Comparative Transcriptome Analysis Reveals Common and Developmental Stage-Specific Genes That Respond to Low Nitrogen in Maize Leaves
Abstract
:1. Introduction
2. Results and Discussion
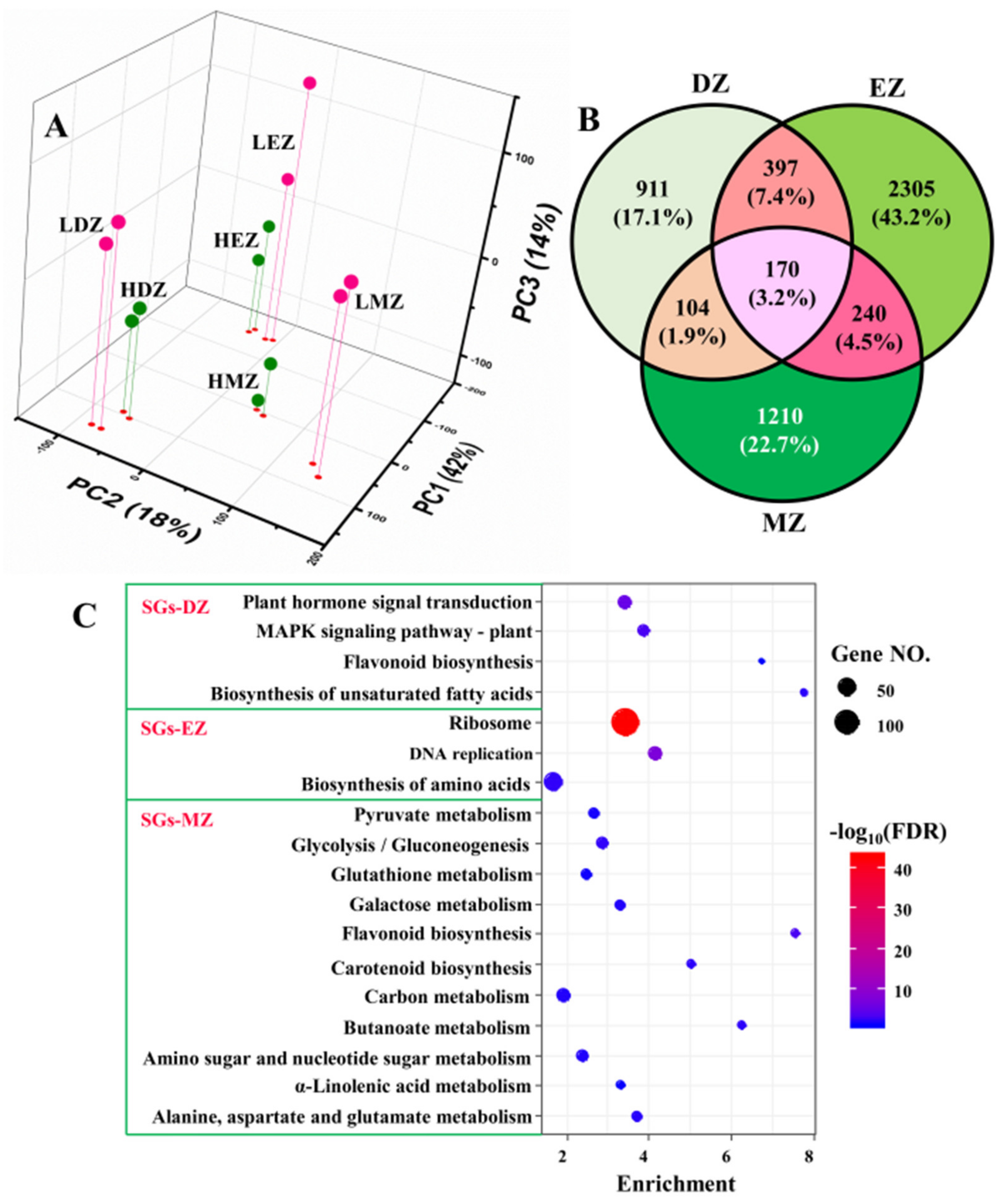
2.1. Identification of LN-Responsive Genes in Different Leaf Regions
2.2. SGs Are Highly Stage-Specific and Involved in Distinct Biological Processes
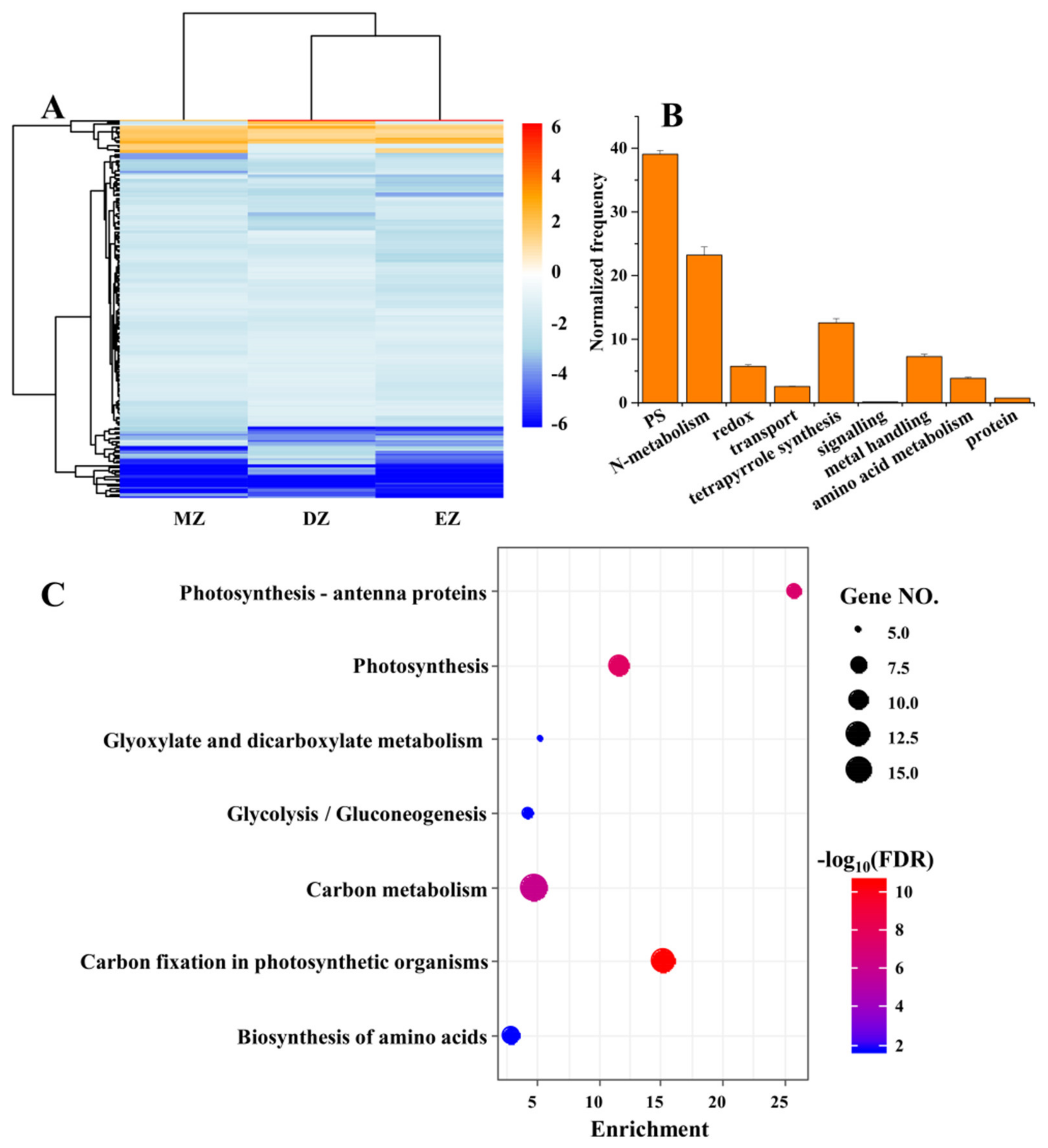
2.3. CGs Are Involved in Key Pathways That Are Important for N Uptake and Metabolism
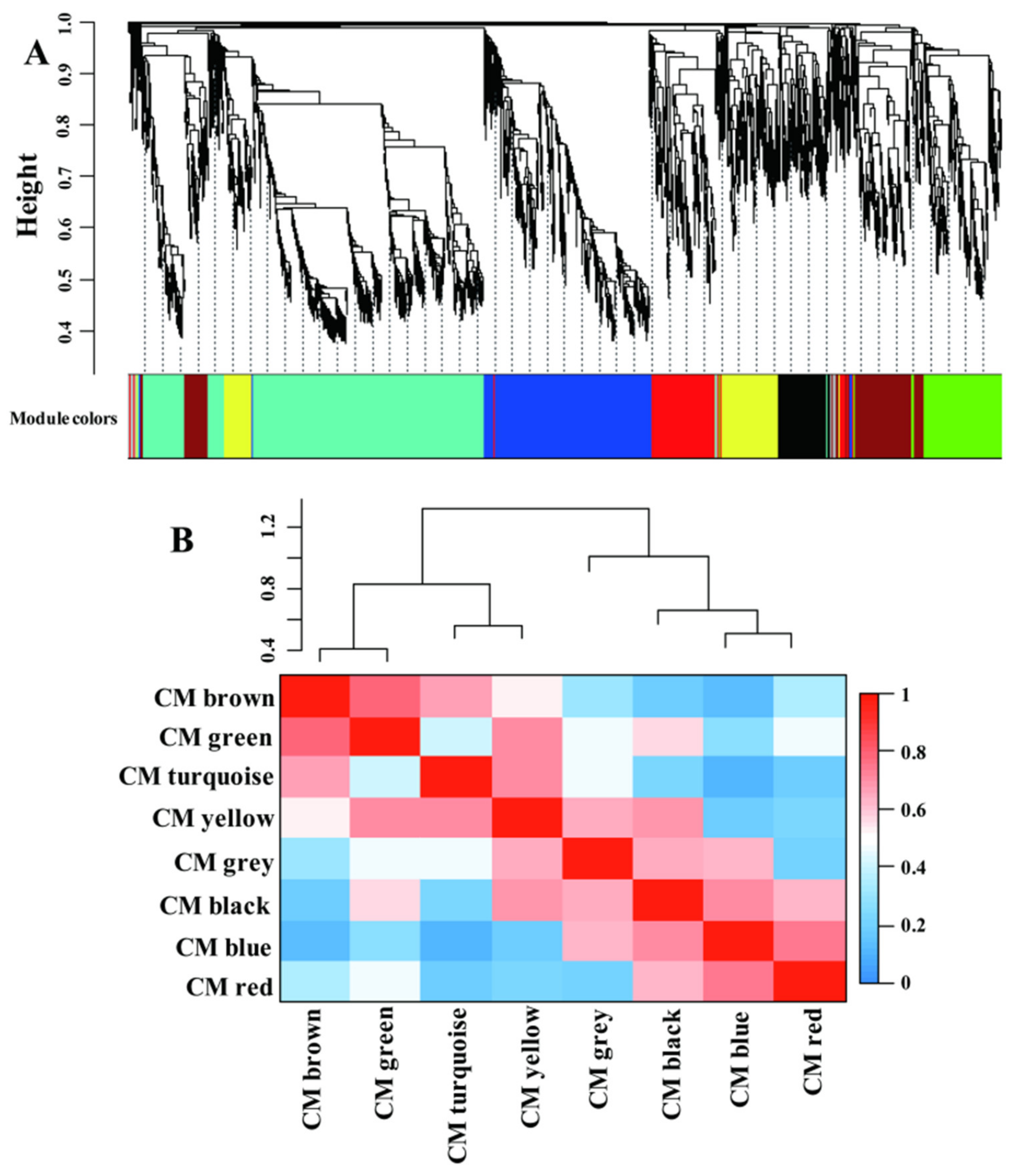
2.4. LN-Responsive Genes Are Highly Coexpressed; CGs and SGs Are Assigned into Different Modules
2.5. CM Blue Plays Essential Roles in Coordinating Multiple Metabolism Processes
3. Materials and Methods
3.1. Plant Material and Growth Condition
3.2. RNA Library Construction and Illumina Sequencing
3.3. Alignment and the Identification of Differentially Expressed Genes (DEG)
3.4. Bioinformation Analysis
3.5. Coexpression Network Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Urban, A.; Rogowski, P.; Wasilewska-Debowska, W.; Romanowska, E. Understanding Maize Response to Nitrogen Limitation inDifferent Light Conditions for the Improvementof Photosynthesis. Plants 2021, 10, 1932. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.H.; Chen, Q.W.; Chen, F.J.; Yuan, L.X.; Mi, G.H. A RNA-Seqanalysis of the response of photosynthetic system to low nitrogen supply in maize leaf. Int. J. Mol. Sci. 2017, 18, 2624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Qin, J.J.; He, F.F.; Li, H.; Liu, T.X.; Polle, A.; Peng, C.H.; Luo, Z.B. Net fluxes of ammonium and nitrate in association with H+ fluxes in fine roots of Populuspopularis. Planta 2013, 237, 919–931. [Google Scholar] [CrossRef] [PubMed]
- Undurraga, S.F.; Ibarra-Henriquez, C.; Fredes, I.; Alvarez, J.M.; Gutierrez, R.A. Nitrate signaling and early responses in Arabidopsis roots. J. Exp. Bot. 2017, 68, 2541–2551. [Google Scholar] [CrossRef]
