Comparative Transcriptome Analysis of Two Kalanchoë Species during Plantlet Formation
Abstract
:1. Introduction
2. Results
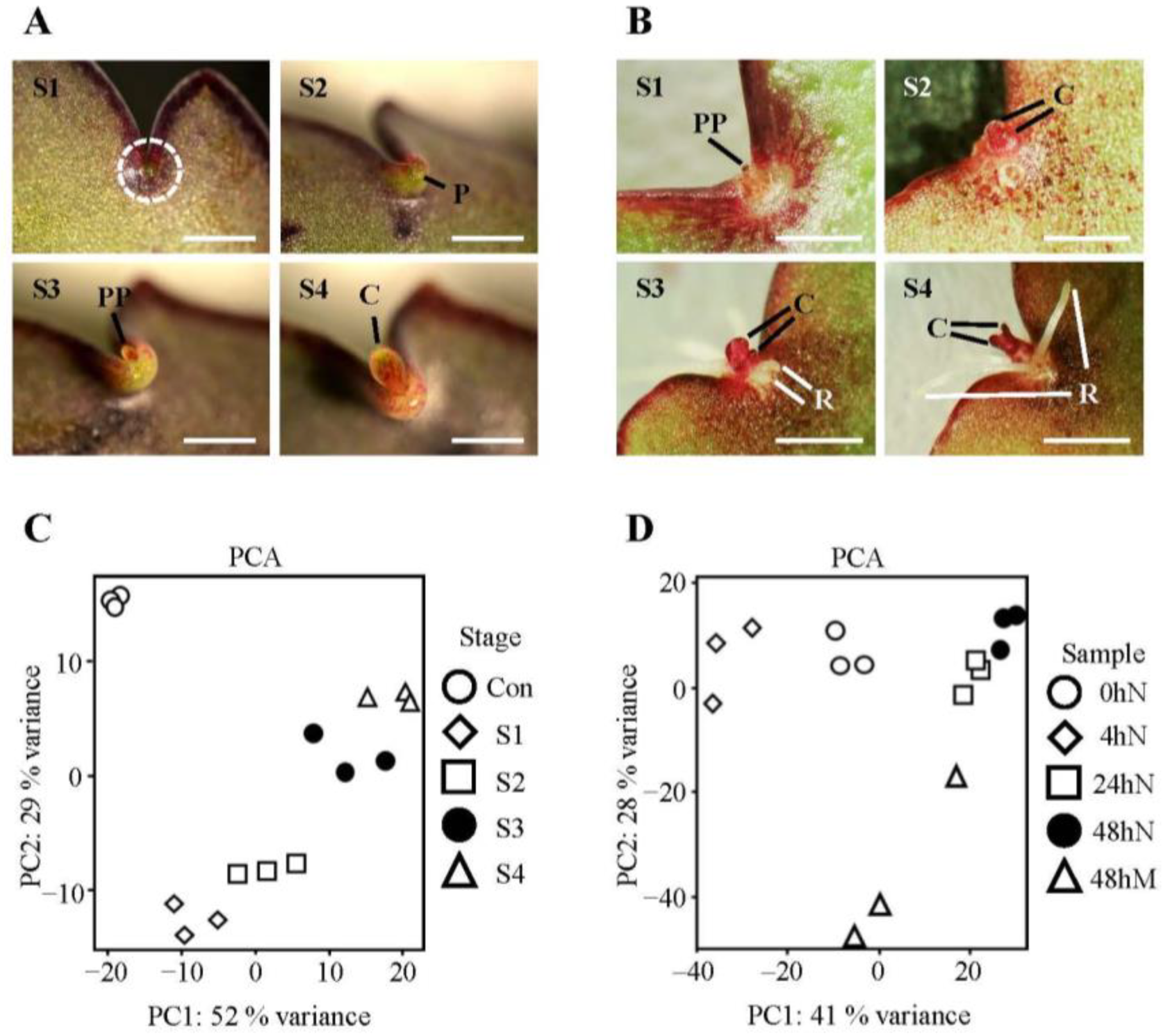
2.1. Morphology of Plantlet Formation and Clustering of Samples from Selected Plantlet Formation Stages and Time Points
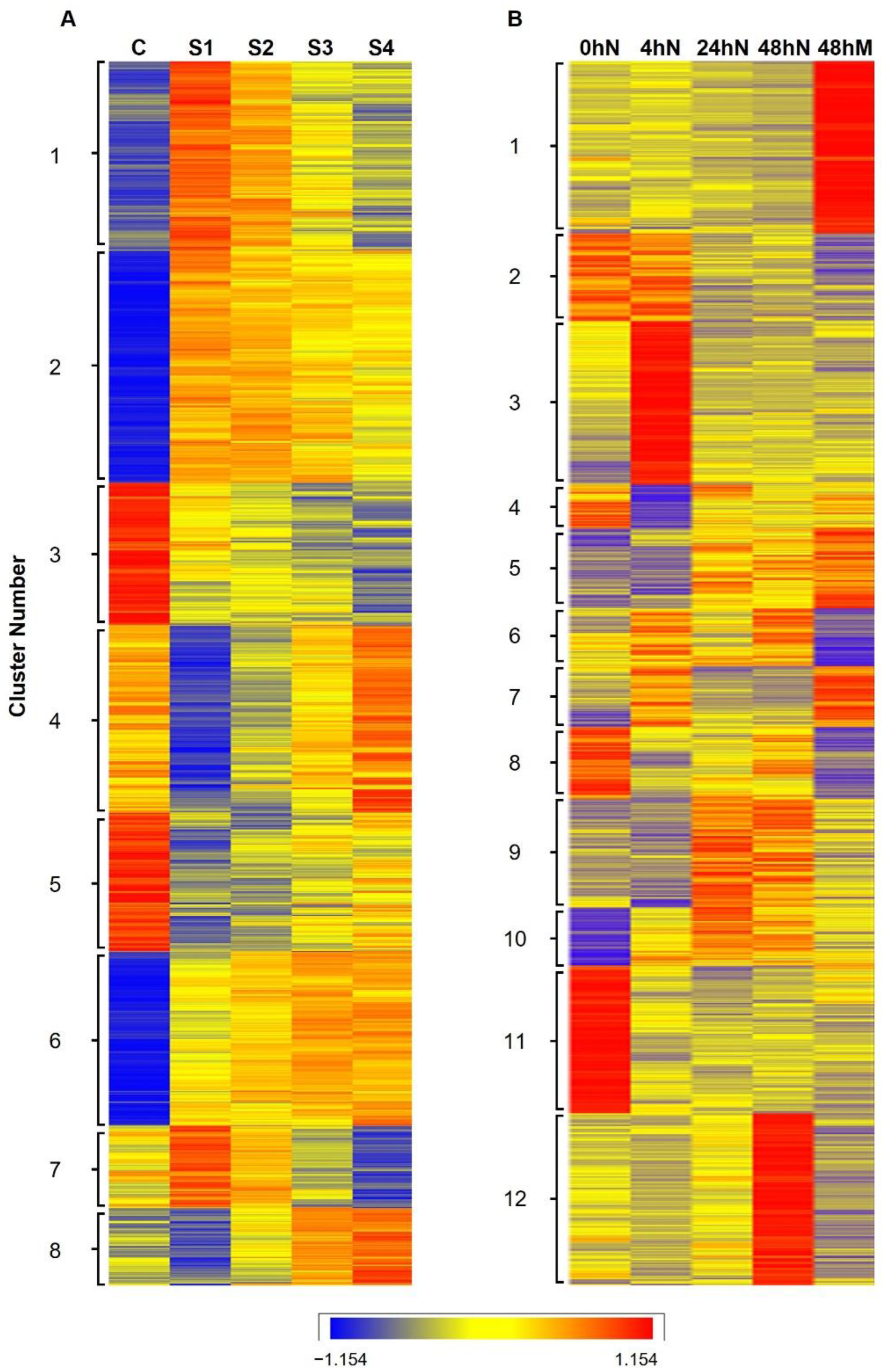
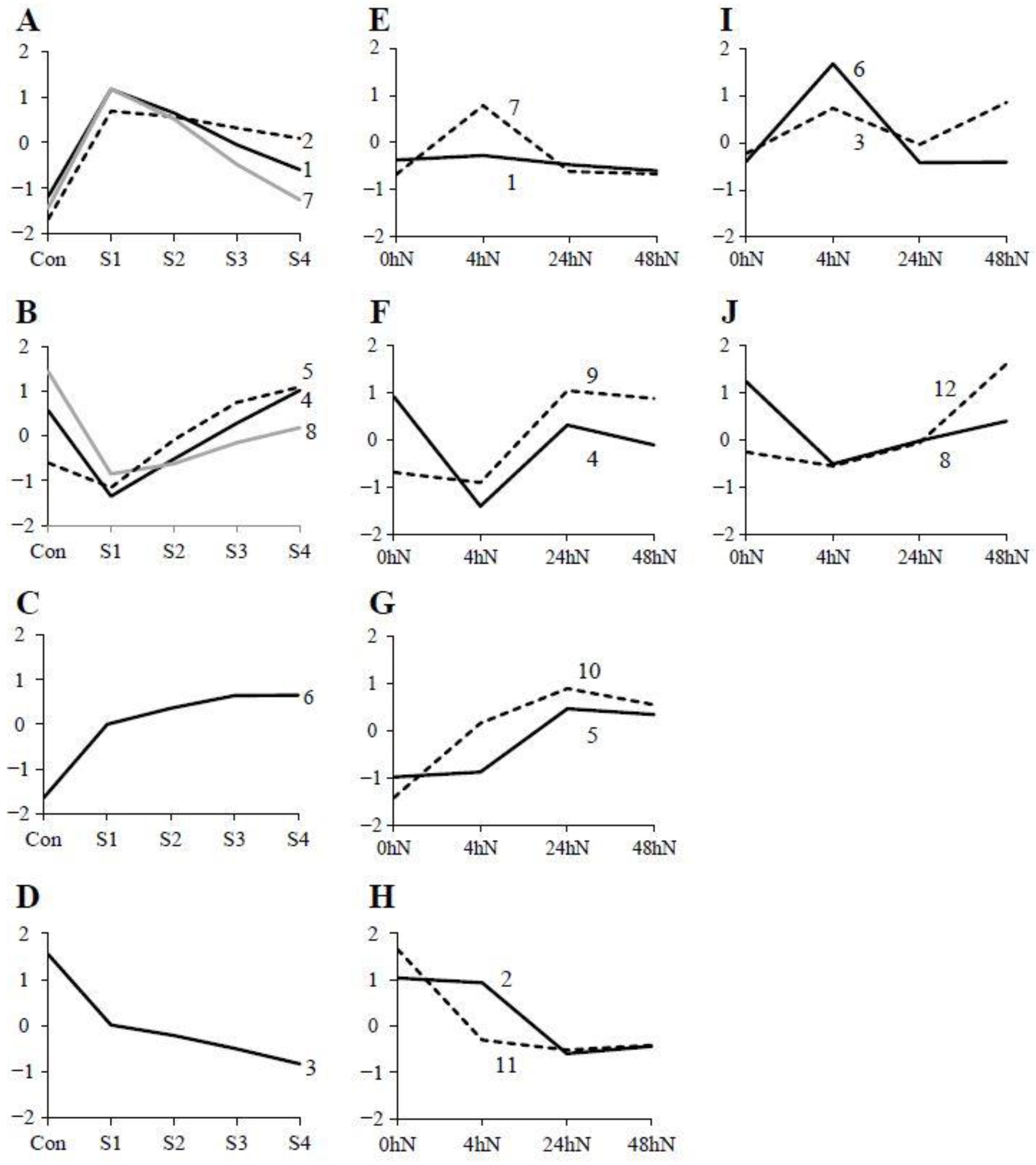
2.2. Heatmap and Graphical Representation of the Expression of Genes in Different Clusters during Plantlet Formation
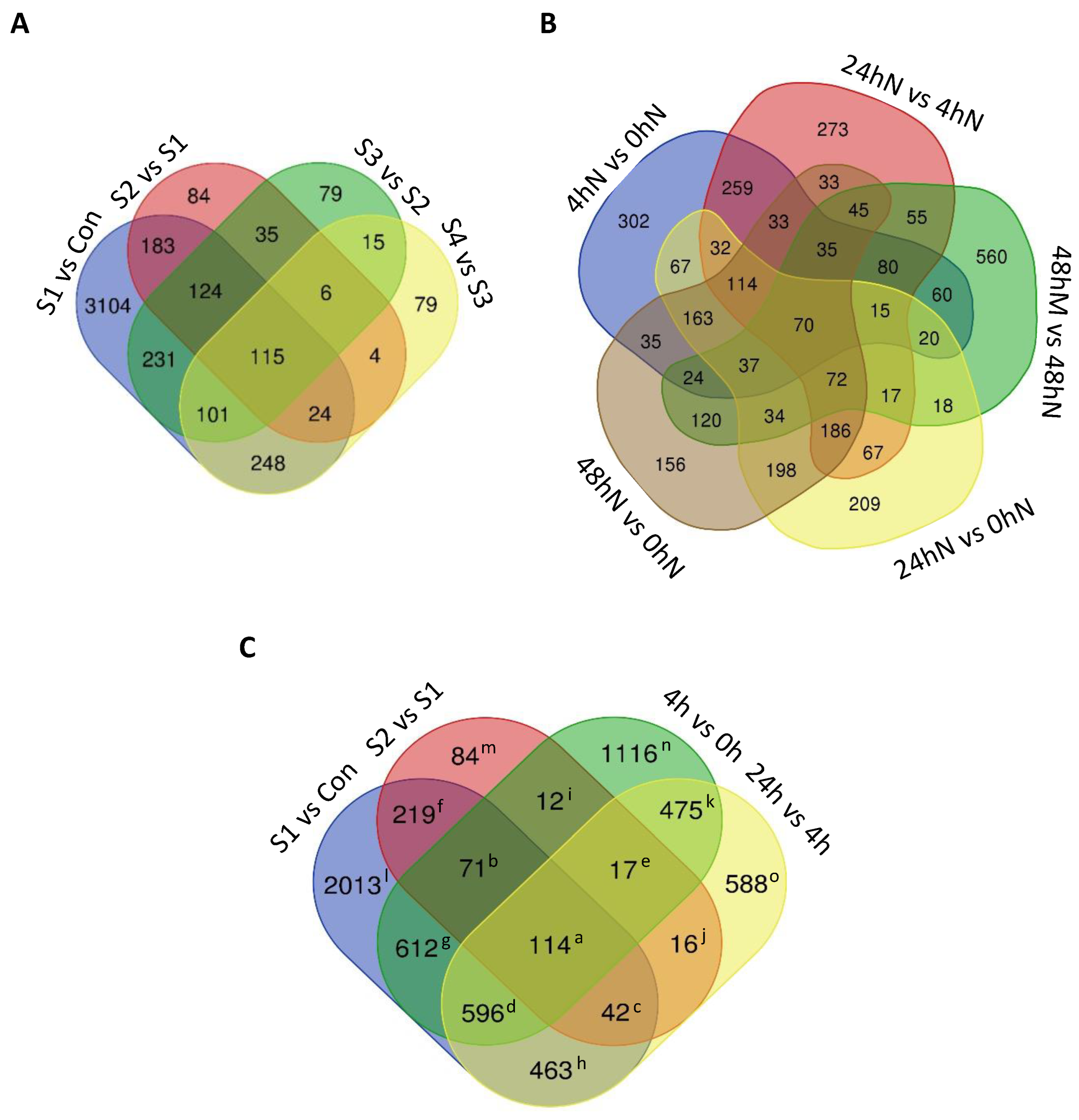
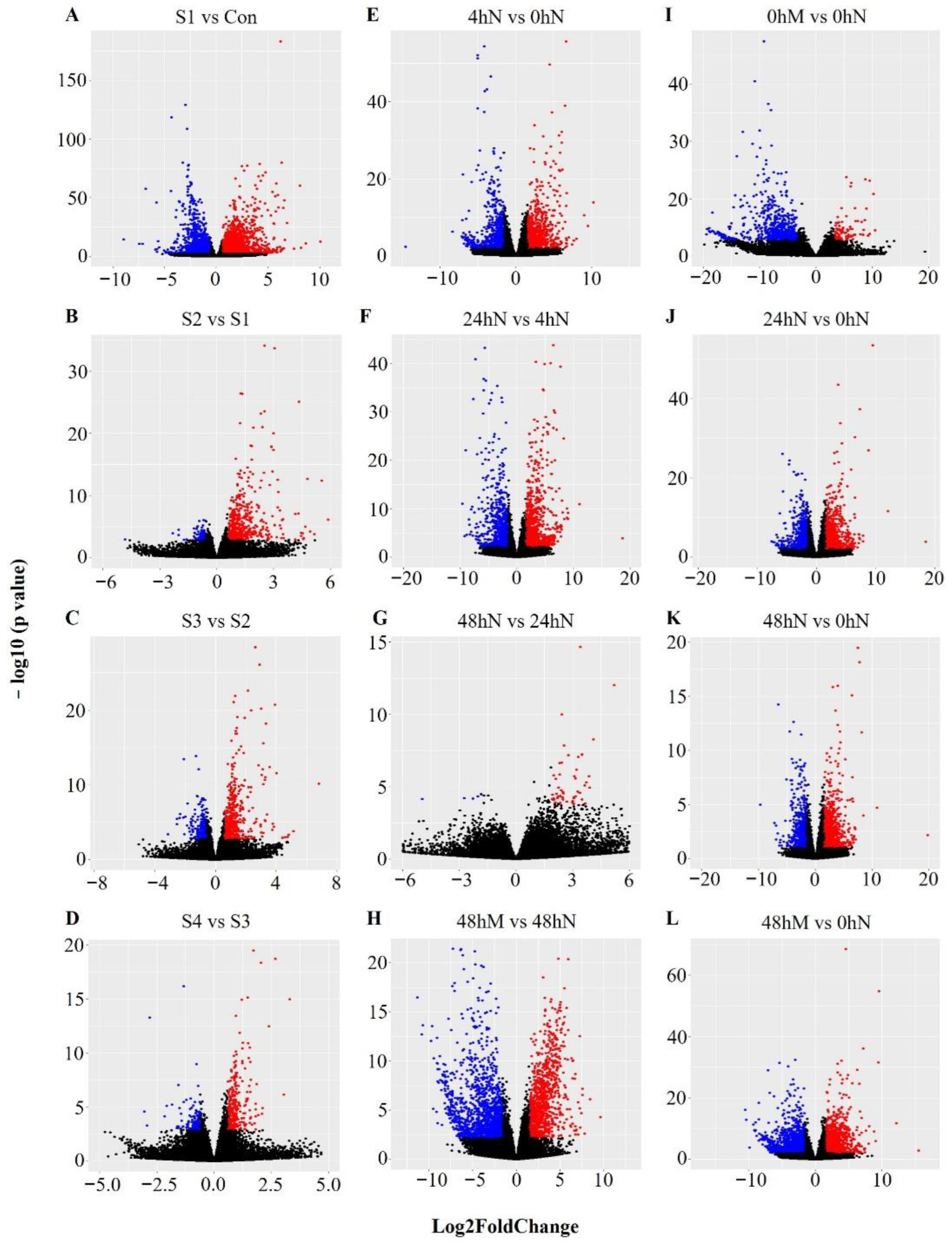
2.3. Number of Significantly Differentially Expressed Genes during Plantlet Formation
2.4. Statistical Significance of Changes in Gene Expression during Plantlet Formation
2.5. Biological Processes during Kalanchoë Plantlet Formation
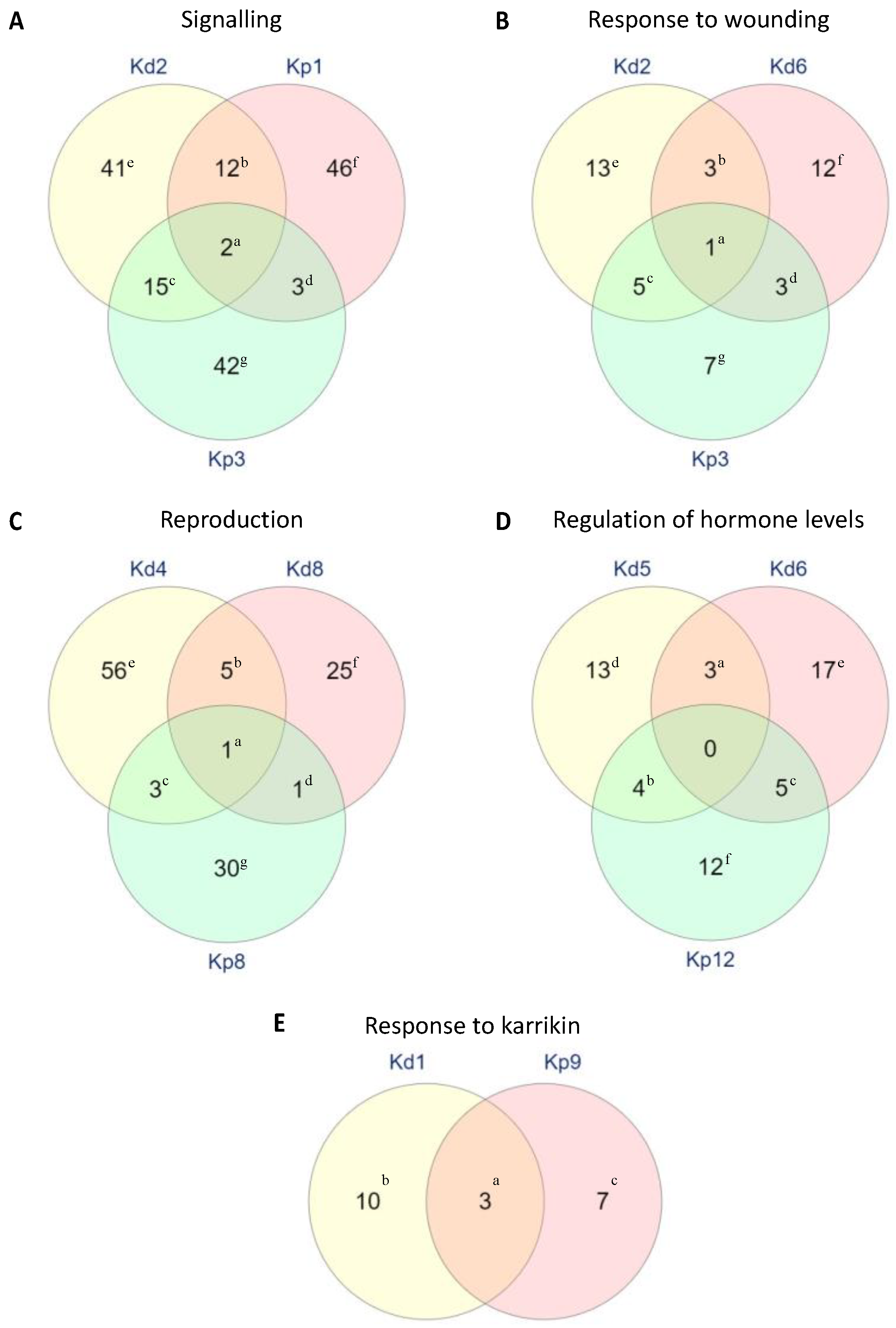
2.6. Specific Functions of DEGs in Selected Biological Processes
2.7. Unique GO Terms in K. daigremontiana and K. pinnata Plantlet Formation
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA Extraction and RNA-Sequencing
4.3. Expression Profiling and Clustering Analysis
4.4. Gene Ontology Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klimeš, L.; Klimešová, J.; Hendriks, R.; van Groenendael, J. Clonal Plant Architecture: A Comparative Analysis of Form and Function. In The Ecology and Evolution of Clonal Plants; Backhuys Publishers: Leiden, The Netherlands, 1997; pp. 1–29. [Google Scholar]
- De Meeûs, T.; Prugnolle, F.; Agnew, P. Asexual reproduction: Genetics and evolutionary aspects. Cell. Mol. Life Sci. 2007, 64, 1355–1372. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Y.; Kim, J.G. The optimal balance between sexual and asexual reproduction in variable environments: A systematic review. J. Ecol. Environ. 2016, 40, 639. [Google Scholar] [CrossRef] [Green Version]
- Barrett, S.C.H. Influences of clonality on plant sexual reproduction. Proc. Natl. Acad. Sci. USA 2015, 112, 8859–8866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, A.; Terada, Y.; Toriba, T.; Kose, K.; Ashikari, M.-Y.; Kyozuka, J. Analysis of Rhizome Development inOryza longistaminata, a Wild Rice Species. Plant Cell Physiol. 2016, 57, 2213–2220. [Google Scholar] [CrossRef] [Green Version]
