Developmental Differences between Anthers of Diploid and Autotetraploid Rice at Meiosis
Abstract
1. Introduction
2. Results
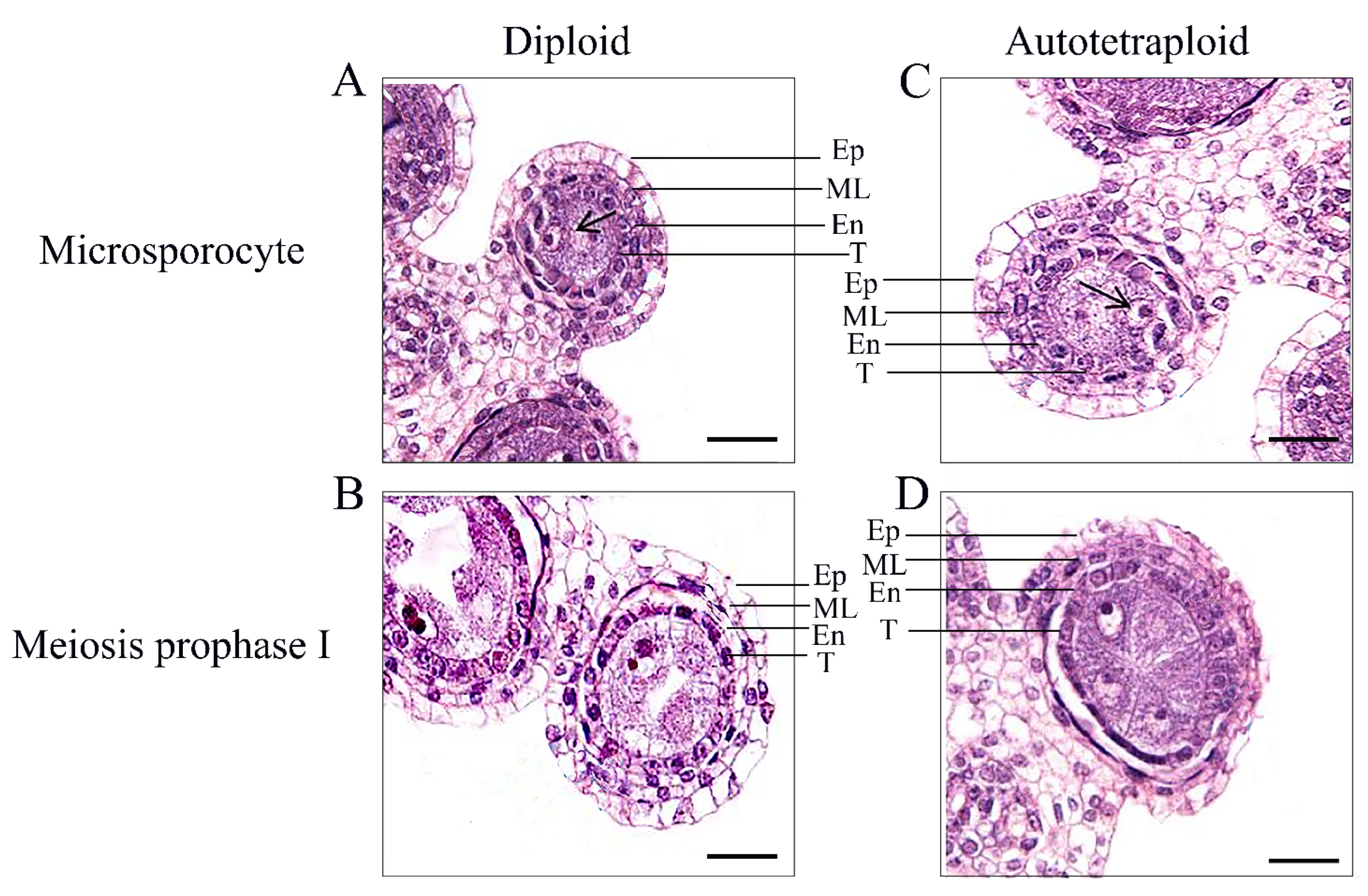
2.1. Cellular Analysis of the Autotetraploid Rice
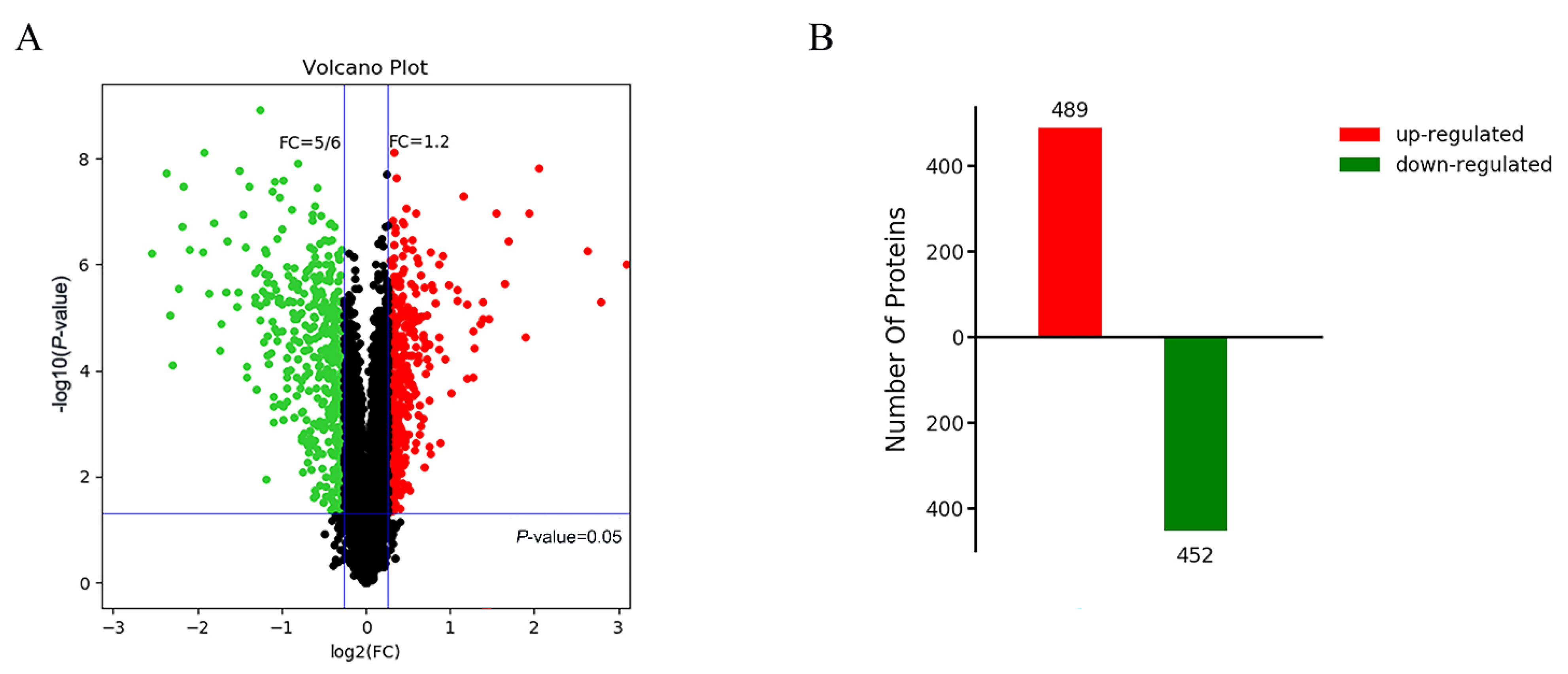
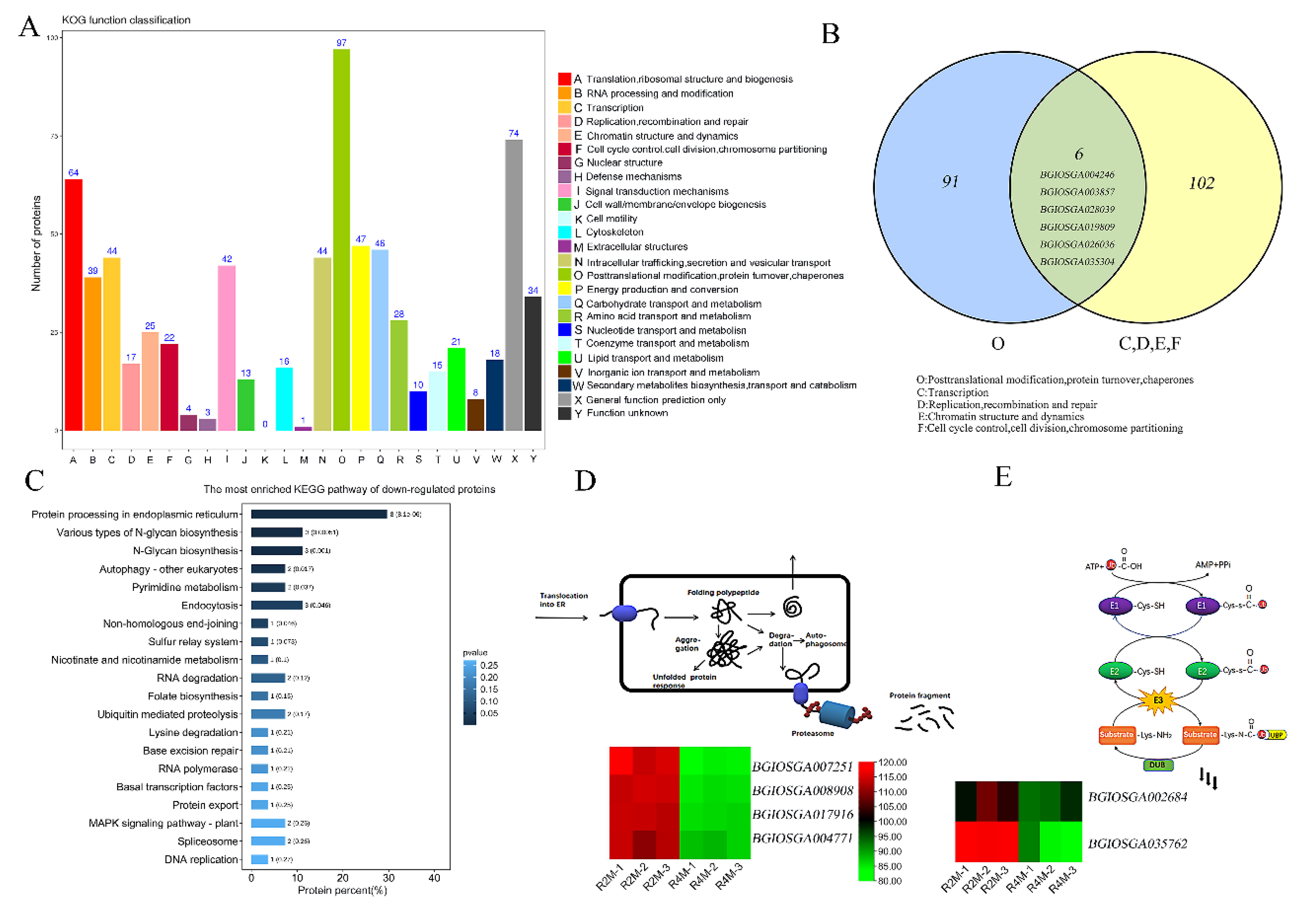
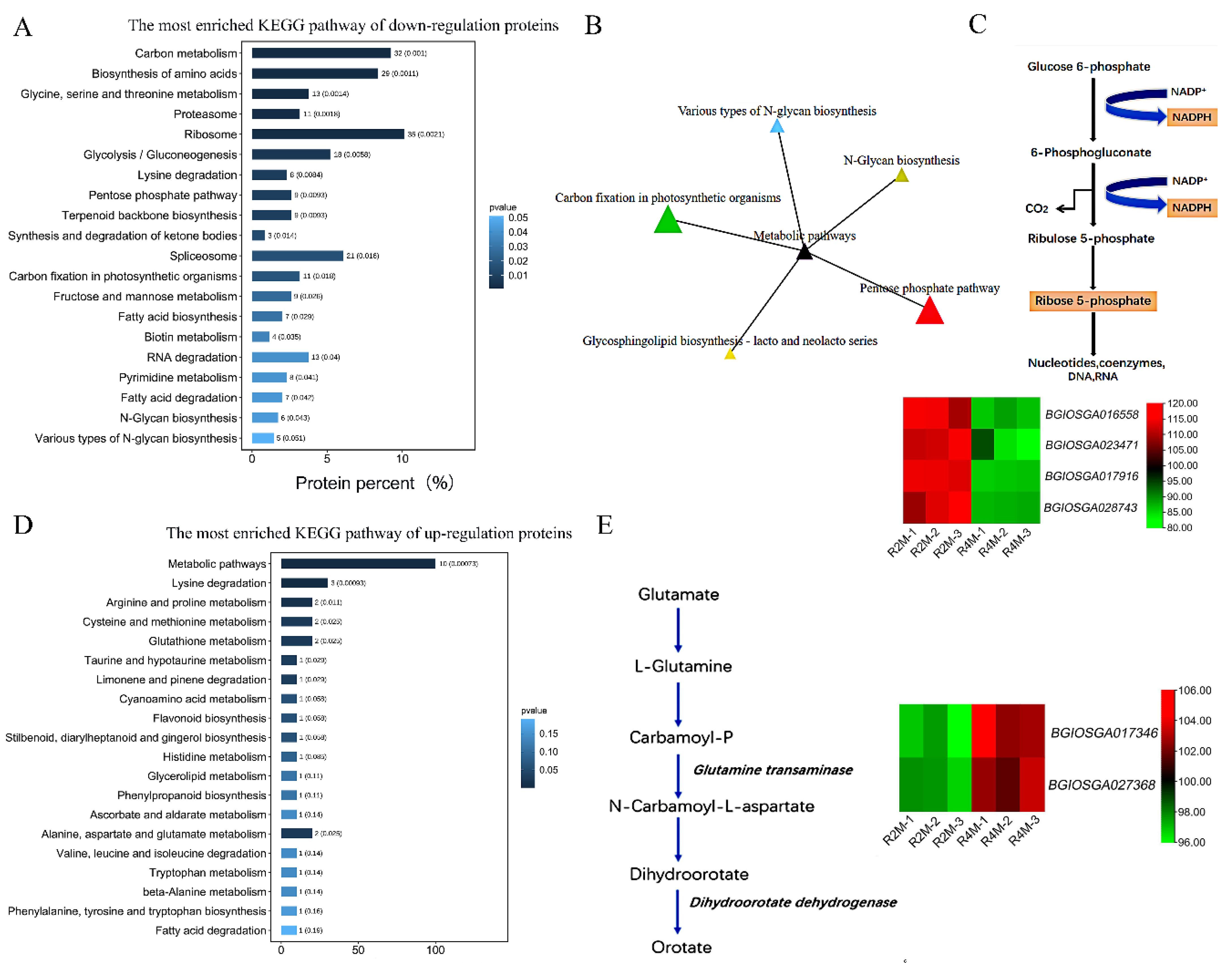
2.2. Post-Translational Modifications and Cell Cycle Regulation Are Related to Abnormal Degradation of Tapetum in Autotetraploid Rice
2.3. Abnormal Phosphopentose Pathway and Decrease of Glutenin in Autotetraploid Rice May Lead to Tapetum Degradation and Affect Pollen Fertility
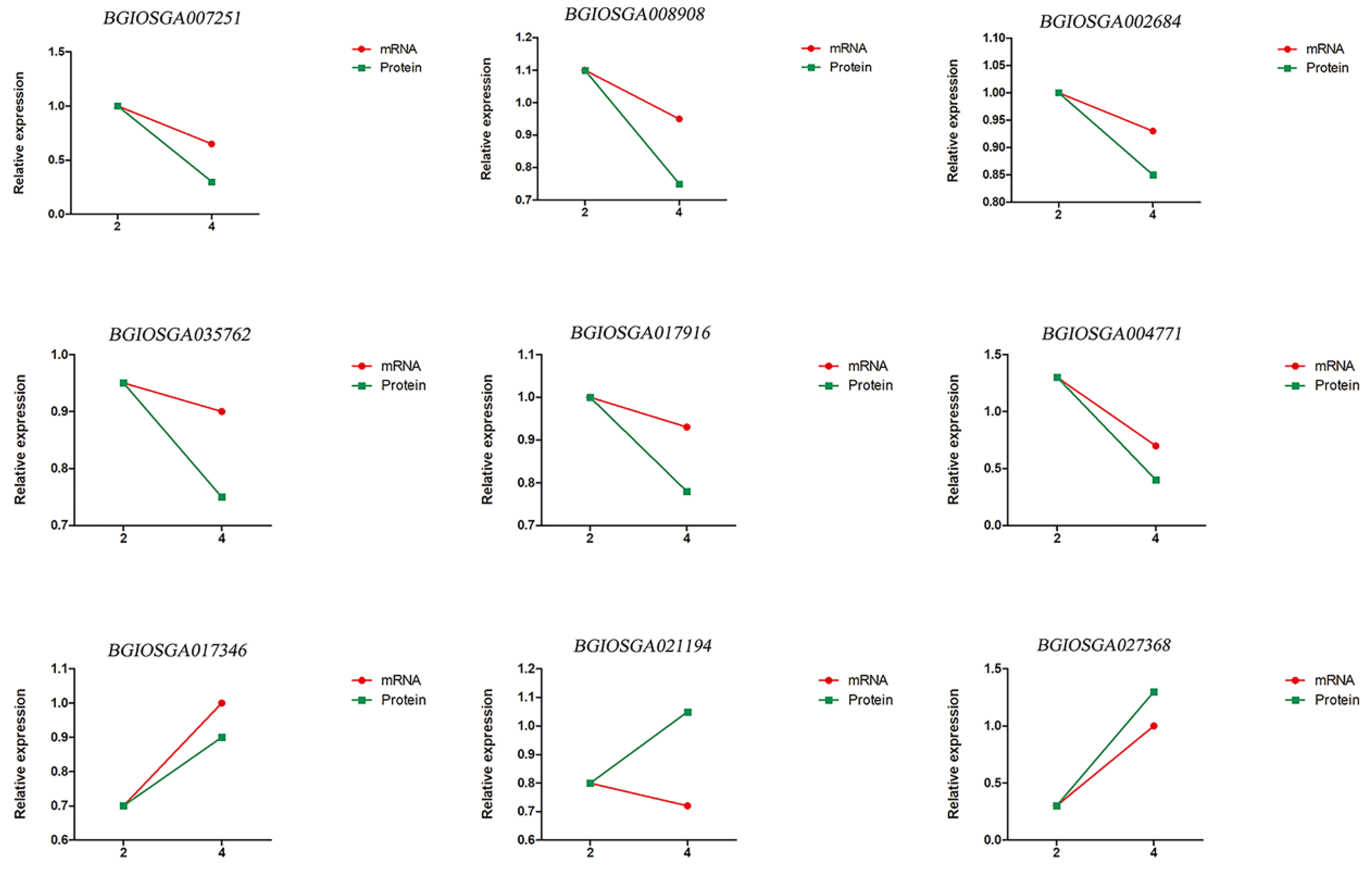
2.4. Verification of Proteomic Results by qRT-PCR (Real-Time Reverse Transcription-PCR)
3. Discussion
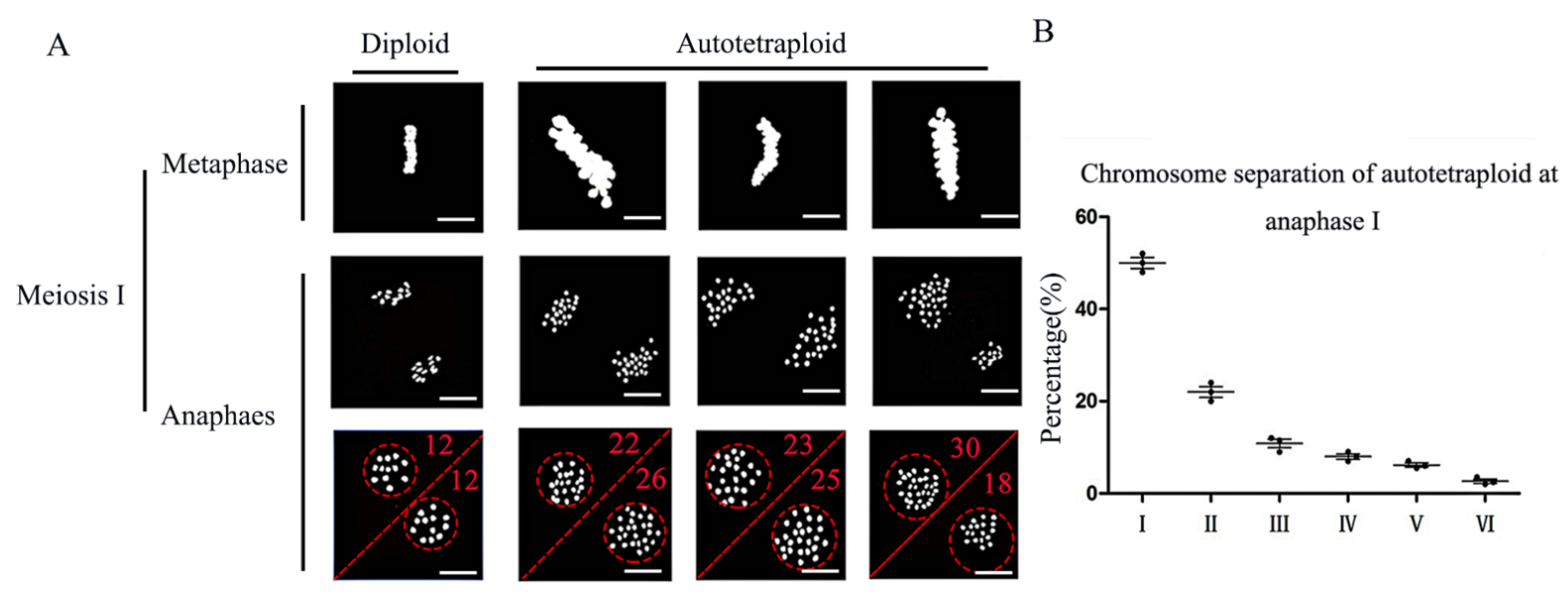
3.1. During Meiosis I, Chromosome Segregation and Degradation of Tapetum Were Abnormal in Autotetraploid Rice Compared with Diploid Rice
3.2. Multi-Pathway Regulation of Degradation of Tapetum
4. Materials and Methods
4.1. Plant Material
4.2. Ploidy Identification and Morphological Observation
4.3. Chromosome Counting
4.4. Pollen Viability Determination
4.5. Paraffin Section
4.6. Observation of Chromosome Behavior during Meiosis
4.7. Determination of Chloroplast Content
4.8. Proteomic Analysis
4.9. Validation Using Quantitative Real-Time PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, R.; Feng, Z.; Zhang, X.; Song, Z.; Cai, D. A New Way of Rice Breeding: Polyploid Rice Breeding. Plants 2021, 10, 422. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Hu, W.; Xing, Y.Z. The history and prospect of rice genetic breeding in China. Hereditas 2018, 40, 841–857. [Google Scholar] [PubMed]
- Koide, Y.; Kuniyoshi, D.; Kishima, Y. Fertile Tetraploids: New Resources for Future Rice Breeding? Front. Plant Sci. 2020, 11, 1231. [Google Scholar] [CrossRef]
