Phylogenetic Analysis and Flower Color Evolution of the Subfamily Linoideae (Linaceae)
Abstract
1. Introduction
2. Results
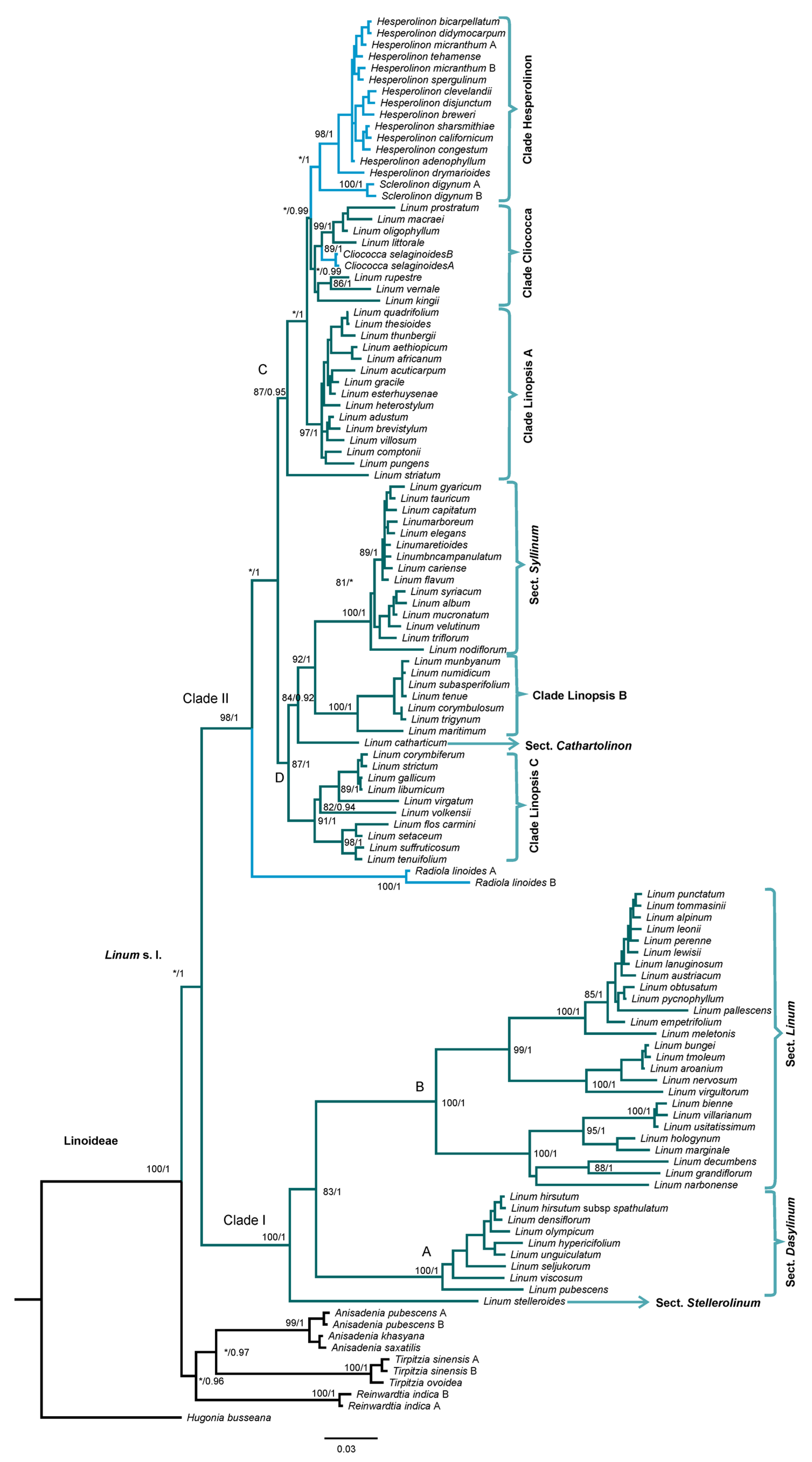
2.1. Phylogenetic Analysis
2.2. Reconstruction of Ancestral Flower Color
3. Discussion
3.1. Phylogenetic Analysis
3.2. Reconstruction of Ancestral Flower Color
4. Material and Methods
4.1. Taxon Sampling
4.2. Phylogenetic Analysis
4.3. Reconstruction of Ancestral Flower Color
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McDill, J.; Simpson, B.B. Molecular phylogenetics of Linaceae with complete generic sampling and data from two plastid genes. Bot. J. Linn. Soc. 2011, 165, 64–83. [Google Scholar] [CrossRef]
- McDill, J.; Repplinger, M.; Simpson, B.B.; Kadereit, W.J. The phylogeny of Linum and Linaceae subfamily Linoideae, with implications for their systematics, biogeography, and evolution of heterostyly. Syst. Bot. 2009, 34, 386–405. [Google Scholar] [CrossRef]
- Dressler, S.M.; Repplinger, M.; Bayer, C. Linaceae. In Flowering Plants. Eudicots, 1st ed.; Kubitzki, K., Ed.; Springer: Heidelberg, Germany, 2014; Volume 10, pp. 237–246. [Google Scholar]
- Melnikova, N.V.; Kudryavtseva, A.V.; Zelenin, A.V.; Lakunina, V.A.; Yurkevich, O.Y.; Speranskaya, A.S.; Snezhkina, A.V. Retrotransposon-based molecular markers for analysis of genetic diversity within the genus. Linum. BioMed Res. Int. 2014, 2014, 231589. [Google Scholar] [CrossRef] [PubMed]
- Rzedowski, J.; Calderón de Rzedowski, G. Linaceae. In Flora del Bajío y de Regiones Adyacentes; Instituto de Ecología A.C.: Pátzcuaro, Mexico, 1992; Volume 6, pp. 1–22. ISSN 0188-5170. [Google Scholar]
- Rzedowski, J.; Calderón de Rzedowski, G. Fascículo 5. Linaceae. In Flora del Valle de Tehuacán-Cuicatlán, 1st ed.; Universidad Nacional Autónoma de México: Mexico City, México, 1994; Volume 5, pp. 5–19. [Google Scholar]
- Touré, A.; Xu, X.M. Flaxseed lignans: Source, biosynthesis, metabolism, antioxidant activity, bio-active components, and health benefits. Compr. Rev. Food Sci. Food Saf. 2010, 9, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Mridula, D.; Rehal, J.; Barnwal, P. Flaxseed: A potential source of food, feed and fiber. Crit. Rev. Food Sci. Nutr. 2011, 51, 210–222. [Google Scholar] [CrossRef]
- Lautié, E.; Quintero, R.; Fliniaux, M.A.; Villarreal, M.L. Selection methodology with scoring system: Application to Mexican plants producing podophyllotoxin related lignans. J. Ethnopharmacol. 2008, 120, 402–412. [Google Scholar] [CrossRef]
- Alonso-Castro, J.A.; Villarreal, M.L.; Sálazar-Olivo, A.L.; Gomez-Sanchez, M.; Dominguez, F.; Garcia-Carranca, A. Mexican medicinal plants used for cancer treatment: Pharmacological phytochemical and ethnobotanical studies. J. Ethnopharmacol. 2011, 133, 945–972. [Google Scholar] [CrossRef]
- Barrera-Robles, P.J.; Burgos-Hernández, M.; Ruíz-Acevedo, A.D.; Castillo-Campos, G. La familia Linaceae en México: Estado actual y perspectivas. Bot. Sci. 2020, 98, 560–572. [Google Scholar] [CrossRef]
- Fu, Y.-B.; Peterson, G.; Diederichsen, A.; Richards, K.W. RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genet. Resour. Crop Evol. 2002, 49, 253–259. [Google Scholar] [CrossRef]
- Ruiz-Martín, J.; Santos-Gally, R.; Escudero, M.; Midgley, J.J.; Pérez-Barrales, R.; Arroyo, J. Style polymorphism in Linum (Linaceae): A case of Mediterranean parallel evolution? J. Plant Biol. 2018, 20, 100–111. [Google Scholar] [CrossRef]
- Burgos-Hernández, M.; Castillo-Campos, G. Contribución al conocimiento del género Linum (Linaceae) en Veracruz, México. Acta Bot. Mex. 2019, 3, 126. [Google Scholar] [CrossRef]
- Planchon, J.E. Sur la famille des Linées. Lond. J. Bot. 1847, 6, 588–603. [Google Scholar]
- Planchon, J.E. Sur la famille des Linées. Lond. J. Bot. 1848, 7, 165–186, 473–501, 507–528. [Google Scholar]
