Genetic Diversity and Structure of Persian Walnut (Juglans regia L.) in Pakistan: Implications for Conservation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Population Sampling
2.2. DNA Extraction, Microsatellite Amplification, and PCR Product Analysis
2.3. Data Analysis
3. Results
3.1. Genetic Diversity
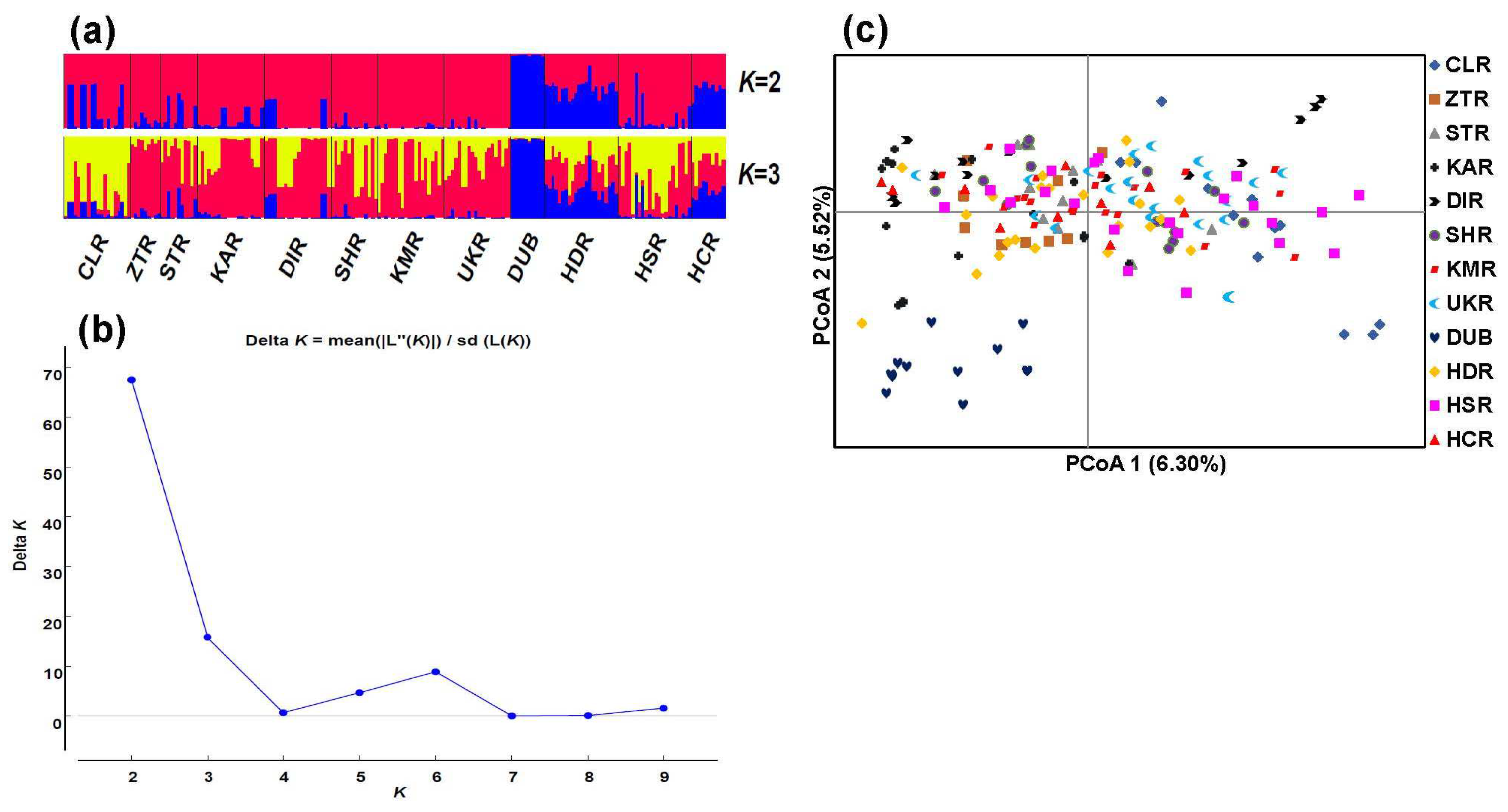
3.2. Population Genetic Structure
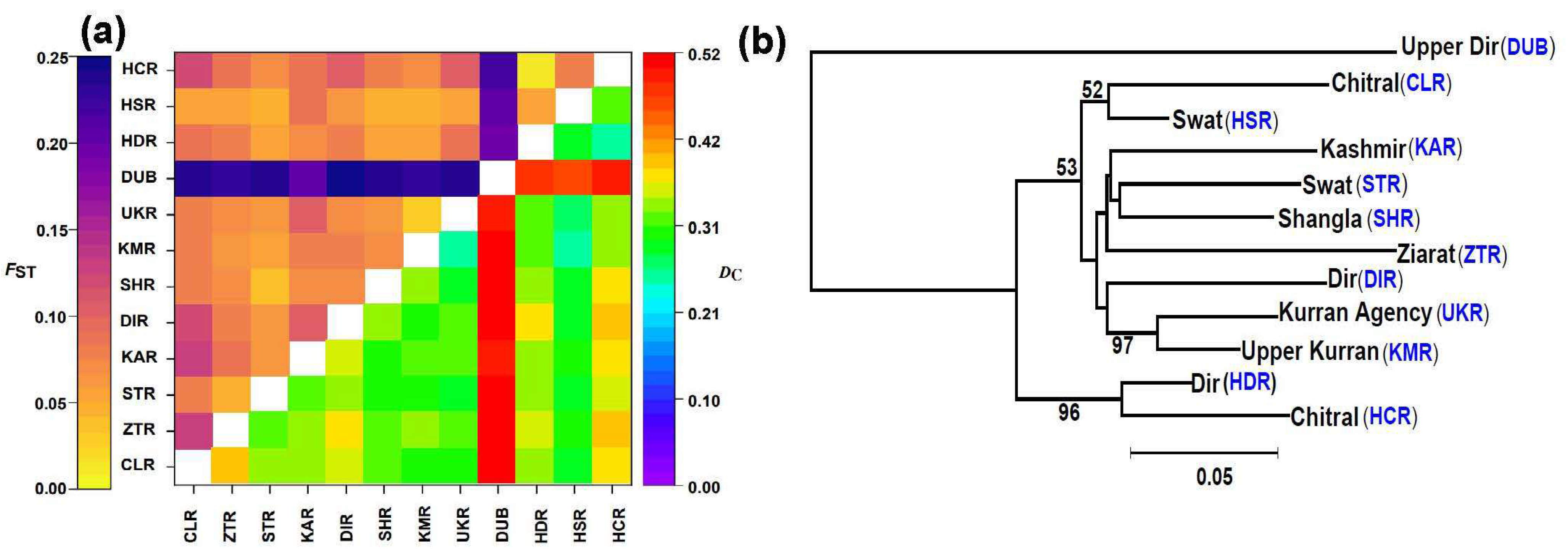
3.2.1. Patterns of Genetic Differentiation
3.2.2. Two Main Genetic Groups
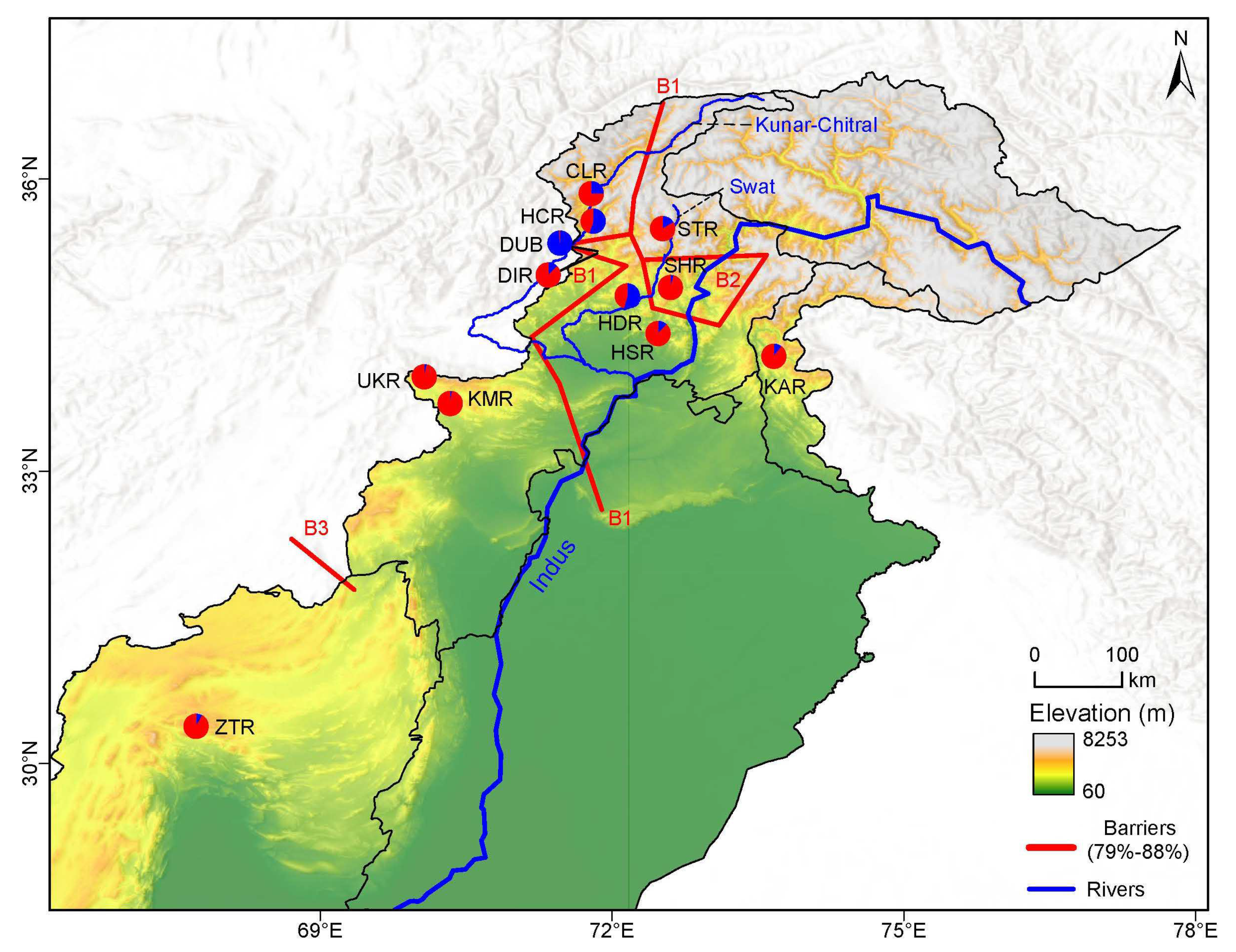
3.2.3. Mantel Test and Genetic Barrier
4. Discussion
4.1. Genetic Diversity of Walnut Populations
4.2. Genetic Structure of Walnut Populations
4.3. Genetic Diversity Hotspots and Conservation Implications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mert, C. Anther and pollen morphology and anatomy in walnut (Juglans regia L.). HortScience 2010, 45, 757–760. [Google Scholar] [CrossRef] [Green Version]
- Luza, J.G.; Polito, V.S. Microsporogenesis and anther differentiation in Juglans regia L.: A developmental basis for heterodichogamy in walnut. Bot. Gaz. 1988, 149, 30–36. [Google Scholar] [CrossRef]
- Hans, A. Chromosome numbers in the Juglandaceae. J. Arnold Arbor. 1970, 51, 534–539. [Google Scholar] [CrossRef]
- Mu, Y.L.; Xi, R. Microsporogenensis studying and karyotype analysis of Juglans regia L. and J. hopeiensis Hu. J. Agric. Univ. Hebei 1988, 11, 48–55. [Google Scholar]
- Tulecke, W.; McGranahan, G.; Ahmadi, H. Regeneration by somatic embryogenesis of triploid plants from endosperm of walnut, Juglans regia L. cv Manregian. Plant Cell Rep. 1988, 7, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Lheureux, F.; Dirlewanger, E. Walnut: Past and future of genetic improvement. Tree Genet. Genomes 2018, 14, 1. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Chen, J. Distinguishing Sichuan walnut cultivars and examining their relationships with Juglans regia and J. sigillata by FISH, early-fruiting gene analysis, and SSR analysis. Front. Plant Sci. 2020, 11, 27. [Google Scholar] [CrossRef]