- Allorge-Boiteau, L. Madagascar centre de speciation et d’origine du genre Kalanchoe (Crassulaceae). Biogeogr. Madag. 1996, 8, 137–145. [Google Scholar]
- Garcês, H.M.; Champagne, C.E.; Townsley, B.T.; Park, S.; Malhó, R.; Pedroso, M.C. Evolution of asexual reproduction in leaves of the genus Kalanchoe. Proc. Natl. Acad. Sci. USA 2007, 104, 15578–15583. [Google Scholar] [CrossRef] [Green Version]
- Gorelick, R. Why Vegetative Propagation of Leaf Cuttings is Possible in Succulent and Semi-Succulent Plants. Haseltonia 2015, 20, 51–57. [Google Scholar] [CrossRef]
- Abdel-Raouf, H.S. Anatomical traits of some species of Kalanchoe (Crassulaceae) and their taxonomic value. Ann. Agric. Sci. 2012, 57, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Garcês, H.; Sinha, N. The ‘Mother of Thousands’ (Kalanchoë daigremontiana): A Plant Model for Asexual Reproduction and CAM Studies. Cold Spring Harb. Protoc. 2009, 2009, pdb.emo133. [Google Scholar] [CrossRef]
- Jaiswal, S.; Sawhney, S. Modulation of TDZ-induced morphogenetic responses by anti-auxin TIBA in bud-bearing foliar explants of Kalanchoe pinnata. Plant Cell Tissue Organ Cult. 2006, 86, 69–76. [Google Scholar] [CrossRef]
- Sawhney, S.; Bansal, A.; Sawhney, N. Epiphyllous bud differentiation in Kalanchoe pinnata: A model developmental system. Indian J. Plant Physiol. 1994, 37, 229–236. [Google Scholar]
- Garcês, H.M.; Koenig, D.; Townsley, B.T.; Kim, M.; Sinha, N.R. Truncation of LEAFY COTYLEDON1 Protein Is Required for Asexual Reproduction in Kalanchoë daigremontiana. Plant Physiol. 2014, 165, 196–206. [Google Scholar] [CrossRef] [Green Version]
- McCready, K.; Spencer, V.; Jácome-Blásquez, F.; Burnett, J.; Sánchez, I.M.V.; Riches, Z.; Kim, M. TARGET OF RAPAMYCIN is essential for asexual vegetative reproduction in Kalanchoë. Plant Physiol. 2021, 189, 248–263. [Google Scholar] [CrossRef]