- Hallier, H. Beitrëge zur Kenntnis der Linaceae (DC. 1819) Dumort. Beih. Bot. Cent. Abt. 1 1921, 39, 1–178. [Google Scholar]
- Small, J.K. Linaceae. N. Am. Flora 1907, 25, 67–87. [Google Scholar]
- Sharsmith, H.K. The genus Hesperolinon (Linaceae). Univ. Calif. Publ. Bot. 1961, 32, 235–314. [Google Scholar]
- Rogers, C.M. Sclerolinon, a new genus in the Linaceae. Madroño 1966, 18, 181–184. [Google Scholar]
- Rogers, C.M.; Mildner, R. The reevaluation of the genus Cliococca (Linaceae) of South America. Rhodora 1971, 73, 560–565. [Google Scholar]
- Lamarck, J.B. Encyclopédie Méthodique, Botanique 1, 1st ed.; Panckoucke: Paris, France, 1791; p. 496. [Google Scholar]
- Babington, C.C. Description of a new genus of Lineae. Trans. Linn. Soc. Lond. 1842, 19, 33–34. [Google Scholar] [CrossRef][Green Version]
- Gray, A. Linum sect. Hesperolinon. Proc. Am. Acad. Arts 1865, 6, 521. [Google Scholar]
- González-Velasco, J.; Burgos-Hernández, M.; Galván-Escobedo, I.G.; Castillo-Campos, G. Taxonomic update of the flax family in Mexico. Phytotaxa 2022, 549, 141–184. [Google Scholar] [CrossRef]
- Schneider, A.C.; Freyman, W.A.; Guilliams, C.M.; Springer, Y.P.; Baldwin, B.G. Pleistocene radiation of the serpentine—Adapted genus Hesperolinon and other divergence times in Linaceae (Malpighiales). Am. J. Bot. 2016, 103, 221–232. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brewer, W.H.; Watson, S.; Gray, A. Botany [of California], 1st ed.; Cambridge University Press: Cambridge, MA, USA, 1876. [Google Scholar]
- Trelease, W. A revision of North American Linaceae. Trans. Acad. Sci. St. Louis 1887, 5, 7–20. [Google Scholar]
- Trelease, W. Linaceae. In Synoptical Flora of North America, 1st ed.; American Book Company: New York, NY, USA, 1897; Volume 2, pp. 344–349. [Google Scholar]
- Winkler, H. Linaceae. In Die Natürlichen Pflanzenfamilien, 19th ed.; Engelmann: Leipzig, Germany, 1931; Volume 10, pp. 82–130. [Google Scholar]
- Ockendon, D.J.; Walters, S.M. Linum. In Flora Europea, 1st ed.; Cambridge University Press: Cambridge, MA, USA, 1968; pp. 206–211. [Google Scholar]
- Maguilla, E.; Escudero, M.; Ruiz-Martín, J.; Arroyo, J. Origin and diversification of flax and their relationship with heterostyly across the range. J. Biogeogr. 2021, 48, 1994–2007. [Google Scholar] [CrossRef]
- Bolsheva, N.L.; Melnikova, N.V.; Dvorianinova, E.M.; Mironova, L.N.; Yurkevich, O.Y.; Amosova, A.V.; Krasnov, G.S.; Dimitriev, A.A.; Muravenko, O.V. Clarification of the position of Linum stelleroides Planch. within the phylogeny of the genus Linum L. Plants. 2022, 11, 652. [Google Scholar] [CrossRef]
- Wesselingh, R.A.; Arnold, M.L. Pollinator behaviour and the evolution of Louisiana iris hybrid zones. J. Evol. Biol. 2000, 13, 171–180. [Google Scholar] [CrossRef]
- Martén-Rodríguez, S.; Fenster, C.B.; Agnarsson, I.; Skog, L.E.; Zimmer, E.A. Evolutionary breakdown of pollination specialization in a Caribbean plant radiation. New Phytol. 2010, 188, 403–441. [Google Scholar] [CrossRef]
- Lehrer, M.; Horridge, G.A.; Zhang, S.W.; Gadagkar, R. Shape vision in bees: Innate preference for flower-like patterns. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995, 347, 123–137. [Google Scholar] [CrossRef]
- Koski, M.H.; Ashman, T.L. Dissecting pollinator responses to a ubiquitous ultraviolet floral pattern in the wild. Funct. Ecol. 2014, 28, 868–877. [Google Scholar] [CrossRef]
- Manning, A. The Effect of Honey-Guides. Behaviour 1956, 9, 114–139. [Google Scholar] [CrossRef]
- Hansen, D.M.; van der Niet, T.; Johnson, S.D. Floral signposts: Testing the significance of visual ‘nectar guides’ for pollinator behaviour and plant fitness. Proc. R. Soc. B 2012, 279, 634–639. [Google Scholar] [CrossRef] [PubMed]