- Zhang, X.; Davidson, E.A.; Mauzerall, D.L.; Searchinger, T.D.; Dumas, P.; Shen, Y. Managing nitrogen for sustainable development. Nature 2015, 528, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.J.; Liu, J.C.; Liu, Z.G.; Chen, Z.; Ren, W.; Gong, X.P.; Wang, L.F.; Cai, H.G.; Pan, Q.C.; Yuan, L.X.; et al. Breeding for high-yield and nitrogen use efficiency in maize: Lessons from comparison between Chinese and US cultivars. Adv. Agron 2021, 166, 251–275. [Google Scholar]
- Zhu, X.G.; Long, S.P.; Ort, D.R. Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 2010, 61, 235–261. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.L.; Wu, D.L.; Mu, X.H.; Xiao, C.X.; Chen, F.J.; Yuan, L.X.; Mi, G.H. Vertical distribution of photosynthetic nitrogen use efficiency and its response to nitrogen in field-grown maize. Crop Sci. 2016, 56, 397–407. [Google Scholar] [CrossRef]
- Mu, X.H.; Chen, Q.W.; Chen, F.J.; Yuan, L.X.; Mi, G.H. Dynamic remobilization of leaf nitrogen components in relation to photosynthetic rate during grain filling in maize. Plant Physiol. Bioch. 2018, 129, 27–34. [Google Scholar] [CrossRef]
- Luo, J.; Li, H.; Liu, T.X.; Polle, A.; Peng, C.H.; Luo, Z.B. Nitrogen metabolism of two contrasting poplar species during acclimation to limiting nitrogen availability. J. Exp. Bot. 2013, 64, 4207–4224. [Google Scholar] [CrossRef] [Green Version]
- Toth, V.R.; Meszaros, I.; Palmer, S.J.; Veres, S.; Precsenyi, I. Nitrogen deprivation induces changes in the leaf elongation zone of maize seedlings. Biol. Plant. 2002, 45, 241–247. [Google Scholar] [CrossRef]
- Vos, J.; Putten, P.E.L.V.D.; Birch, C.J. Effect of nitrogen supply on leaf appearance, leaf growth, leaf nitrogen economy and photosynthetic capacity in maize (Zea mays L.). Field Crop Res. 2005, 93, 64–73. [Google Scholar] [CrossRef]
- Kavanová, M.; Lattanzi, F.A.; Schnyder, H. Nitrogen deficiency inhibits leaf blade growth in Loliumperenne by increasing cell cycle duration and decreasing mitotic and post-mitotic growth rates. Plant Cell Environ. 2008, 31, 727–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polyn, S.; Willems, A.; De Veylder, L. Cell cycle entry, maintenance, and exit during plant development. Curr. Opin. Plant Biol. 2015, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, H.; Rymen, B.; Jikumaru, Y.; Demuynck, K.; Van