- Endrizzi, K.; Moussian, B.; Haecker, A.; Levin, J.; Laux, T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996, 10, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Scofield, S.; Dewitte, W.; Murray, J.A. STM sustains stem cell function in theArabidopsisshoot apical meristem and controlsKNOXgene expression independently of the transcriptional repressor AS1. Plant Signal. Behav. 2014, 9, e28934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, J.A.; Moan, E.I.; Medford, J.I.; Barton, M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 1996, 379, 66–69. [Google Scholar] [CrossRef]
- Arick, M.; Hsu, C.-Y. Differential Gene Expression Analysis of Plants. Methods Mol. Biol. 2018, 1783, 279–298. [Google Scholar]
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Luque, F.; Perez, M.D.L.O.L.; Leterrier, M.; Corpas, F.J.; Barroso, J.B. Differential Transcriptomic Analysis by RNA-Seq of GSNO-Responsive Genes Between Arabidopsis Roots and Leaves. Plant Cell Physiol. 2014, 55, 1080–1095. [Google Scholar] [CrossRef]
- Garg, R.; Jain, M. RNA-Seq for Transcriptome Analysis in Non-model Plants. Methods Mol. Biol. 2013, 1069, 43–58. [Google Scholar]
- Zeng, F.; Biligetu, B.; Coulman, B.; Schellenberg, M.P.; Fu, Y.-B. RNA-Seq Analysis of Plant Maturity in Crested Wheatgrass (Agropyron cristatum L.). Genes 2017, 8, 291. [Google Scholar] [CrossRef]
- Khalil-Ur-Rehman, M.; Sun, L.; Li, C.-X.; Faheem, M.; Wang, W.; Tao, J.-M. Comparative RNA-seq based transcriptomic analysis of bud dormancy in grape. BMC Plant Biol. 2017, 17, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Wang, B.; Xi, J.; Zhang, Y.; He, C.; Zheng, J.; Gao, J.; Chen, H.; Zhang, S.; Wu, W.; et al. Transcriptome Comparison Reveals Distinct Selection Patterns in Domesticated and Wild Agave Species, the Important CAM Plants. Int. J. Genom. 2018, 2018, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Kulka, R.G. Hormonal control of root development on epiphyllous plantlets of Bryophyllum (Kalanchoe) marnierianum: Role of auxin and ethylene. J. Exp. Bot. 2008, 59, 2361–2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.; Zhong, T.; Zhu, C.; Zeng, H. Analysis of gene expression in Kalanchoe daigremontiana leaves during plantlet formation under drought stress. Electron. J. Biotechnol. 2013, 16, 4. [Google Scholar] [CrossRef]
- Hershey, D.R. Using the Kalanchoe daigremontiana Plant to Show the Effects of Photoperiodism on Plantlet Formation. Sci. Act. Classr. Proj. Curric. Ideas 2002, 39, 30–34. [Google Scholar] [CrossRef]