- Streisfeld, M.A.; Rausher, M.D. Altered trans-regulatory control of gene expression in multiple anthocyanin genes contributes to adaptive flower color evolution in Mimulus aurantiacus. Mol. Biol. Evol. 2009, 26, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.D.; Rausher, M.D. Gene loss and parallel evolution contribute to specie difference in flower color. Mol. Biol. Evol. 2011, 28, 2799–2810. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, C.; Yao, X.; Wang, F.; Wu, J.; King, G.J.; Liu, F. Disruption of a carotenoid cleavage dioxygenase 4 gene converts flower colour from white to yellow in Brassica species. New Phytol. 2015, 206, 1513–1526. [Google Scholar] [CrossRef] [PubMed]
- Eaton, D.A.R.; Fenster, C.B.; Hereford, J.; Huang, S.; Ree, R.H. Floral diversity a community structure in Pedicularis (Orobanchaceae). Ecology 2012, 93, 182–193. [Google Scholar] [CrossRef]
- Lawson, D.A.; Rands, S.A. The evolution of floral guides: Using a genetic algorithm to investigate the evolution of floral cue arrangements. Biol. J. Linn. Soc. Lond. 2018, 123, 739–753. [Google Scholar] [CrossRef]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef]
- Haq, B.U.; Hardenbol, J.A.N.; Vail, P.R. Chronology of fluctuating sea levels since the Triassic. Science 1987, 235, 1156–1167. [Google Scholar] [CrossRef]
- Raven, P.H.; Axelrod, D.I. Origin and relationships of the California flora. Univ. Calif. Publ. Bot. 1978, 72, 1–134. [Google Scholar]
- Anacker, B.L.; Whittall, J.B.; Goldberg, E.E.; Harrison, S.P. Origins and consequences of serpentine endemism in the California flora. Evolution 2011, 65, 365–376. [Google Scholar] [CrossRef]
- Springer, Y.P. Do extreme environments provide a refuge from pathogens? A phylogenetic test using serpentine flax. Am. J. Bot. 2009, 96, 2010–2021. [Google Scholar] [CrossRef] [PubMed]
- Yuzepchuk, S.V. Genus Linum-Linaceae Dumort. In Flora SSSR (Flora of the Soviet Union); Shishkin, B.K., Bobrov, E.G., Eds.; Izdatel’stvo Akademii Nauk SSSR: Moscow, Russia, 1949; Volume 14, pp. 84–146. [Google Scholar]
- Bolsheva, N.L.; Melnikova, N.V.; Kirov, I.V.; Speranskaya, A.S.; Krinitsina, A.A.; Dmitriev, A.A.; Muravenko, O.V. Evolution of blue-flowered species of genus Linum based on high-throughput sequencing of ribosomal RNA genes. BMC Evol. Biol. 2017, 17, 253. [Google Scholar] [CrossRef] [PubMed]
- Martzenitzina, K.K. The chromosomes of some species of the genus Linum L. Bull. Appl. Bot. Genet. Plant Breed. 1927, 17, 25–264. [Google Scholar]
- Petrova, A.V. IOPB Chromosome number reports XXXV. Taxon 1927, 21, 161–166. [Google Scholar]
- Rogers, C.M. The systematics of Linum sect. Linopsis. Plant Syst. Evol. 1982, 140, 225–234. [Google Scholar] [CrossRef]
- Castañeda-Alvarez, N.P.; Khoury, C.K.; Achicanoy, H.A.; Bernau, V.; Dempewolf, H.; Eastwood, R.J.; Toll, J. Global conservation priorities for crop wild relatives. Nat. Plants 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Von Wettberg, E.J.; Chang, P.L.; Başdemir, F.; Carrasquila-Garcia, N.; Korbu, L.B.; Moenga, S.M.; Bedada, G.; Greenlon, A.; Moriuchi, K.S.; Singh, V.; et al. Ecology and genomics of an important crop wild relative as a prelude to agricultural innovation. Nat. Commun. 2018, 9, 649. [Google Scholar] [CrossRef]
- Nair, K.P. Utilizing crop wild relatives to combat global warming. Adv. Agron. 2019, 153, 175–258. [Google Scholar] [CrossRef]
- Vincent, H.; Hole, D.; Maxted, N. Congruence between global crop wild relative hotspots and biodiversity hotspots. Biol. Conserv. 2022, 265, 109432. [Google Scholar] [CrossRef]
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of crop wild relatives: Expanding the gene pool for crop improvement. Plant Biotechnol. J. 2016, 14, 1070–1085. [Google Scholar] [CrossRef]