Lijsebettens, M.; Kamiya, Y.; Inze, D.; Beemster, G.T. A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr. Biol. 2012, 22, 1183–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA gene family in plants: Molecular structure, regulation, and function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wirén, N.V. Integration of nutrient and water availabilities via auxininto the root developmental program. Curr. Opin. Plant Biol. 2022, 65, 102117. [Google Scholar] [CrossRef]
- Yu, P.; Eggert, K.; von Wiren, N.; Li, C.J.; Hochholdinger, F. Cell type-specific gene expression analyses by rna sequencing reveal local high nitrate-triggered lateral root initiation in shoot-borne roots of maize by modulating auxin-related cell cycle regulation. Plant Physiol. 2015, 169, 690–704. [Google Scholar] [CrossRef]
- Pan, X.Y.; Hasan, M.M.; Li, Y.Q.; Liao, C.S.; Zheng, H.Y.; Liu, R.Y.; Li, X.X. Asymmetric transcriptomic signatures between the cob and florets in the maize ear under optimal- and low-nitrogen conditions at silking, and functional characterization of amino acid transporters ZmAAP4 and ZmVAAT3. J. Exp. Bot. 2015, 66, 6149–6166. [Google Scholar] [CrossRef]
- Colcombet, J.; Hirt, H. Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 2008, 413, 217–226. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, S.Q. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.H.; Lee, I.; Moradi, E.; Hung, N.J.; Johnson, A.W.; Marcotte, E.M. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009, 7, e1000213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nybakken, L.; Lie, M.H.; Julkunen-Tiitto, R.; Asplund, J.; Ohlson, M. Fertilization Changes Chemical Defense in Needles of Mature Norway Spruce (Piceaabies). Front. Plant Sci. 2018, 9, 770. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.-J. Growth performance, photosynthesis, and root characteristics are associated with nitrogen use efficiency in six poplar species. Environ. Exp. Bot. 2019, 164, 40–51. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.-J.; Masclaux-Daubresse, C.; Wang, N.; Wang, H.; Zheng, B. Morphological and physiological responses to contrasting nitrogen regimes in Populuscathayana is linked to resources allocation and carbon/nitrogen partition. Environ. Exp. Bot. 2019, 162, 247–255. [Google Scholar] [CrossRef]