- Chen, F.; Chen, J.; Wang, Z.; Li, J.; Lin, M.; Song, Y.; Zhong, L. Genomics: Cracking the mysteries of walnuts. J. Genet. 2019, 98, 33. [Google Scholar] [CrossRef]
- Gupta, A.; Behl, T.; Panichayupakaranan, P. A review of phytochemistry and pharmacology profile of Juglans regia. Obes. Med. 2019, 16, 100142. [Google Scholar] [CrossRef]
- Ali, M.; Ullah, A.; Ullah, H.; Khan, F.; Ibrahim, S.M.; Ali, L.; Ahmad, S. Fruit properties and nutritional composition of some walnut cultivars grown in Pakistan. Pak. J. Nutr. 2010, 9, 240–244. [Google Scholar] [CrossRef] [Green Version]
- Aune, D.; Keum, N.; Giovannucci, E.; Fadnes, L.T.; Boffetta, P.; Greenwood, D.C.; Tonstad, S.; Vatten, L.J.; Riboli, E.; Norat, T. Nut consumption and risk of cardiovascular disease, total cancer, all-cause and cause-specific mortality: A systematic review and dose-response meta-analysis of prospective studies. BMC Med. 2016, 14, 207. [Google Scholar] [CrossRef] [Green Version]
- FAO. FAO Statistics Division 2020. Statistics|Food and Agriculture Organization of the United Nations (fao.org). Available online: http://www.fao.org/ag/againfo/resources/en/pubs_sap.html (accessed on 30 November 2021).
- Beer, R.; Kaiser, F.; Schmidt, K.; Ammann, B.; Carraro, G.; Grisa, E.; Tinner, W. Vegetation history of the walnut forests in Kyrgyzstan (Central Asia): Natural or anthropogenic origin? Quat. Sci. Rev. 2008, 27, 621–632. [Google Scholar] [CrossRef]
- Martínez-García, P.J.; Crepeau, M.W.; Puiu, D.; Gonzalez-Ibeas, D.; Whalen, J.; Stevens, K.A.; Paul, R.; Butterfield, T.S.; Britton, M.T.; Reagan, R.L. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 2016, 87, 507–532. [Google Scholar] [CrossRef] [Green Version]