- Eyres, L. Flaxseed fibre—A functional superfood? Food N. Z. 2015, 15, 24. [Google Scholar]
- Kaur, P.; Waghmare, R.; Kumar, V.; Rasane, P.; Kaur, S.; Gat, Y. Recent advances in utilization of flaxseed as potential source for value addition. OCL 2018, 25, A304. [Google Scholar] [CrossRef]
- Sheidai, M.; Darini, S.; Talebi, S.M.; Koohdar, F.; Ghasemzadeh-Baraki, S. Molecular systematic study in the genus Linum (Linaceae) in Iran. Acta Bot. Hung. 2019, 61, 421–434. [Google Scholar] [CrossRef]
- Tammes, T. The genetics of the genus Linum. Bibliogr Genet. 1928, 4, 1–36. [Google Scholar]
- Allaby, R.G.; Peterson, G.W.; Merriwether, A.; Fu, Y.B. Evidence of the domestication history of flax (Linum usitatissimum) from genetic diversity of the sad2 locus. Theor. Appl. Genet. 2005, 112, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Maxted, N.; Scholten, M.; Codd, R.; Ford-Lloyd, B. Creation and use of a national inventory of crop wild relatives. Biol. Conserv. 2007, 140, 142–159. [Google Scholar] [CrossRef]
- Cullis, J.O. Diagnosis and management of anaemia of chronic disease: Current status. Br. J. Haematol. 2011, 154, 289–300. [Google Scholar] [CrossRef]
- Tork, D.G.; Anderson, N.O.; Wyse, D.L.; Betts, K.J. Domestication of perennial flax using an ideotype approach for oilseed, cut flower, and garden performance. Agronomy 2019, 9, 707. [Google Scholar] [CrossRef]
- Mohammed, M.M.; Christensen, L.P.; Ibrahim, N.A.; Awad, N.E.; Zeid, I.F.; Pedersen, E.B.; Jensen, K.B.; Colla, P.L. Anti-HIV-1 activities of the extracts from the medicinal plant Linum grandiflorum Desf. In Proceedings of 4th Conference on Research and Development of Pharmaceutical Industries (Current Challenges). Med. Aromat. Plant Sci. Biotechnol. 2009, 3, 37–41. [Google Scholar]
- Rogers, C.M. Relationships of Hesperolinon and Linum (Linaceae). Madroño 1975, 23, 153–159. [Google Scholar]
- Bradshaw, H.D.; Wilbert, S.M.; Otto, K.G.; Schemske, D.W. Genetic mapping of floral traits associated with reproductive isolation in monkey flowers (Mimulus). Nature 1995, 376, 762–765. [Google Scholar] [CrossRef]
- Van der Niet, T.; Johnson, S.D. Phylogenetic evidence for pollinator driven diversification of angiosperms. Trends Ecol. Evol. 2012, 27, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.G.; Fairbanks, R.G.; Mountain, G.S. Tertiary oxygen isotope synthesis, sea level history and continental margin erosion. Paleoceanogr. Paleoclimatol. 1987, 2, 1–19. [Google Scholar] [CrossRef]
- Tiffney, B.H.; Manchester, S.R. The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the Northern Hemisphere Tertiary. Int. J. Plant Sci. 2001, 162, S3–S17. [Google Scholar] [CrossRef]
- Bush, M.; Flenley, J.; Gosling, W. Tropical Rainforest Responses to Climatic Change, 2nd ed.; Springer: Berlin, Germany, 2011; pp. 1–34, 85–123. [Google Scholar] [CrossRef]
- Herbert, T.D.; Lawrence, K.T.; Tzanova, A.; Peterson, L.C.; Caballero-Gill, R.; Kelly, C.S. Late Miocene global cooling and the rise of modern ecosystems. Nat. Geosci. 2016, 11, 843–847. [Google Scholar] [CrossRef]
- Milne, R.I.; Abbott, R.J. The origin and evolution of tertiary relict floras. Adv. Bot. Res. 2002, 38, 281–314. [Google Scholar] [CrossRef]
- Weijermars, R. Neogene tectonics in the Western Mediterranean may have caused the Messinian Salinity Crisis and an associated glacial event. Tectonophysics 1988, 148, 211–219. [Google Scholar] [CrossRef]
- Casimiro-Soriguer, R.; Talavera, M.; Balao, F.; Terrab, A.; Herrera, J.; Talavera, S. Phylogeny and genetic structure of Erophaca (Leguminosae), a East-West Mediterranean disjunct genus from the Tertiary. Mol. Phylogenet. Evol. 2010, 56, 441–450. [Google Scholar] [CrossRef]