- Cai, D.; Chen, J.; Chen, D.; Dai, B.; Zhang, W.; Song, Z.; Yang, Z.; Du, C.; Tang, Z.; He, Y.; et al. The breeding of two polyploid rice lines with the characteristic of polyploid meiosis stability. Sci. China Ser. C Life Sci. 2007, 50, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Shahid, M.Q.; Chen, L.; Chen, Z.; Wang, L.; Liu, X.; Lu, Y. Polyploidy Enhances F1 Pollen Sterility Loci Interactions That Increase Meiosis Abnormalities and Pollen Sterility in Autotetraploid Rice. Plant Physiol. 2015, 169, 2700–2717. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Guo, H.B.; Chen, S.L.; Yang, H.J.; Ghouri, F.; Shahid, M.Q. Comparative study on cytogenetics and transcriptome between diploid and autotetraploid rice hybrids harboring double neutral genes. PLoS ONE 2020, 15, e0239377. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Xia, E.H.; Yao, Q.Y.; Liu, X.D.; Gao, L.Z. Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, E7022–E7029. [Google Scholar] [CrossRef]
- He, Y.C.; Ge, J.; Wei, Q.; Jiang, A.M.; Gan, L.; Song, Z.J.; Cai, D.T. Using a polyploid meiosis stability (PMeS) line as a parent improves embryo development and the seed set rate of a tetraploid rice hybrid. Can. J. Plant Sci. 2011, 91, 325–335. [Google Scholar] [CrossRef]
- Zhang, X.; Zuo, B.; Song, Z.; Wang, W.; He, Y.; Liu, Y.; Cai, D. Breeding and study of two new photoperiod- and thermo-sensitive genic male sterile lines of polyploid rice (Oryza sativa L.). Sci. Rep. 2017, 7, 14744. [Google Scholar] [CrossRef]
- Shahid, M.Q.; Xu, H.M.; Lin, S.Q.; Chen, Z.X.; Liu, A. Genetic analysis and hybrid vigor study of grain yield and other quantitative traits in autotetraploid rice. Pak J. Bot 2011, 44, 237–246. [Google Scholar]
- Luan, L.; Wang, X.; Luan, L.; Long, W.B.; Liu, Y.H.; Tu, S.B.; Zhao, Z.P.; Kong, F.L.; Yu, M.Q. Microsatellite analysis of genetic variation and population genetic differentiation in autotetraploid and diploid rice. Biochem. Genet. 2008, 46, 248–266. [Google Scholar] [CrossRef] [PubMed]
- Nakamori, E. On the Occurrence of the Tetraploid Plant of Rice, Oryza sativa L. Proc. Imp. Acad. 2008, 9, 340–341. [Google Scholar] [CrossRef]
- Nayar, N.M. Origin and Cytogenetics of Rice. Adv. Genet. 1973, 17, 153–292. [Google Scholar]