- Fehér, A. Somatic embryogenesis—Stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta—Gene Regul. Mech. 2015, 1849, 385–402. [Google Scholar] [CrossRef]
- Castander-Olarieta, A.; Pereira, C.; Sales, E.; Meijón, M.; Arrillaga, I.; Cañal, M.J.; Goicoa, T.; Ugarte, M.D.; Moncaleán, P.; Montalbán, I.A. Induction of Radiata Pine Somatic Embryogenesis at High Temperatures Provokes a Long-Term Decrease in DNA Methylation/Hydroxymethylation and Differential Expression of Stress-Related Genes. Plants 2020, 9, 1762. [Google Scholar] [CrossRef]
- Krishnan, S.R.S.; Siril, E.A. Auxin and nutritional stress coupled somatic embryogenesis in Oldenlandia umbellata L. Physiol. Mol. Biol. Plants 2017, 23, 471–475. [Google Scholar] [CrossRef] [Green Version]
- Nic-Can, G.I.; Loyola-Vargas, V.M. The Role of the Auxins during Somatic Embryogenesis. In Somatic Embryogenesis: Fundamental Aspects and Applications; Springer International Publishing: Cham, Switzerland, 2016; pp. 171–182. [Google Scholar]
- Pandey, D.K.; Chaudhary, B. Oxidative Stress Responsive SERK1 Gene Directs the Progression of Somatic Embryogenesis in Cotton (Gossypium hirsutum L. cv. Coker 310). Am. J. Plant Sci. 2014, 5, 80–102. [Google Scholar] [CrossRef] [Green Version]
- Batygina, T.B.; Bragina, E.A.; Titova, G.E. Morphogenesis of propagules in viviparous species Bryophyllum daigremontianum and B. calycinum. Acta Soc. Bot. Pol. 2014, 65, 127–133. [Google Scholar] [CrossRef]
- Guo, J.; Liu, H.; He, Y.; Cui, X.; Du, X.; Zhu, J. Origination of asexual plantlets in three species of Crassulaceae. Protoplasma 2015, 252, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abat, J.K.; Deswal, R. Nitric Oxide Modulates the Expression of Proteins and Promotes Epiphyllous Bud Differentiation in Kalanchoe pinnata. J. Plant Growth Regul. 2012, 32, 92–101. [Google Scholar] [CrossRef]
- Luo, Y.L.; Su, Z.L.; Cui, X.L.; Lan, Q.Y. Water loss prevention plays a greater role than ROS scavenging in dehydration tolerance of Kalanchoe tubiflora epiphyllous buds. Isr. J. Plant Sci. 2015, 62, 153–159. [Google Scholar] [CrossRef]