- Fernández-Mazuecos, M.; Jiménez-Mejías, P.; Rotllan-Puig, X.; Vargas, P. Perspectives in plant ecology, evolution and systematics narrow endemics to Mediterranean islands: Moderate genetic diversity but narrow climatic niche of the ancient, critically endangered Naufraga (Apiaceae). Perspect. Plant Ecol. Evol. Syst. 2014, 16, 190–202. [Google Scholar] [CrossRef]
- García-Castaño, J.L.; Terrab, A.; Ortiz, M.A.; Stuessy, T.F.; Talavera, S. Patterns of phylogeography and vicariance of Chamaerops humilis L. (Palmae). Turk. J. Bot. 2014, 38, 1132–1146. [Google Scholar] [CrossRef]
- Zhang, Z.; Ramstein, G.; Schuster, M.; Li, C.; Contoux, C.; Yan, Q. Aridification of the Sahara Desert caused by Tethys Sea shrinkage during the Late Miocene. Nature 2014, 513, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Medail, F.; Diadema, K. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J. Biogeogr. 2009, 36, 1333–1345. [Google Scholar] [CrossRef]
- Popov, S.; Rögl, F.; Rozanov, A.; Steininger, F.F.; Shcherba, I.; Kovac, M. Lithological-paleogeographic maps of Paratethys. Late Eocene to Pliocene. Cour. Forschungsinst. Senckenberg 2004, 250, 1–46. [Google Scholar]
- Rögl, F. Mediterranean and Paratethys. Facts and hypotheses of an Oligocene to Miocene paleogeography (short overview). Geol. Carpath. 1999, 50, 339–349. [Google Scholar]
- Donoghue, M.J.; Smith, S.A. Patterns in the assembly of temperate forests around the Northern Hemisphere. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 1633–1644. [Google Scholar] [CrossRef]
- Grímsson, F.; Denk, T. Fagus from the Miocene of Iceland: Systematics and biogeographical considerations. Rev. Palaeobot. Palynol. 2005, 134, 27–54. [Google Scholar] [CrossRef]
- Grímsson, F.; Denk, T. Floristic turnover in Iceland from 15 to 6 Ma extracting biogeographical signals from fossil floral assemblages. J. Biogeogr. 2007, 34, 1490–1504. [Google Scholar] [CrossRef]
- Grímsson, F.; Denk, T.; Símonarson, L.A. Middle Miocene floras of Iceland—The early colonization of an island? Rev. Palaeobot. Palynol. 2007, 144, 181–219. [Google Scholar] [CrossRef]
- Grímsson, F.; Denk, T.; Zetter, R. Pollen, fruits, and leaves of Tetracentron (Trochodendraceae) from the Cainozoic of Iceland and western North America and their palaeobiogeographic implications. Grana 2008, 47, 1–14. [Google Scholar] [CrossRef]
- Denk, T.; Grimsson, F.; Zetter, R.; Símonarson, L. Late Cainozoic Floras of Iceland: 15 Million Years of Vegetation and Climate History in the Northern North Atlantic; Springer Science & Business Media: New York, NY, USA, 2011; Volume 35. [Google Scholar] [CrossRef]
- Akhmetiev, M.A.; Bratzeva, G.M.; Giterman, R.E.; Golubeva, L.V.; Moiseyeva, A.I. Late Cainozoic Stratigraphy and Flora of Iceland; Academy of Sciences of the USSR: Moscow, Russia, 1978. [Google Scholar]
- Denk, T.; Grímsson, F.; KvaČek, Z. The Miocene floras of Iceland and their significance for late Cainozoic North Atlantic biogeography. Bot. J. Linn. Soc. 2005, 149, 369–417. [Google Scholar] [CrossRef]
- Milne, R.I. Phylogeny and biogeography of Rhododendron subsection Pontica, a group with a tertiary relict distribution. Mol. Phylogenet. Evol. 2004, 33, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D. Plant Evolution in the Mediterranean: Insights for Conservation, 2nd ed.; Oxford University Press: New York, NY, USA, 2020; pp. 10–20. [Google Scholar]
- Fenster, C.B.; Armbruster, W.S.; Wilson, P.; Dudash, M.R.; Thomson, J.D. Pollination syndromes and floral specialization. Annu. Rev. Ecol. Evol. Syst. 2004, 35, 375–403. [Google Scholar] [CrossRef]
- Williams, I.H.; Martin, A.P.; Clark, S.J. Pollination requirements of linseed (Linum usitatissimum). J. Agric. Sci. 1990, 115, 347–352. [Google Scholar] [CrossRef]
- Kearnsaf1, C.A.; Inouye, D.W. Fly pollination of Linum lewish (Linaceae). Am. J. Bot. 1994, 81, 1091–1095. [Google Scholar] [CrossRef]