- Huang, Q.; Sun, J. Study on Reproductive Characters of Autotetraploid Rice. Sci. Agric. Sin. 1999, 32, 14–17. [Google Scholar]
- Huang, Q.C.; Dai, X.M.; Feng, L.Y. Studies on Polyploidizing Induction Technique in Rice. Hybrid Rice 2005, 5, 54–56. [Google Scholar]
- Yang, Q.; Lin, Q.; Ke, L.; Tu, S. Analysis of Genetic Variation and DNA Methylation Alteration in Autotetraploid and Diploid Rice. Chin. J. Appl. Environ. Biol. 2013, 19, 630–636. [Google Scholar] [CrossRef]
- Guo, H.B.; Mendrikahy, J.N.; Xie, L.; Deng, J.F.; Lu, Z.J.; Wu, J.W.; Li, X.; Shahid, M.Q.; Liu, X.D. Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci. Rep. 2017, 7, 40139. [Google Scholar] [CrossRef]
- Liu, X.; Song, Z.; Wei, W.; Jin, J.; Zhang, X.; Liao, S.; Cai, D. Cytomorphological Characterization of the Backcross Progeny of Synthetic Amphiploid Rice (AABB) and Tetraploid Oryza sativa (AAAA). J. Agric. Sci. 2013, 5, 850–861. [Google Scholar]
- Chen, L.; Yuan, Y.; Wu, J.; Chen, Z.; Liu, X. Carbohydrate metabolism and fertility related genes high expression levels promote heterosis in autotetraploid rice harboring double neutral genes. Rice 2019, 12, 1–20. [Google Scholar] [CrossRef]
- Kitada, K.; Kurata, N.; Satoh, H.; Omura, T. Genetic control of meiosis in rice, Oryza sativa L. I. Classification of meiotic mutants induced by MNU and their cytogenetical characteristics. Jpn. J. Genet. 1983, 58, 231–240. [Google Scholar] [CrossRef]
- Xie, X.; Zhang, Z.; Zhao, Z.; Xie, Y.; Li, H.; Ma, X.; Liu, Y.-G.; Chen, L. The mitochondrial aldehyde dehydrogenase OsALDH2b negatively regulates tapetum degeneration in rice. J. Exp. Bot. 2020, 71, 2551–2560. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, Y.-C.; Liao, J.-Y.; Yu, Y.; Zhou, Y.-F.; Feng, Y.-Z.; Yang, Y.-W.; Lei, M.-Q.; Bai, M.; Wu, H.; et al. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 2019, 15, e1008120. [Google Scholar] [CrossRef] [PubMed]