- Pollegioni, P.; Woeste, K.; Chiocchini, F.; Del Lungo, S.; Ciolfi, M.; Olimpieri, I.; Tortolano, V.; Clark, J.; Hemery, G.E.; Mapelli, S. Rethinking the history of common walnut (Juglans regia L.) in Europe: Its origins and human interactions. PLoS ONE 2017, 12, e0172541. [Google Scholar] [CrossRef]
- Avanzato, D.; McGranahan, G.H.; Vahdati, K.; Botu, M.; Iannamico, L.; Assche, J.V. Following Walnut Footprints (Juglans regia L.): Cultivation and Culture, Folklore and History, Traditions and Uses; International Society for Horticultural Science (ISHS): Leuven, Belgium, 2014. [Google Scholar]
- Muhammad, S.; Karadenİz, T. Walnut production status in Pakistan. Bahçe 2017, 46, 113–115. [Google Scholar]
- Khan, M.W.; Khan, I.; Ahmad, H.; Ali, H.; Ghafoor, S.; Afzal, M.; Khan, F.; Shah, M.; Afridi, S. Estimation of genetic diversity in walnut. Pak. J. Bot. 2010, 42, 1791–1796. [Google Scholar]
- Rahman, K.U.; Rab, A.; Ali, N.; Sajid, M.; Nabi, G.; Khan, A.; Rauf, M.A.; Alam, R. RAPD based estimation of genetic diversity in walnut genotypes growing in Malakund division, Pakistan. Pak. J. Agric. Sci. 2015, 52, 997–1003. [Google Scholar]
- Rabadán, A.; Pardo, J.E.; Pardo-Giménez, A.; Álvarez-Ortí, M. Effect of genotype and crop year on the nutritional value of walnut virgin oil and defatted flour. Sci. Total Environ. 2018, 634, 1092–1099. [Google Scholar] [CrossRef]
- Zeneli, G.; Kola, H.; Dida, M. Phenotypic variation in native walnut populations of Northern Albania. Sci. Hortic. 2005, 105, 91–100. [Google Scholar] [CrossRef]
- Thiel, T.; Michalek, W.; Varshney, R.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Mathur, S.; Lawson, S.S.; LaBonte, N.R.; Lorch, A.; Coggeshall, M.V.; Woeste, K.E. Microsatellite Borders and Micro-sequence Conservation in Juglans. Sci. Rep. 2019, 9, 3748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.C.; Jin, Y.C.; Milne, R.I.; Xiahou, Z.Y.; Qin, H.T.; Ye, L.J.; Gao, L.M.; Liu, J.; Li, D.Z. Development of 32 novel microsatellite loci in Juglans sigillata using genomic data. Appl. Plant Sci. 2020, 8, e11328. [Google Scholar] [CrossRef] [Green Version]