- Gürbüz, B. Determination of cross-pollination in flax (Linum usitatissimum) using different experimental designs. J. Agric. Sci. 1999, 133, 31–35. [Google Scholar] [CrossRef]
- Lebel, M.; Obolski, U.; Hadany, L.; Sapir, Y. Pollinator-mediated selection on floral size and tube color in Linum pubescens: Can differential behavior and preference in different times of the day maintain dimorphism? Ecol. Evol. 2018, 8, 1096–1106. [Google Scholar] [CrossRef]
- Whittall, J.B.; Hodges, S.A. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature 2007, 447, 706–709. [Google Scholar] [CrossRef]
- Campbell, D.R.; Bischoff, M.; Lord, J.M.; Robertson, A.W. Flower color influences insect visitation in alpine New Zealand. Ecology 2010, 91, 2638–2649. [Google Scholar] [CrossRef]
- Armbruster, W.S. Can indirect selection and genetic context contribute to trait diversification? A transition-probability study of blossom-colour evolution in two genera. J. Evol. Biol. 2002, 15, 468–486. [Google Scholar] [CrossRef]
- Cooley, A.M.; Carvallo, G.; Willis, J.H. Is floral diversification associated with pollinator divergence? Flower shape, flower colour and pollinator preference in Chilean Mimulus. Ann. Bot. 2008, 101, 641–650. [Google Scholar] [CrossRef]
- Smith, C.I.; Godsoe, W.K.; Tank, S.; Yoder, J.B.; Pellmyr, O. Distinguishing coevolution from covicariance in an obligate pollination mutualism: Asynchronous divergence in Joshua tree and its pollinators. Evolution 2008, 62, 2676–2687. [Google Scholar] [CrossRef] [PubMed]
- Paget-Seekins, J. Ribes (Grossulariaceae) Pollination in Northern California: Strong Overlap in Visitor Assemblages Despite Floral Diversity. Master’s Thesis, The Faculty of Humboldt State University, Arcata, CA, USA, 2012. [Google Scholar]
- Muchhala, N.; Johnsen, S.; Smith, S.D. Competition for hummingbird pollination shapes flower color variation in Andean Solanaceae. Evolution 2014, 68, 2275–2286. [Google Scholar] [CrossRef] [PubMed]
- Song, B.M.; Lee, C.H. Toward a mechanistic understanding of color vision in insects. Front. Neural Circuits 2018, 12, 16. [Google Scholar] [CrossRef]
- Koski, M.H. Macroevolution of flower color patterning: Biased transition rates and correlated evolution with flower size. Front. Plant Sci. 2020, 945. [Google Scholar] [CrossRef]
- Van der Kooi, C.J.; Stavenga, D.G.; Arikawa, K.; Belušič, G.; Kelber, A. Evolution of insect color vision: From spectral sensitivity to visual ecology. Annu. Rev. Entomol. 2021, 66, 435–461. [Google Scholar] [CrossRef]
- Smith, S.D.; Miller, R.E.; Otto, S.P.; FitzJohn, R.G.; Rausher, M.D. The effects of flower color transitions on diversification rates in morning glories (Ipomoea subg. Quamoclit, Convolvulaceae). In Darwin’s Heritage Today, Proceedings of the Darwin 200 Beijing International Conference, Beijing, China, 24–26 October 2009; Higher Education Press: Beijing, China, 2010; pp. 202–226. [Google Scholar]
- Sobel, J.M.; Streisfeld, M.A. Flower color as a model system for studies of plant evo-devo. Front. Plant Sci. 2013, 4, 321. [Google Scholar] [CrossRef]
- Ng, J.; Smith, S.D. How traits shape trees: New approaches for detecting character state-dependent lineage diversification. J. Evol. Biol. 2014, 27, 2035–2045. [Google Scholar] [CrossRef]
- Waser, N.M.; Campbell, D.R. Ecological speciation in flowering plants. In Adaptive Speciation, 1st ed.; Cambridge University Press: Cambridge, UK, 2004; pp. 264–277. [Google Scholar]
- The Angiosperm Phylogeny Group; Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S.; et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Müller, J.; Müller, K.; Neinhuis, C.; Quandt, D. PhyDE-Phylogenetic Data Editor. 2005. Available online: http://www.phyde.de (accessed on 22 January 2021).