- Petre-Lazar, B.; Livera, G.; Moreno, S.G.; Trautmann, E.; Duquenne, C.; Hanoux, V.; Habert, R.; Coffigny, H. The role of p63 in germ cell apoptosis in the developing testis. J. Cell. Physiol. 2006, 210, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.J.; Guo, X.T.; Huang, Z.Y.; Xia, J.; Li, X.; Wu, J.W.; Yu, H.; Shahid, M.Q.; Liu, X.D. Transcriptome and Gene Editing Analyses Reveal MOF1a Defect Alters the Expression of Genes Associated with Tapetum Development and Chromosome Behavior at Meiosis Stage Resulting in Low Pollen Fertility of Tetraploid Rice. Int. J. Mol. Sci. 2020, 21, 7489. [Google Scholar] [CrossRef]
- Ding, L.; Li, S.C.; Wang, S.Q.; Deng, Q.M.; Zhang, J.; Zheng, A.P.; Wang, L.X.; Chu, M.G.; Zhu, J.; Li, P. Phenotypic characterization and genetic mapping of a new gene required for male and female gametophyte development in rice. Mol. Breed. 2010, 29, 1–12. [Google Scholar] [CrossRef]
- Wang, T.; Li, Y.; Song, S.; Qiu, M.; Zhang, L.; Li, C.; Dong, H.; Li, L.; Wang, J. EMBRYO SAC DEVELOPMENT 1 affects seed setting rate in rice by controlling embryo sac development. Plant Physiol. 2021, 186, 1060–1073. [Google Scholar] [CrossRef]
- Sahebi, M.; Hanafi, M.M.; Rafii, M.Y.; Mahmud, T.M.M.; Azizi, P.; Osman, M.; Abiri, R.; Taheri, S.; Kalhori, N.; Shabanimofrad, M.; et al. Improvement of Drought Tolerance in Rice (Oryza sativa L.): Genetics, Genomic Tools, and the WRKY Gene Family. Biomed. Res. Int. 2018, 2018, 3158474. [Google Scholar] [CrossRef]
- Bai, S.W.; Yu, H.; Wang, B.; Li, J.Y. Retrospective and perspective of rice breeding in China. J. Genet. Genom. 2018, 45, 603–612. [Google Scholar] [CrossRef]
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef]
- Burstin, J.; Devienne, D.; Dubreuil, P.; Damerval, C. Molecular Markers and Protein Quantities as Genetic Descriptors in Maize.1. Genetic Diversity among 21 Inbred Lines. Theor. Appl. Genet. 1994, 89, 943–950. [Google Scholar] [CrossRef]
- Coate, J.E.; Bar, H.; Doyle, J.J. Extensive Translational Regulation of Gene Expression in an Allopolyploid (Glycine dolichocarpa). Plant Cell 2014, 26, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.C.; Liu, Z.M.; Liu, Y.H.; Noriyuki, K.; Hua, Y.C.; Shi, H.R.; Zhao, W.L.; Han, Y.Q.; Toshio, Y.; Chen, W.F.; et al. iTRAQ-Based Proteomics Investigation of Critical Response Proteins in Embryo and Coleoptile During Rice Anaerobic Germination. Rice Sci. 2021, 28, 391–401. [Google Scholar]