- Wambulwa, M.C.; Fan, P.-Z.; Milne, R.; Wu, Z.-Y.; Luo, Y.-H.; Wang, Y.-H.; Wang, H.; Gao, L.-M.; Xiahou, Z.-Y.; Jin, Y.-C.; et al. Genetic analysis of walnut cultivars from southwest China: Implications for germplasm improvement. Plant Divers. 2021; in press. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Zarei, A.; Lawson, S.; Woeste, K.E.; Smulders, M. Genetic diversity and genetic structure of Persian walnut (Juglans regia) accessions from 14 European, African, and Asian countries using SSR markers. Tree Genet. Genomes 2016, 12, 114. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Zarei, A.; McKenna, J.R.; Bujdoso, G.; Woeste, K.E. Genetic diversity of Persian walnut (Juglans regia) in the cold-temperate zone of the United States and Europe. Sci. Hortic. 2017, 220, 36–41. [Google Scholar] [CrossRef]
- Foroni, I.; Woeste, K.; Monti, L.; Rao, R. Identification of ‘Sorrento’walnut using simple sequence repeats (SSRs). Genet. Resour. Crop Evol. 2007, 54, 1081–1094. [Google Scholar] [CrossRef]
- Torokeldiev, N.; Ziehe, M.; Gailing, O.; Finkeldey, R. Genetic diversity and structure of natural Juglans regia L. populations in the southern Kyrgyz Republic revealed by nuclear SSR and EST-SSR markers. Tree Genet. Genomes 2019, 15, 5. [Google Scholar] [CrossRef]
- Vahdati, K.; Mohseni Pourtaklu, S.; Karimi, R.; Barzehkar, R.; Amiri, R.; Mozaffari, M.; Woeste, K. Genetic diversity and gene flow of some Persian walnut populations in southeast of Iran revealed by SSR markers. Plant Syst. Evol. 2014, 301, 691–699. [Google Scholar] [CrossRef]