- Edgar, R.C. MUSCLE: Multiple sequence aligment with high accurancy and high troughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Farris, J.S.; Kallersjo, M.; Kluge, A.G.; Bult, C. Constructing a significance test for incongruence. Syst. Biol. 1995, 44, 570–572. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP* Phylogenetic Analysis Using Parsimony Version 4.0a168. Available online: http://paup.sc.fsu.edu (accessed on 25 June 2021).
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, A.S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 3 May 2021).
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Xi, Z.; Ruhfel, B.R.; Schaefer, H.; Amorim, A.M.; Sugumaran, M.; Wurdack, K.J.; Endress, P.K.; Matthews, M.L.; Stevens, P.F.; Mathews, S.; et al. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc. Natl. Acad. Sci. USA 2012, 109, 17519–17524. [Google Scholar] [CrossRef]
- Punt, W.; Den Breejen, P. Linaceae. Rev. Palaeobot. Palynol. 1981, 33, 75–115. [Google Scholar] [CrossRef]
- Cavagnetto, C.; Anadón, P. Preliminary palynological data on floristic and climatic changes during the Middle Eocene-Early Oligocene of the eastern Ebro Basin, northeast Spain. Rev. Palaeobot. Palynol. 1996, 92, 281–305. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Virus Evol. 2018, 67, 901. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A. TreeAnnotator v.2.4.3. 2016. Available online: http://beast.community/treeannotator (accessed on 12 May 2021).
- Rogers, C.M. Yellow flowered species of Linum in Eastern North America. Brittonia 1963, 15, 97–122. [Google Scholar] [CrossRef]
- Rogers, C.M. Yellow-flowered Linum (Linaceae) in Texas. Sida 1964, 1, 328–336. [Google Scholar]
- Rogers, C.M. Yellow-flowered species of Linum in Central America and western North America. Brittonia 1968, 20, 107–135. [Google Scholar] [CrossRef]
- POWO, Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. Available online: http://www.plantsoftheworldonline.org/ (accessed on 1 May 2022).
- Yan, Y.; Harris, A.J.; He, X. S-DIVA (Statistical Dispersal-Vicariance Analysis): A tool for inferring biogeographic histories. Mol. Phylogenet. Evol. 2010, 56, 848–850. [Google Scholar] [CrossRef]