- Zhao, D.L.; Reddy, K.R.; Kakani, V.G.; Reddy, V.R. Nitrogen deficiency effects on plant growth, leaf photosynthesis, and hyperspectral reflectance properties of sorghum. Eur. J. Agron. 2005, 22, 391–403. [Google Scholar] [CrossRef]
- Ghannoum, O.; Evans, J.R.; Chow, W.S.; Andrews, T.J.; Conroy, J.P.; von Caemmerer, S. Faster rubisco is the key to superior nitrogen-use efficiency in NADP-malic enzyme relative to NAD-malic enzyme C(4) grasses. Plant Physiol. 2005, 137, 638–650. [Google Scholar] [CrossRef] [Green Version]
- Ciompi, S.; Gentili, E.; Guidi, L.; Soldatini, G.F. The effect of nitrogen deficiency on leaf gas exchange and chlorophyll fluorescence parameters in sunflower. Plant Sci. 1996, 118, 177–184. [Google Scholar] [CrossRef]
- Makino, A.; Sakuma, H.; Sudo, E.; Mae, T. Differences between maize and rice in N-use efficiency for photosynthesis and protein allocation. Plant Cell Physiol. 2003, 44, 952–956. [Google Scholar] [CrossRef] [Green Version]
- Have, M.; Marmagne, A.; Chardon, F.; Masclaux-Daubresse, C. Nitrogen remobilization during leaf senescence: Lessons from Arabidopsis to crops. J. Exp. Bot. 2017, 68, 2513–2529. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.; Li, H.; Shi, W.G.; Polle, A.; Lu, M.Z.; Sun, X.M.; Luo, Z.B. Global poplar root and leaf transcriptomes reveal links between growth and stress responses under nitrogen starvation and excess. Tree Physiol. 2015, 35, 1283–1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, X.; Luo, J. Evolutionary analyses of NIN-like proteins in plants and their roles in nitrate signaling. Cell Mol. Life Sci. 2019, 76, 3753–3764. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Álvarez, J.M.; Moyano, T.C.; Gutiérrez, R.A. Transcriptional networks in the nitrate response of Arabidopsis thaliana. Curr. Opin. Plant Biol. 2015, 27, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.-R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [Green Version]
- Ho, C.-H.; Tsay, Y.-F. Nitrate, ammonium, and potassium sensing and signaling. Curr. Opin. Plant Biol. 2010, 13, 604–610. [Google Scholar] [CrossRef]