- Nonomura, K.I.; Miyoshi, K.; Eiguchi, M.; Suzuki, T.; Miyao, A.; Hirochika, H.; Kurata, N. The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 2003, 15, 1728–1739. [Google Scholar] [CrossRef]
- Wu, J.W.; Shahid, M.Q.; Guo, H.B.; Yin, W.; Chen, Z.X.; Wang, L.; Liu, X.D.; Lu, Y.G. Comparative cytological and transcriptomic analysis of pollen development in autotetraploid and diploid rice. Plant Reprod. 2014, 27, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Garriga, J.; Jayaraman, A.L.; Limon, A.; Jayadeva, G.; Sotillo, E.; Truongcao, M.; Patsialou, A.; Wadzinski, B.E.; Grana, X. A dynamic equilibrium between CDKs and PP2A modulates phosphorylation of pRB, p107 and p130. Cell Cycle 2004, 3, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Kucukali, C.I.; Salman, B.; Yuceer, H.; Ulusoy, C.; Abaci, N.; Ekmekci, S.S.; Tuzun, E.; Bilgic, B.; Hanagasi, H.A. Small ubiquitin-related modifier (SUMO) 3 and SUMO4 gene polymorphisms in Parkinson’s disease. Neurol Res. 2020, 42, 451–457. [Google Scholar] [CrossRef]
- Baarends, W.M.; Hoogerbrugge, J.W.; Roest, H.P.; Ooms, M.; Vreeburg, J.; Hoeijmakers, J.H.; Grootegoed, J. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 1999, 207, 322–333. [Google Scholar] [CrossRef]
- Pratt, W.B.; Toft, D.O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 2003, 228, 111–133. [Google Scholar] [CrossRef]
- Raviol, H.; Sadlish, H.; Rodriguez, F.; Mayer, M.P.; Bukau, B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. Embo. J. 2006, 25, 2510–2518. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Bio. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Peng, J.M.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.M.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Raines, C.A. The Calvin cycle revisited. Photosynth. Res. 2003, 75, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Murata, N. Interruption of the Calvin cycle inhibits the repair of Photosystem II from photodamage. Biochim. Biophys. Acta Bioenerg. 