- Kulka, R.G. Cytokinins inhibit epiphyllous plantlet development on leaves of Bryophyllum (Kalanchoe) marnierianum. J. Exp. Bot. 2006, 57, 4089–4098. [Google Scholar] [CrossRef]
- Kefu, Z.; Hai, F.; San, Z.; Jie, S. Study on the salt and drought tolerance of Suaeda salsa and Kalanchoe claigremontiana under iso-osmotic salt and water stress. Plant Sci. 2003, 165, 837–844. [Google Scholar] [CrossRef]
- Malwattage, N.R.M.; Garcia, T.M.; Cushman, J.C.; Wone, B.W.M. Enhancing Abiotic Stress Response via a Kalanchoe fedtschenkoi NF-Y Transcription Factor in C 3 Plants. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Yarbrough, J.A. Anatomical and Developmental Studies of the Foliar Embryos of Bryophyllum calycinum. Am. J. Bot. 1932, 19, 443. [Google Scholar] [CrossRef]
- Howe, M.D. A Morphological Study of the Leaf Notches of Bryophyllum calycinum. Am. J. Bot. 1931, 18, 387–390. [Google Scholar] [CrossRef]
- Gouda, M.H.B.; Zhang, C.; Wang, J.; Peng, S.; Chen, Y.; Luo, H.; Yu, L. ROS and MAPK Cascades in the Post-harvest Senescence of Horticultural Products. J. Proteom. Bioinform. 2020, 13, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Jibran, R.; Hunter, D.; Dijkwel, P.P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013, 82, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Pogson, B.J.; Morris, S.C. Postharvest Senescence of Vegetables and its Regulation. In Plant Cell Death Processes; Elsevier: Amsterdam, The Netherlands, 2004; pp. 319–329. [Google Scholar]
- Tsuwamoto, R.; Fukuoka, H.; Takahata, Y. GASSHO1 and GASSHO2 encoding a putative leucine-rich repeat transmembrane-type receptor kinase are essential for the normal development of the epidermal surface in Arabidopsis embryos. Plant J. 2007, 54, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Xing, Q.; Creff, A.; Waters, A.; Tanaka, H.; Goodrich, J.; Ingram, G. ZHOUPI controls embryonic cuticle formation via a signalling pathway involving the subtilisin protease ABNORMAL LEAF-SHAPE1 and the receptor kinases GASSHO1 and GASSHO2. Development 2013, 140, 770–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, B.; Xue, X.-Y.; Hu, W.-L.; Wang, L.-J.; Chen, X.-Y. An ABC Transporter Gene of Arabidopsis thaliana, AtWBC11, is Involved in Cuticle Development and Prevention of Organ Fusion. Plant Cell Physiol. 2007, 48, 1790–1802. [Google Scholar] [CrossRef] [Green Version]