- Roor, W.; Konrad, H.; Mamadjanov, D.; Geburek, T. Population differentiation in common Walnut (Juglans regia L.) across major parts of its native range—Insights from molecular and morphometric data. J. Hered. 2017, 108, 391–404. [Google Scholar] [CrossRef] [Green Version]
- Doyle, D.A. Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Photochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Liu, J.; Gao, L. Comparative analysis of three different methods of total DNA extraction used in Taxus. Guangxi Zhiwu/Guihaia 2011, 31, 244–249. [Google Scholar]
- Qi, J.; Wang, K.; Wu, C.; Wang, W.; Hao, Y.; Leng, P. Development of EST-SSR markers in Juglans regia. J. Agric. Biotechnol. 2009, 17, 872–876. [Google Scholar]
- Zhang, Z.; Han, J.; Jin, Q.; Wang, Y.; Pang, X.; Li, Y. Development and characterization of new microsatellites for walnut (Juglans regia). Genet. Mol. Res. 2013, 12, 4723–4734. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, T.; Gao, X.-X.; Wang, Y.; Zhang, Q.; Zhou, H.-J.; Zhao, G.-F.; Wang, M.-L.; Woeste, K.E.; Zhao, P. De novo assembly and characterization of the leaf, bud, and fruit transcriptome from the vulnerable tree Juglans mandshurica for the development of 20 new microsatellite markers using Illumina sequencing. Mol. Genet. Genom. 2016, 291, 849–862. [Google Scholar] [CrossRef]
- Topçu, H.; Ikhsan, A.S.; Sütyemez, M.; Çoban, N.; Güney, M.; Kafkas, S. Development of 185 polymorphic simple sequence repeat (SSR) markers from walnut (Juglans regia L.). Sci. Hortic. 2015, 194, 160–167. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel: Population genetic software for teaching and research—An update. Bioinformatics 2012, 6, 288–295. [Google Scholar] [CrossRef] [Green Version]
- Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices, Version 2.9.3. Available online: http://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 20 January 2022).