2005, 1708, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Mujahid, H.; Zhang, Y.; Peng, X.; Redoña, E.D.; Wang, C.; Peng, Z. Comprehensive Analysis of the Lysine Succinylome and Protein Co-modifications in Developing Rice Seeds. Mol. Cell. Proteom. 2019, 18, 2359–2372. [Google Scholar] [CrossRef]
- Yamagata, H.; Sugimoto, T.; Tanaka, K.; Kasai, Z. Biosynthesis of Storage Proteins in Developing Rice Seeds. Plant Physiol. 1982, 70, 1094–1100. [Google Scholar] [CrossRef]
- Xian, L.; Long, Y.X.; Yang, M.; Chen, Z.X.; Wu, J.W.; Liu, X.D.; Wang, L. iTRAQ-based quantitative glutelin proteomic analysis reveals differentially expressed proteins in the physiological metabolism process during endosperm development and their impacts on yield and quality in autotetraploid rice. Plant Sci. 2021, 306, 110859. [Google Scholar] [CrossRef]
- Luan, L.; Wang, X.; Long, W.B.; Liu, Y.H.; Tu, S.B.; Xiao, X.Y.; Kong, F.L. A comparative cytogenetic study of the rice (Oryza sativa L.) autotetraploid restorers and hybrids. Russ. J. Genet. 2009, 45, 1074–1081. [Google Scholar] [CrossRef]
- Macaulay, A.D.; Allais, A.; FitzHarris, G. Chromosome dynamics and spindle microtubule establishment in mouse embryos. FASEB J. 2020, 34, 8057–8067. [Google Scholar] [CrossRef]
- Elting, M.W.; Hueschen, C.L.; Udy, D.B.; Dumont, S. Force on spindle microtubule minus ends moves chromosomes. J. Cell Biol. 2014, 206, 245–256. [Google Scholar] [CrossRef]
- Oita, S.; Hayashi, T.; Ohnishi-Kameyama, M. Degradation of Epitope Peptides of Wheat Gliadin and Glutenin for Atopic Dermatitis by Crude Proteases from Germinated Wheat Seeds. Food Sci. Technol. Res. 2009, 15, 639–644. [Google Scholar] [CrossRef][Green Version]