- Panikashvili, D.; Savaldi-Goldstein, S.; Mandel, T.; Yifhar, T.; Franke, R.B.; Höfer, R.; Schreiber, L.; Chory, J.; Aharoni, A. The ArabidopsisDESPERADO/AtWBC11Transporter Is Required for Cutin and Wax Secretion. Plant Physiol. 2007, 145, 1345–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bird, D.; Beisson, F.; Brigham, A.; Shin, J.; Greer, S.; Jetter, R.; Kunst, L.; Wu, X.; Yephremov, A.; Samuels, L. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion†. Plant J. 2007, 52, 485–498. [Google Scholar] [CrossRef]
- Panikashvili, D.; Shi, J.X.; Bocobza, S.; Franke, R.B.; Schreiber, L.; Aharoni, A. The Arabidopsis DSO/ABCG11 Transporter Affects Cutin Metabolism in Reproductive Organs and Suberin in Roots. Mol. Plant 2010, 3, 563–575. [Google Scholar] [CrossRef]
- Nonogaki, H. Repression of transcription factors by microRNA during seed germination and postgerminaiton. Plant Signal. Behav. 2008, 3, 65–67. [Google Scholar] [CrossRef] [Green Version]
- Weitbrecht, K.; Müller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar] [CrossRef] [Green Version]
- Campos, M.; Kang, J.-H.; Howe, G.A. Jasmonate-Triggered Plant Immunity. J. Chem. Ecol. 2014, 40, 657–675. [Google Scholar] [CrossRef]
- Singh, P.; Dave, A.; Vaistij, F.E.; Worrall, D.; Holroyd, G.H.; Wells, J.G.; Kaminski, F.; Graham, I.A.; Roberts, M.R. Jasmonic acid-dependent regulation of seed dormancy following maternal herbivory in Arabidopsis. New Phytol. 2017, 214, 1702–1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [Green Version]
- Pignocchi, C.; Minns, G.E.; Nesi, N.; Koumproglou, R.; Kitsios, G.; Benning, C.; Lloyd, C.W.; Doonan, J.H.; Hills, M.J. ENDOSPERM DEFECTIVE1 Is a Novel Microtubule-Associated Protein Essential for Seed Development inArabidopsis. Plant Cell 2009, 21, 90–105. [Google Scholar] [CrossRef] [Green Version]
- Dilworth, L.L.; Riley, C.K.; Stennett, D.K. Plant Constituents. In Pharmacognosy; Elsevier: Amsterdam, The Netherlands, 2017; pp. 61–80. [Google Scholar]