- Kalinowski, S. hp-rare 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Goudet, J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef] [Green Version]
- Chapuis, M.-P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef] [Green Version]
- Cavalli-Sforza, L.L.; Edwards, A.W. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 1967, 19, 233. [Google Scholar]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Takezaki, N.; Nei, M.; Tamura, K. POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol. Biol. Evol. 2010, 27, 747–752. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. FigTree 1.4. 2 Software; Institute of Evolutionary Biology, University Edinburgh: Edinburgh, UK, 2014. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Ersts, P. Geographic Distance Matrix Generator (Version 1.2.3). In American Museum of Natural History; Center for Biodiversity and Conservation: New York, NY, USA, 2011; pp. 1–4. [Google Scholar]
- Manni, F.; Guérard, E.; Heyer, E. Geographic patterns of (genetic, morphologic, linguistic) variation: How barriers can be detected by using Monmonier’s algorithm. Hum. Biol. 2004, 76, 173–190. [Google Scholar] [CrossRef]
- Monmonier, M.S. Maximum-difference barriers: An alternative numerical regionalization method. Geogr. Anal. 1973, 5, 245–261. [Google Scholar] [CrossRef]
- Dieringer, D.; Schlötterer, C. MICROSATELLITE ANALYSER (MSA): A platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes 2003, 3, 167–169. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Azhar, N.; Khan, M.; Riaz, M.; Bibi, A.; Ali, G.; Begums, S.; Swati, J. Genetic relationship analysis of walnut (Juglans regia L.) grown in western himalayian in azad kashmir, pakistan. J. Anim. Plant Sci. 2019, 29, 500–509. [Google Scholar]
- González, A.V.; Gómez-Silva, V.; Ramírez, M.J.; Fontúrbel, F.E. Meta-analysis of the differential effects of habitat fragmentation and degradation on plant genetic diversity. Conserv. Biol. 2020, 34, 711–720. [Google Scholar] [CrossRef]
- Honnay, O.; Jacquemyn, H. Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv. Biol. 2007, 21, 823–831. [Google Scholar] [CrossRef]
- Poudel, R.C.; Möller, M.; Liu, J.; Gao, L.M.; Baral, S.R.; Li, D.Z. Low genetic diversity and high inbreeding of the endangered yews in Central Himalaya: Implications for conservation of their highly fragmented populations. Divers. Distr. 2014, 20, 1270–1284. [Google Scholar] [CrossRef]
- Cristóbal-Pérez, E.J.; Fuchs, E.J.; Martén-Rodríguez, S.; Quesada, M. Habitat fragmentation negatively affects effective gene flow via pollen, and male and female fitness in the dioecious tree, Spondias purpurea (Anacardiaceae). Biol. Conserv. 2021, 256, 109007. [Google Scholar] [CrossRef]
- Hamadeh, B.; Chalak, L.; Coppens d’Eeckenbrugge, G.; Benoit, L.; Joly, H.I. Evolution of almond genetic diversity and farmer practices in Lebanon: Impacts of the diffusion of a graft-propagated cultivar in a traditional system based on seed-propagation. BMC Plant Biol. 2018, 18, 155. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wang, X.; Yao, J.; Li, Q.; Liu, F.; Yotsukura, N.; Krupnova, T.N.; Duan, D. Effect of domestication on the genetic diversity and structure of Saccharina japonica populations in China. Sci. Rep. 2017, 7, 42158. [Google Scholar] [CrossRef] [Green Version]