- Chen, Q.; Liu, R.; Wu, Y.; Wei, S.; Wang, Q.; Zheng, Y.; Xia, R.; Shang, X.; Yu, F.; Yang, X.; et al. ERAD-related E2 and E3 enzymes modulate the drought response by regulating the stability of PIP2 aquaporins. Plant Cell 2021, 33, 2883–2898. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Kato, A.; Kobayashi, J.; Yanagihara, H.; Sakamoto, S.; Oliveira, D.N.P.; Shimada, M.; Tauchi, H.; Suzuki, H.; Tashiro, S.; et al. Regulation of Homologous Recombination by RNF20-Dependent H2B Ubiquitination. Mol. Cell 2011, 41, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Fisher, E.A.; Khanna, N.A.; McLeod, R.S. Ubiquitination regulates the assembly of VLDL in HepG2 cells and is the committing step of the apoB-100 ERAD pathway. J. Lipid Res. 2011, 52, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Mao, Q.D.Z.; Jia, J.L.; Wang, Z.Y.; Liu, Y.; Liu, T.T.; Luo, B.W.; Zhang, Z.R. Pentose Phosphate Pathway Regulates Tolerogenic Apoptotic Cell Clearance and Immune Tolerance. Front. Immunol. 2022, 12, 797091. [Google Scholar] [CrossRef]
- Sharkey, T.D. Pentose Phosphate Pathway Reactions in Photosynthesizing Cells. Cells 2021, 10, 1547. [Google Scholar] [CrossRef]
- Cherepanova, O.A.; Byzova, T.V. Pentose phosphate pathway drives vascular maturation. Nat. Metab. 2022, 4, 15–16. [Google Scholar] [CrossRef]
- Liu, J.X.; Zhang, J.W.; Zhu, G.R.; Zhu, D.; Yan, Y.M. Effects of water deficit and high N fertilization on wheat storage protein synthesis, gluten secondary structure, and breadmaking quality. Crop J. 2022, 10, 216–223. [Google Scholar] [CrossRef]
- Udachan, I.; Sahoo, A.K. Quality evaluation of gluten free protein rich broken rice pasta. J. Food Meas. Charact. 2017, 11, 1378–1385. [Google Scholar] [CrossRef]
- Shim, S.H.; Mahong, B.; Lee, S.K.; Kongdin, M.; Lee, C.; Kim, Y.J.; Qu, G.R.; Zhang, D.B.; Cairns, J.R.K.; Jeon, J.S. Rice beta-glucosidase Os12BGlu38 is required for synthesis of intine cell wall and pollen fertility. J. Exp. Bot. 2022, 73, 784–800. [Google Scholar] [CrossRef]
- Ji, C.H.; Li, H.Y.; Chen, L.B.; Xie, M.; Wang, F.P.; Chen, Y.L.; Liu, Y.G. A Novel Rice bHLH Transcription Factor, DTD, Acts Coordinately with TDR in Controlling Tapetum Function and Pollen Development. Mol. Plant 2013, 6, 1715–1718. [Google Scholar] [CrossRef]