- Yu, Y.; Blair, C.; He, X.J. RASP 4: Ancestral State Reconstruction Tool for Multiple Genes and Characters. Mol. Biol. Evol. 2020, 37, 604–606. [Google Scholar] [CrossRef] [PubMed]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 2.75. 2011. Available online: http://mesquiteproject.org (accessed on 22 January 2022).
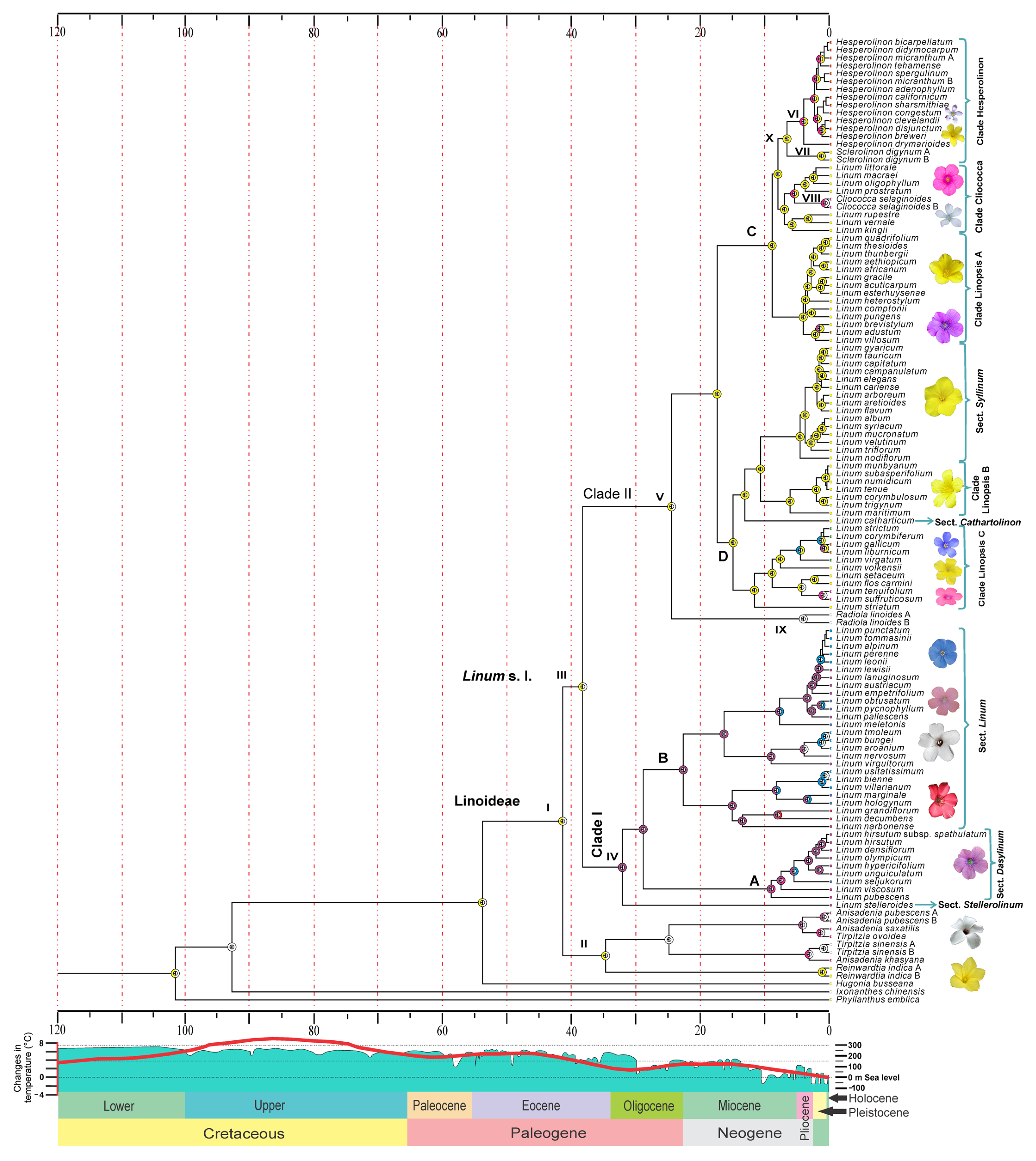
| Code | Node | Mean (Ma) | 95% HPD (Ma) | AC | P | Color Code |
|---|---|---|---|---|---|---|
| I | Linoideae | 41.44 | 36.95–47.45 | AC | 0.25 | A = Yellow C = White D = Purple AC = Yellow + White AD = Yellow + Purple AF = Yellow + Pink CD = White + Purple CF = White + Pink |
| CD | 0.15 | |||||
| II | Anisadenia + Reinwardtia + Tirpitzia | 34.77 | 20.96–46.61 | AC | 0.30 | |
| C | 0.20 | |||||
| III | Linum s.l. | 38.32 | 35.64–42.91 | AC | 0.16 | |
| C | 0.15 | |||||
| IV | Subclade I (sections Dasylinum + Linum + Stellerolinum) | 32.16 | 26.33–37.7 | D | 0.32 | |
| CD | 0.18 | |||||
| V | Subclade II (sections Linopsis + Syllinum + Cathartolinum + segregated genera) | 24.49 | 16.29–33.68 | AC | 0.62 | |
| A | 0.15 | |||||
| VI | Hesperolinon | 3.75 | 1.95–5.78 | AF | 0.86 | |
| AD | 0.14 | |||||
| VII | Sclerolinon | 1.19 | 0.25–2.36 | A | 1.00 | |
| VIII | Cliococca | 0.22 | 0–0.61 | CF | 1.00 | |
| IX | Radiola | 4.01 | 1.15–8.11 | C | 1.00 | |
| X | Clade Hesperolinon (Hesperolinon + Sclerolinon) | 6.55 | 4.14–9.36 | A | 0.84 | |
| AF | 0.15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villalvazo-Hernández, A.; Burgos-Hernández, M.; González, D. Phylogenetic Analysis and Flower Color Evolution of the Subfamily Linoideae (Linaceae). Plants 2022, 11, 1579. https://doi.org/10.3390/plants11121579
Villalvazo-Hernández A, Burgos-Hernández M, González D. Phylogenetic Analysis and Flower Color Evolution of the Subfamily Linoideae (Linaceae). Plants. 2022; 11(12):1579. https://doi.org/10.3390/plants11121579
Chicago/Turabian StyleVillalvazo-Hernández, Alejandra, Mireya Burgos-Hernández, and Dolores González. 2022. "Phylogenetic Analysis and Flower Color Evolution of the Subfamily Linoideae (Linaceae)" Plants 11, no. 12: 1579. https://doi.org/10.3390/plants11121579
APA StyleVillalvazo-Hernández, A., Burgos-Hernández, M., & González, D. (2022). Phylogenetic Analysis and Flower Color Evolution of the Subfamily Linoideae (Linaceae). Plants, 11(12), 1579. https://doi.org/10.3390/plants11121579