- Maeda, Y.; Konishi, M.; Kiba, T.; Sakuraba, Y.; Sawaki, N.; Kurai, T.; Ueda, Y.; Sakakibara, H.; Yanagisawa, S. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 2018, 9, 1376. [Google Scholar] [CrossRef] [Green Version]
- Medici, A.; Marshall-Colon, A.; Ronzier, E.; Szponarski, W.; Wang, R.; Gojon, A.; Crawford, N.M.; Ruffel, S.; Coruzzi, G.M.; Krouk, G. AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 2015, 6, 6274. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Xia, W.; Cao, P.; Xiao, Z.a.; Zhang, Y.; Liu, M.; Zhan, C.; Wang, N. Integrated transcriptome analysis reveals plant hormones jasmonic acid and salicylic acid coordinate growth and defense responses upon fungal infection in poplar. Biomolecules 2019, 9, 12. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Li, H.; Luo, J.; Ma, C.; Li, S.; Qu, L.; Gai, Y.; Jiang, X.; Janz, D.; Polle, A. A transcriptomic network underlies microstructural and physiological responses to cadmium in Populus× canescens. Plant Physiol. 2013, 162, 424–439. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Liang, Z.; Wu, M.; Mei, L. Genome-wide identification of BOR genes in poplar and their roles in response to various environmental stimuli. Environ. Exp. Bot. 2019, 164, 101–113. [Google Scholar] [CrossRef]
- Campos, F.G.; Vieira, M.A.R.; Amaro, A.C.E.; delaCruz-Chacón, I.; Marques, M.O.M.; Ferreira, G.; Boaro, C.S.F. Nitrogen in the defense system of Annonaemarginata (Schltdl.) H. Rainer. PLoS ONE 2019, 14, e0217930. [Google Scholar] [CrossRef] [PubMed]
- Suárez-López, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116. [Google Scholar] [CrossRef]
- Lin, M.-K.; Belanger, H.; Lee, Y.-J.; Varkonyi-Gasic, E.; Taoka, K.-I.; Miura, E.; Xoconostle-Cázares, B.; Gendler, K.; Jorgensen, R.A.; Phinney, B. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 2007, 19, 1488–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Oosterom, E.J.; Borrell, A.K.; Chapman, S.C.; Broad, I.J.; Hammer, G.L. Functional dynamics of the nitrogen balance of sorghum: I. N demand of vegetative plant parts. Field Crop Res. 2010, 115, 19–28. [Google Scholar] [CrossRef]