- Chybicki, I.J.; Burczyk, J. Simultaneous estimation of null alleles and inbreeding coefficients. J. Hered. 2009, 100, 106–113. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Pan, G.; Ma, Q.; Zhang, J.; Pei, D. The genetic diversity and introgression of Juglans regia and Juglans sigillata in Tibet as revealed by SSR markers. Tree Genet. Genomes 2015, 11, 1–11. [Google Scholar] [CrossRef]
- Abbott, R.J.; Smith, L.C.; Milne, R.I.; Crawford, R.M.; Wolff, K.; Balfour, J. Molecular analysis of plant migration and refugia in the Arctic. Science 2000, 289, 1343–1346. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Gunn, B.F.; Aradhya, M.; Salick, J.M.; Miller, A.J.; Yongping, Y.; Lin, L.; Xian, H. Genetic variation in walnuts (Juglans regia and J. sigillata; Juglandaceae): Species distinctions, human impacts, and the conservation of agrobiodiversity in Yunnan, China. Am. J. Bot. 2010, 97, 660–671. [Google Scholar] [CrossRef] [Green Version]
- Taira, H.; Tsumura, Y.; Tomaru, Y.; Ohba, K. Regeneration system and genetic diversity of Cryptomeria japonica growing at different altitudes. Can. J. For. Res. 1997, 27, 447–452. [Google Scholar] [CrossRef]
- Muriira, N.G.; Muchugi, A.; Yu, A.; Xu, J.; Liu, A. Genetic diversity analysis reveals genetic differentiation and strong population structure in Calotropis plants. Sci. Rep. 2018, 8, 7832. [Google Scholar] [CrossRef]
- Chen, L.; Dong, R.; Ma, Q.; Zhang, Y.; Xu, S.; Ning, D.; Chen, Q.; Pei, D. Precocious genotypes and homozygous tendency generated by self-pollination in walnut. BMC Plant Biol. 2018, 18, 323. [Google Scholar] [CrossRef]
- Lusini, I.; Velichkov, I.; Pollegioni, P.; Chiocchini, F.; Hinkov, G.; Zlatanov, T.; Cherubini, M.; Mattioni, C. Estimating the genetic diversity and spatial structure of Bulgarian Castanea sativa populations by SSRs: Implications for conservation. Conserv. Genet. 2014, 15, 283–293. [Google Scholar] [CrossRef]
- Meng, J.; He, S.-L.; Li, D.-Z.; Yi, T.-S. Nuclear genetic variation of Rosa odorata var. gigantea (Rosaceae): Population structure and conservation implications. Tree Genet. Genomes 2016, 12, 1–14. [Google Scholar] [CrossRef]
- Pollegioni, P.; Woeste, K.E.; Chiocchini, F.; Olimpieri, I.; Tortolano, V.; Clark, J.; Hemery, G.E.; Mapelli, S.; Malvolti, M.E. Landscape genetics of Persian walnut (Juglans regia L.) across its Asian range. Tree Genet. Genomes 2014, 10, 1027–1043. [Google Scholar] [CrossRef]
- Francisco-Ortega, J.; Santos-Guerra, A.; Kim, S.C.; Crawford, D.J. Plant genetic diversity in the Canary Islands: A conservation perspective. Am. J. Bot. 2000, 87, 909–919. [Google Scholar] [CrossRef] [Green Version]

| Sampling Site | ID | N | Latitude | Longitude | Elevation (m) | Habitat |
|---|---|---|---|---|---|---|
| Chitral | CLR | 20 | 35.7713 | 71.7418 | 2753 | Mountainous region |
| Ziarat | ZTR | 9 | 30.3814 | 67.7179 | 2543 | Mountainous region |
| Swat | STR | 11 | 35.506 | 72.5714 | 2075 | Mountainous region |
| Kashmir | KAR | 20 | 34.1809 | 73.6642 | 1152 | Mountainous region |
