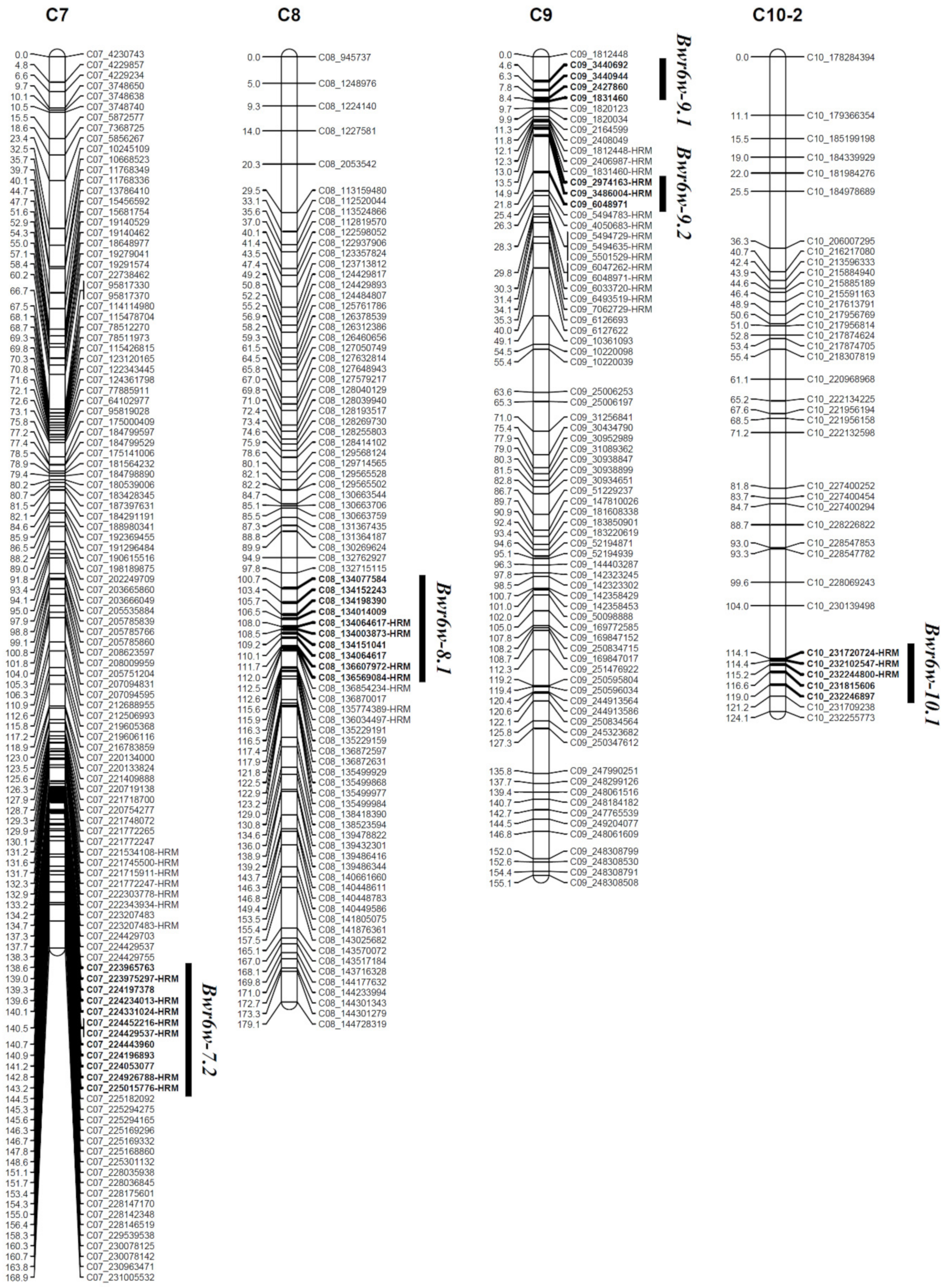
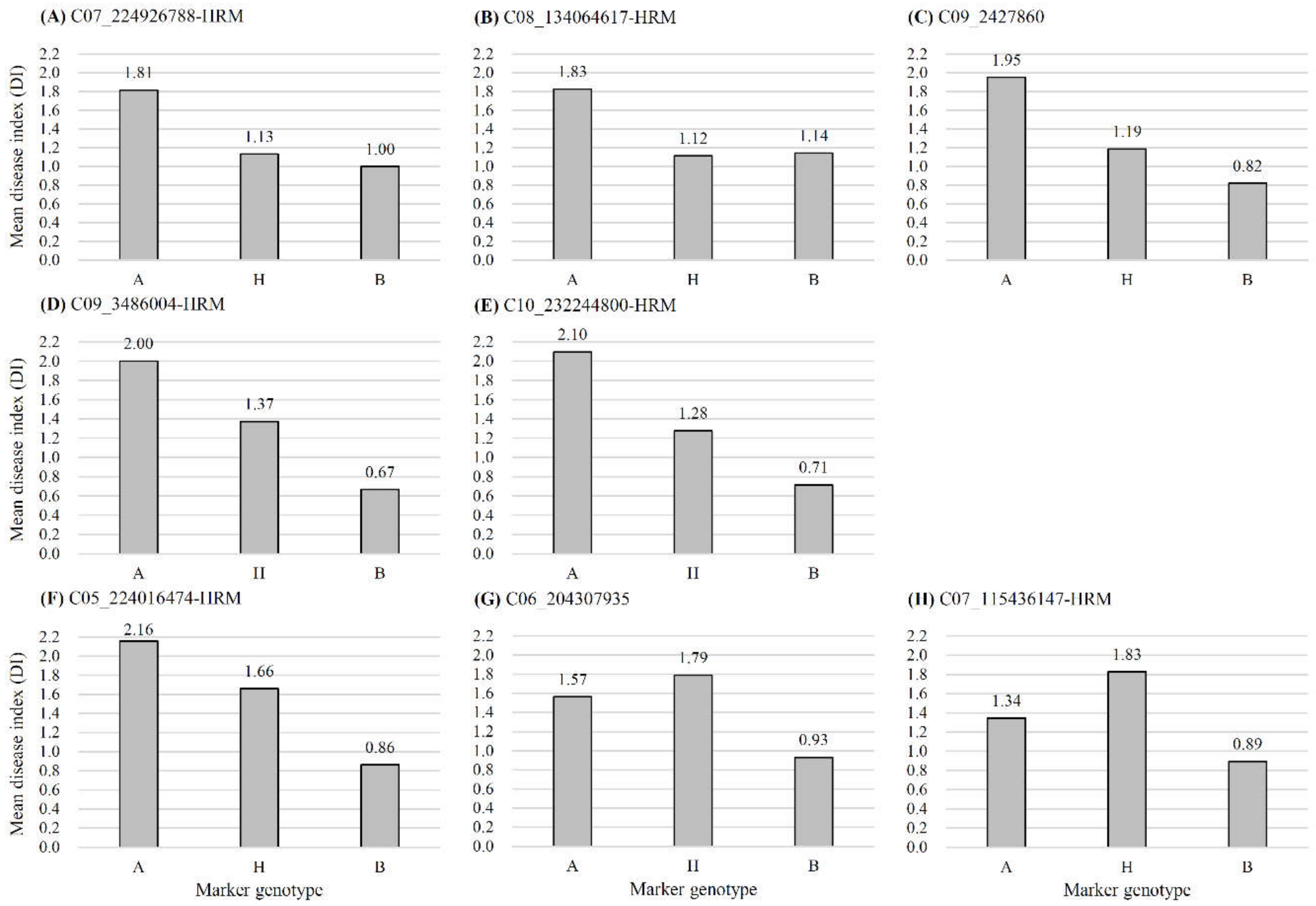
| Trait | QTL | Chr. | QTL Peak Position (cM) | Marker Interval | The Closest HRM Marker | R2 (%) | Additive | Dominant | LOD | LOD Threshold |
|---|---|---|---|---|---|---|---|---|---|---|
| ‘HS’ resistance | Bwr6w-7.2 | 7 | 141.2 | C07_223965763-C07_225015776-HRM | C07_224926788-HRM | 13.05 | 0.4251 | −0.5439 | 3.28 | 3.1 |
| Bwr6w-8.1 | 8 | 105.7 | C08_134077584-C08_136569084-HRM | C08_134064617-HRM | 12.67 | 0.3439 | −0.6937 | 3.28 | 3.1 | |
| Bwr6w-9.1 | 9 | 7.3 | C09_3440692-C09_1831460 | - | 15.07 | 0.6336 | −0.2142 | 3.53 | 3.1 | |
| Bwr6w-9.2 | 9 | 14.9 | C09_2974163-HRM-C09_6048971 | C09_3486004-HRM | 10.46 | 0.6555 | −0.0027 | 3.37 | 3.1 | |
| Bwr6w-10.1 | 10 | 117.7 | C10_231720724-HRM-C10_232246897 | C10_232244800-HRM | 9.69 | 0.6061 | −0.0337 | 3.12 | 3.1 | |
| ‘HWA’ resistance | Bwr6w-5.1 | 5 | 120.4 | C05_224044238-C05_224047094 | C05_224016474-HRM | 19.67 | 0.7266 | 0.0500 | 5.89 | 3.0 |
| Bwr6w-6.1 | 6 | 93.2 | C06_200031568-C06_203415605 | - | 16.50 | 0.4528 | 0.4926 | 3.78 | 3.0 | |