- Trachsel, S.; Burgueno, J.; Suarez, E.A.; San Vicente, F.M.; Rodriguez, C.S.; Dhliwayo, T. Interrelations among Early Vigor, Flowering Time, Physiological Maturity, and Grain Yield in Tropical Maize (Zea mays L.) under Multiple Abiotic Stresses. Crop Sci. 2017, 57, 229–242. [Google Scholar] [CrossRef]
- Ó’Maoiléidigh, D. The White Stripes featuring ALBOSTRIANS, a chloroplast-localized CCT domain protein. Plant Cell 2019, 31, 1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Hensel, G.; Mascher, M.; Melzer, M.; Budhagatapalli, N.; Rutten, T.; Himmelbach, A.; Beier, S.; Korzun, V.; Kumlehn, J. Leaf variegation and impaired chloroplast development caused by a truncated CCT domain gene in albostrians barley. Plant Cell 2019, 31, 1430–1445. [Google Scholar] [CrossRef] [Green Version]
- Ohmiya, A.; Oda-Yamamizo, C.; Kishimoto, S. Overexpression of CONSTANS-like 16 enhances chlorophyll accumulation in petunia corollas. Plant Sci. 2019, 280, 90–96. [Google Scholar] [CrossRef]
- Ohmiya, A.; Sasaki, K.; Nashima, K.; Oda-Yamamizo, C.; Hirashima, M.; Sumitomo, K. Transcriptome analysis in petals and leaves of chrysanthemums with different chlorophyll levels. BMC Plant Biol. 2017, 17, 202. [Google Scholar] [CrossRef] [Green Version]
- Gao, K.; Chen, F.; Yuan, L.; Zhang, F.; Mi, G. A comprehensive analysis of root morphological changes and nitrogen allocation in maize in response to low nitrogen stress. Plant Cell Environ. 2015, 38, 740–750. [Google Scholar] [CrossRef]
- Mu, X.H.; Chen, Q.W.; Wu, X.Y.; Chen, F.J.; Yuan, L.X.; Mi, G.H. Gibberellins synthesis is involved in the reduction of cell flux and elemental growth rate in maize leaf under low nitrogen supply. Environ. Exp. Bot. 2018, 150, 198–208. [Google Scholar] [CrossRef]
- Gu, R.L.; Duan, F.Y.; An, X.; Zhang, F.S.; von Wiren, N.; Yuan, L.X. Characterization of AMT-Mediated High-Affinity Ammonium Uptake in Roots of Maize (Zea mays L.). Plant Cell Physiol. 2013, 54, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Oliveros, J.; Venny, C. An Interactive Tool for Comparing Lists with Venn’s Diagrams. 2007–2015. 2016. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 12 May 2022).