- Zhao, Y. Auxin Biosynthesis and Its Role in Plant Development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estornell, L.H.; Landberg, K.; Cierlik, I.; Sundberg, E. SHI/STY Genes Affect Pre- and Post-meiotic Anther Processes in Auxin Sensing Domains in Arabidopsis. Front. Plant Sci. 2018, 9, 150. [Google Scholar] [CrossRef] [Green Version]
- Kovtun, Y.; Chiu, W.-L.; Zeng, W.; Sheen, J. Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 1998, 395, 716–720. [Google Scholar] [CrossRef]
- Nelson, D.C.; Riseborough, J.-A.; Flematti, G.R.; Stevens, J.; Ghisalberti, E.L.; Dixon, K.W.; Smith, S.M. Karrikins Discovered in Smoke Trigger Arabidopsis Seed Germination by a Mechanism Requiring Gibberellic Acid Synthesis and Light. Plant Physiol. 2008, 149, 863–873. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Waters, M.T. Perception of karrikins by plants: A continuing enigma. J. Exp. Bot. 2019, 71, 1774–1781. [Google Scholar] [CrossRef]
- Wu, R.; Citovsky, V. Adaptor proteins GIR1 and GIR2. I. Interaction with the repressor GLABRA2 and regulation of root hair development. Biochem. Biophys. Res. Commun. 2017, 488, 547–553. [Google Scholar] [CrossRef]
- Zheng, Z.; Xia, Q.; Dauk, M.; Shen, W.; Selvaraj, G.; Zou, J. Arabidopsis AtGPAT1, a Member of the Membrane-Bound Glycerol-3-Phosphate Acyltransferase Gene Family, Is Essential for Tapetum Differentiation and Male Fertility. Plant Cell 2003, 15, 1872–1887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dotto, M.; Casati, P. Developmental reprogramming by UV-B radiation in plants. Plant Sci. 2017, 264, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Vanhaelewyn, L.; Van Der Straeten, D.; De Coninck, B.; Vandenbussche, F. Ultraviolet Radiation From a Plant Perspective: The Plant-Microorganism Context. Front. Plant Sci. 2020, 11, 597642. [Google Scholar] [CrossRef] [PubMed]
- Boisnard-Lorig, C.; Colon-Carmona, A.; Bauch, M.; Hodge, S.; Doerner, P.; Bancharel, E.; Dumas, C.; Haseloff, J.; Berger, F. Dynamic Analyses of the Expression of the HISTONE::YFP Fusion Protein in Arabidopsis Show That Syncytial Endosperm Is Divided in Mitotic Domains. Plant Cell 2001, 13, 495–509. [Google Scholar] [CrossRef] [Green Version]