| Bwr6w-7.1 | 7 | 76.0 | C07_77877210-C07_78012481-HRM | C07_115436147-HRM | 12.56 | 0.3109 | 0.7013 | 3.27 | 3.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Chakma, N.; Joung, S.; Lee, J.M.; Lee, J. QTL Mapping for Resistance to Bacterial Wilt Caused by Two Isolates of Ralstonia solanacearum in Chili Pepper (Capsicum annuum L.). Plants 2022, 11, 1551. https://doi.org/10.3390/plants11121551
Lee S, Chakma N, Joung S, Lee JM, Lee J. QTL Mapping for Resistance to Bacterial Wilt Caused by Two Isolates of Ralstonia solanacearum in Chili Pepper (Capsicum annuum L.). Plants. 2022; 11(12):1551. https://doi.org/10.3390/plants11121551
Chicago/Turabian StyleLee, Saeyoung, Nidhi Chakma, Sunjeong Joung, Je Min Lee, and Jundae Lee. 2022. "QTL Mapping for Resistance to Bacterial Wilt Caused by Two Isolates of Ralstonia solanacearum in Chili Pepper (Capsicum annuum L.)" Plants 11, no. 12: 1551. https://doi.org/10.3390/plants11121551
APA StyleLee, S., Chakma, N., Joung, S., Lee, J. M., & Lee, J. (2022). QTL Mapping for Resistance to Bacterial Wilt Caused by Two Isolates of Ralstonia solanacearum in Chili Pepper (Capsicum annuum L.). Plants, 11(12), 1551. https://doi.org/10.3390/plants11121551