| Dir 1 | DIR | 20 | 35.1975 | 71.8681 | 1487 | Mountainous region |
| Shangla | SHR | 14 | 34.8873 | 72.6009 | 1904 | Mountainous region |
| Kurram Agency | KMR | 20 | 33.6959 | 70.3368 | 1197 | River side |
| Upper Kurram | UKR | 20 | 33.9702 | 70.0701 | 2964 | Mountainous region |
| Dir upper | DUB | 10 | 35.2119 | 71.8725 | 1514 | Mountainous region |
| Dir 2 | HDR | 22 | 34.8012 | 72.1575 | 1245 | Hilly areas |
| Swat | HSR | 22 | 34.4147 | 72.4735 | 980 | Hilly areas |
| Chitral | HCR | 10 | 35.5688 | 71.8067 | 1359 | Hilly areas |
| POP | NT | NA | HO | HE | UHE | AR | FIS |
|---|---|---|---|---|---|---|---|
| CLR | 112 | 3.613 | 0.564 | 0.541 | 0.555 | 2.200 | −0.016 |
| ZTR | 113 | 3.645 | 0.649 | 0.594 | 0.630 | 2.360 | −0.033 |
| STR | 116 | 3.742 | 0.554 | 0.575 | 0.609 | 2.340 | 0.090 |
| KAR | 128 | 4.129 | 0.628 | 0.591 | 0.606 | 2.320 | −0.038 |
| DIR | 99 | 3.194 | 0.509 | 0.521 | 0.535 | 2.110 | 0.049 |
| SHR | 122 | 3.935 | 0.610 | 0.580 | 0.602 | 2.320 | −0.014 |
| KMR | 120 | 3.871 | 0.563 | 0.568 | 0.583 | 2.230 | 0.036 |
| UKR | 121 | 3.903 | 0.520 | 0.521 | 0.534 | 2.140 | 0.029 |
| DUB | 83 | 2.677 | 0.523 | 0.440 | 0.465 | 1.930 | −0.134 |
| HDR | 142 | 4.581 | 0.542 | 0.614 | 0.629 | 2.390 | 0.141 |
| HSR | 133 | 4.290 | 0.514 | 0.569 | 0.582 | 2.260 | 0.120 |
| HCR | 122 | 3.935 | 0.575 | 0.578 | 0.609 | 2.340 | 0.060 |
| Total | - | - | - | - | - | - | - |
| Mean | 117.583 | 3.793 | 0.563 | 0.558 | 0.578 | - | ─ |
| Source of Variation | Degree of Freedom | Sum of Squares | Variance Components | Percentage Variation (%) | p Value |
|---|---|---|---|---|---|
| Among populations | 11 | 383.111 | 0.588 | 9 | <0.001 |
| Among individuals | 186 | 1577.53 | 0.106 | 2 | <0.001 |
| within populations | |||||
| Within individuals | 198 | 1593.5 | 6.280 | 89 | <0.001 |
| Total | 395 | 3554.14 | 6.974 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magige, E.A.; Fan, P.-Z.; Wambulwa, M.C.; Milne, R.; Wu, Z.-Y.; Luo, Y.-H.; Khan, R.; Wu, H.-Y.; Qi, H.-L.; Zhu, G.-F.; et al. Genetic Diversity and Structure of Persian Walnut (Juglans regia L.) in Pakistan: Implications for Conservation. Plants 2022, 11, 1652. https://doi.org/10.3390/plants11131652
Magige EA, Fan P-Z, Wambulwa MC, Milne R, Wu Z-Y, Luo Y-H, Khan R, Wu H-Y, Qi H-L, Zhu G-F, et al. Genetic Diversity and Structure of Persian Walnut (Juglans regia L.) in Pakistan: Implications for Conservation. Plants. 2022; 11(13):1652. https://doi.org/10.3390/plants11131652
Chicago/Turabian StyleMagige, Ephie A., Peng-Zhen Fan, Moses C. Wambulwa, Richard Milne, Zeng-Yuan Wu, Ya-Huang Luo, Raees Khan, Hong-Yu Wu, Hai-Ling Qi, Guang-Fu Zhu, and et al. 2022. "Genetic Diversity and Structure of Persian Walnut (Juglans regia L.) in Pakistan: Implications for Conservation" Plants 11, no. 13: 1652. https://doi.org/10.3390/plants11131652
APA StyleMagige, E. A., Fan, P.-Z., Wambulwa, M. C., Milne, R., Wu, Z.-Y., Luo, Y.-H., Khan, R., Wu, H.-Y., Qi, H.-L., Zhu, G.-F., Maity, D., Khan, I., Gao, L.-M., & Liu, J. (2022). Genetic Diversity and Structure of Persian Walnut (Juglans regia L.) in Pakistan: Implications for Conservation. Plants, 11(13), 1652. https://doi.org/10.3390/plants11131652