- Qin, Q.B.; Wang, Y.D.; Wang, J.; Dai, J.; Liu, Y.; Liu, S.J. Abnormal chromosome behavior during meiosis in the allotetraploid of Carassius auratus red var. (female) x Megalobrama amblycephala (male). BMC Genet. 2014, 15, 95. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Qu, G.; Guo, J.; Wei, F.; Wang, R. Rational design of geranylgeranyl diphosphate synthase enhances carotenoid production and improves photosynthetic efficiency in Nicotiana tabacum. Sci. Bull. 2022, 67, 315–327. [Google Scholar] [CrossRef]
- Shu, L.B.; Ding, W.; Wu, J.H.; Feng, F.J.; Luo, L.J.; Mei, H.W. Proteomic Analysis of Rice Leaves Shows the Different Regulations to Osmotic Stress and Stress Signals. J. Integr. Plant Biol. 2010, 52, 981–995. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Tian, Q.Y.; Zhou, S.Q.; Mao, D.D.; Chen, L.B. A quantitative proteomic analysis of the molecular mechanism underlying fertility conversion in thermo-sensitive genetic male sterility line AnnongS-1′. BMC Plant Biol. 2019, 19, 65. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ku, T.; Gu, H.; Li, Z.; Tian, B.; Xie, Z.; Shi, G.; Chen, W.; Wei, F.; Cao, G. Developmental Differences between Anthers of Diploid and Autotetraploid Rice at Meiosis. Plants 2022, 11, 1647. https://doi.org/10.3390/plants11131647
Ku T, Gu H, Li Z, Tian B, Xie Z, Shi G, Chen W, Wei F, Cao G. Developmental Differences between Anthers of Diploid and Autotetraploid Rice at Meiosis. Plants. 2022; 11(13):1647. https://doi.org/10.3390/plants11131647
Chicago/Turabian StyleKu, Tianya, Huihui Gu, Zishuang Li, Baoming Tian, Zhengqing Xie, Gongyao Shi, Weiwei Chen, Fang Wei, and Gangqiang Cao. 2022. "Developmental Differences between Anthers of Diploid and Autotetraploid Rice at Meiosis" Plants 11, no. 13: 1647. https://doi.org/10.3390/plants11131647
APA StyleKu, T., Gu, H., Li, Z., Tian, B., Xie, Z., Shi, G., Chen, W., Wei, F., & Cao, G. (2022). Developmental Differences between Anthers of Diploid and Autotetraploid Rice at Meiosis. Plants, 11(13), 1647. https://doi.org/10.3390/plants11131647