- Durand, M.; Mainson, D.; Porcheron, B.; Maurousset, L.; Lemoine, R.; Pourtau, N. Carbon source–sink relationship in Arabidopsis thaliana: The role of sucrose transporters. Planta 2017, 247, 587–611. [Google Scholar] [CrossRef] [Green Version]
- Breia, R.; Conde, A.; Badim, H.; Fortes, A.M.; Gerós, H.; Granell, A. Plant SWEETs: From sugar transport to plant–pathogen interaction and more unexpected physiological roles. Plant Physiol. 2021, 186, 836–852. [Google Scholar] [CrossRef]
- Powikrowska, M.; Oetke, S.; Jensen, P.E.; Krupinska, K. Dynamic composition, shaping and organization of plastid nucleoids. Front. Plant Sci. 2014, 5, 424. [Google Scholar] [CrossRef]
- Aichinger, E.; Kornet, N.; Friedrich, T.; Laux, T. Plant Stem Cell Niches. Annu. Rev. Plant Biol. 2012, 63, 615–636. [Google Scholar] [CrossRef]
- Lup, S.D.; Tian, X.; Xu, J.; Pérez-Pérez, J.M. Wound signaling of regenerative cell reprogramming. Plant Sci. 2016, 250, 178–187. [Google Scholar] [CrossRef]
- Perez-Garcia, P.; Moreno-Risueno, M.A. Stem cells and plant regeneration. Dev. Biol. 2018, 442, 3–12. [Google Scholar] [CrossRef]
- Reinhardt, D.; Pesce, E.-R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Traas, J.; Friml, J.; Kuhlemeier, C. Regulation of phyllotaxis by polar auxin transport. Nature 2003, 426, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zeng, J.; Wu, H.; Tian, Z.; Zhao, Z. A Molecular Framework for Auxin-Controlled Homeostasis of Shoot Stem Cells in Arabidopsis. Mol. Plant 2018, 11, 899–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jácome-Blásquez, F.; Ooi, J.P.; Zeef, L.; Kim, M. Comparative Transcriptome Analysis of Two Kalanchoë Species during Plantlet Formation. Plants 2022, 11, 1643. https://doi.org/10.3390/plants11131643
Jácome-Blásquez F, Ooi JP, Zeef L, Kim M. Comparative Transcriptome Analysis of Two Kalanchoë Species during Plantlet Formation. Plants. 2022; 11(13):1643. https://doi.org/10.3390/plants11131643
Chicago/Turabian StyleJácome-Blásquez, Francisco, Joo Phin Ooi, Leo Zeef, and Minsung Kim. 2022. "Comparative Transcriptome Analysis of Two Kalanchoë Species during Plantlet Formation" Plants 11, no. 13: 1643. https://doi.org/10.3390/plants11131643