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef] [Green Version]
- Provart, N.; Zhu, T. A browser-based functional classification SuperViewer for Arabidopsis genomics. Curr. Comput. Mol. Biol. 2003, 2003, 271–272. [Google Scholar]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. Cluster Profiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Kolde, R.; Kolde, M.R. Package ‘pheatmap’. R Package 2015, 1, 790. [Google Scholar]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Locus Name | Name * | Closet AGI | Symbol of AGI | Description | DZ | EZ | MZ |
|---|---|---|---|---|---|---|---|
| Amino acid and N metabolism | |||||||
| Zm00001d050694 | ZmEMB1075 | AT1G43710 | EMB1075 | embryo defective 1075 | −2.1 | −3.0 | −1.9 |
| Zm00001d052247 | ZmSK1a | AT2G21940 | SK1 | shikimate kinase 1 | −1.6 | −1.1 | −1.2 |
| Zm00001d018061 | ZmSK1b | AT2G21940 | SK1 | shikimate kinase 1 | −1.4 | −1.6 | −1.6 |
| Zm00001d002880 | ZmIDM2 | AT1G80560 | IMD2 | isopropylmalate dehydrogenase 2 | −1.1 | −1.8 | −1.3 |
| Zm00001d026501 | ZmGS2 | AT5G35630 | GS2 | glutamine synthetase 2 | −1.1 | −1.8 | −1.4 |
| Zm00001d018161 | ZmNIR1 | AT2G15620 | NIR1 | nitrite reductase 1 | −5.8 | −5.6 | −2.9 |
| Zm00001d049995 | ZmNIA1 | AT1G77760 | NIA1 | nitrate reductase 1 | −2.8 | −2.5 | −2.5 |
| Protein synthesis and degradation | |||||||
| Zm00001d047186 | ZmRPS9C | AT5G39850 | RPS9C | 40S ribosomal protein S9 | −1.3 | −2.6 | −1.2 |
| Zm00001d038084 | ZmEMB3126 | AT3G63490 | EMB3126 | embryo defective 3126 | −1.1 | −1.4 | −1.0 |
| Zm00001d016072 | ZmRPL21C | AT1G35680 | RPL21C | chloroplast ribosomal protein l21 | −1.9 | −1.5 | −1.4 |
| Zm00001d014488 | ZmRPL24 | AT5G54600 | RPL24 | plastid ribosomal protein l24 | −1.4 | −1.2 | −1.4 |
| Zm00001d044130 | ZmEMB2184 | AT1G75350 | EMB2184 | embryo defective 2184 | −1.0 | −1.6 | −1.3 |
| Zm00001d018412 | ZmRPL9 | AT3G44890 | RPL9 | ribosomal protein l9 | −1.7 | −1.3 | −1.6 |
| Zm00001d015628 | ZmGHS1 | AT3G27160 | GHS1 | glucose hypersensitive 1 | −1.8 | −2.2 | −1.9 |
| Zm00001d043195 | ZmMC5 | AT1G79330 | MC5 | metacaspase 5 | −1.1 | −1.0 | −2.0 |
| Zm00001d046952 | ZmAAH | AT4G20070 | AAH | allantoateamidohydrolase | 1.3 | 1.4 | 1.8 |
| Transport related to N | |||||||
| Zm00001d012231 | ZmAAP8 | AT1G10010 | AAP8 | amino acid permease 8 | −4.3 | −4.3 | −3.3 |
| Zm00001d044529 | ZmNPF6.2 | AT2G26690 | NPF6.2 | NRT1/ PTR family 6.2 | −6.8 | −8.4 | −8.3 |
| Zm00001d042684 | ZmNPF5.16 | AT1G22550 | NPF5.16 | NRT1/ PTR family 5.16 | −6.1 | −8.4 | −8.3 |
| Zm00001d051525 | ZmOPT4 | AT5G64410 | OPT4 | oligopeptide transporter 4 | 1.8 | 2.3 | 2.7 |
| Zm00001d045519 | ZmOPT6 | AT4G10770 | OPT7 | oligopeptide transporter 7 | 2.3 | 2.3 | 2.2 |
| Zm00001d015702 | ZmCLCG | AT5G33280 | CLCG | chloride channel G | −2.1 | −1.1 | −1.7 |
| Zm00001d046919 | ZmCLC-Aa | AT5G40890 | CLC-A | chloride channel A | −1.8 | −2.9 | −1.5 |
| Zm00001d015700 | ZmCLC-Ab | AT5G49890 | CLC-A | chloride channel A | −2.6 | −1.8 | −1.6 |
| Locus Name | Name * | Closet AGI | Symbol of AGI | Description | DZ | EZ | MZ |
|---|---|---|---|---|---|---|---|
| Zm00001d034160 | ZmHHO5 | AT4G37180 | HHO5 | HRS1 homolog5 | −4.8 | −5.2 | −2.7 |
| Zm00001d013202 | ZmHHO4a | AT2G03500 | HHO4 | HRS1 homolog4 | −5.6 | −5.5 | −4.8 |
| Zm00001d030891 | ZmHHO4b | AT2G03500 | HHO4 | HRS1 homolog4 | −2.0 | −1.8 | −3.1 |
| Zm00001d007962 | ZmHHO4c | AT2G03500 | HHO4 | HRS1 homolog4 | −3.2 | −1.5 | −1.8 |
| Zm00001d023402 | ZmHHO2 | AT1G68670 | HHO2; NIGT1.2 | HRS1 homolog2 | −3.5 | −7.3 | −8.1 |
| Zm00001d051749 | ZmHB-1 | AT3G01470 | HB-1 | homeobox 1 | 1.3 | 1.0 | −1.2 |
| Zm00001d029601 | ZmLBD37a | AT5G67420 | LBD37 | LOB domain-containing protein 37 | −3.8 | −7.4 | −5.5 |
| Zm00001d021995 | ZmLBD37b | AT5G67420 | LBD37 | LOB domain-containing protein 37 | −6.0 | −5.7 | −8.1 |
| Zm00001d018255 | ZmNF-YA5 | AT1G54160 | NF-YA5 | nuclear factor Y, subunit A5 | 1.5 | 2.7 | 2.0 |
| Zm00001d015201 | ZmNLP6 | AT1G64530 | NLP6 | NIN-like protein 6 | −3.9 | −4.9 | −4.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Arshad, A.; Yang, L.; Qin, Y.; Mu, X.; Mi, G. Comparative Transcriptome Analysis Reveals Common and Developmental Stage-Specific Genes That Respond to Low Nitrogen in Maize Leaves. Plants 2022, 11, 1550. https://doi.org/10.3390/plants11121550
Guo S, Arshad A, Yang L, Qin Y, Mu X, Mi G. Comparative Transcriptome Analysis Reveals Common and Developmental Stage-Specific Genes That Respond to Low Nitrogen in Maize Leaves. Plants. 2022; 11(12):1550. https://doi.org/10.3390/plants11121550
Chicago/Turabian StyleGuo, Song, Adnan Arshad, Lan Yang, Yusheng Qin, Xiaohuan Mu, and Guohua Mi. 2022. "Comparative Transcriptome Analysis Reveals Common and Developmental Stage-Specific Genes That Respond to Low Nitrogen in Maize Leaves" Plants 11, no. 12: 1550. https://doi.org/10.3390/plants11121550







