Multilocus Data Analysis Reveal the Diversity of Cryptic Species in the Tillandsia ionantha (Bromeliaceae: Tillansiodeae) Complex
Abstract
:1. Introduction
2. Results
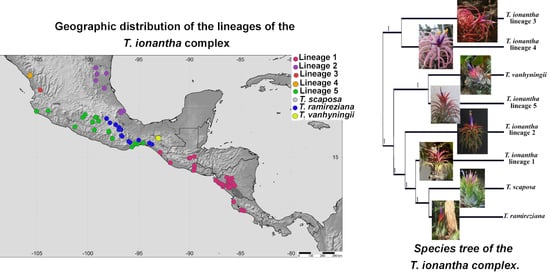
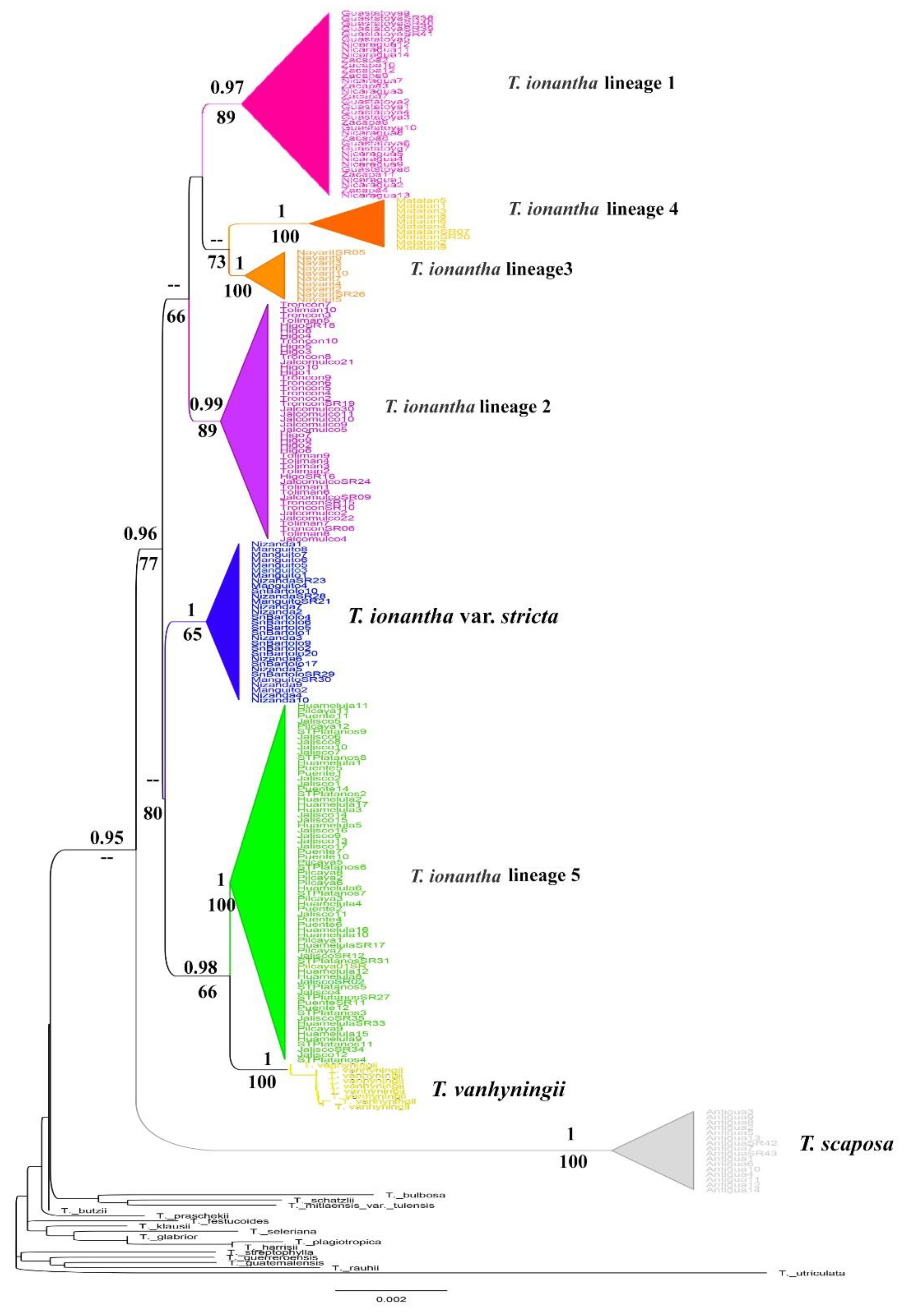
2.1. Phylogenetic Analysis
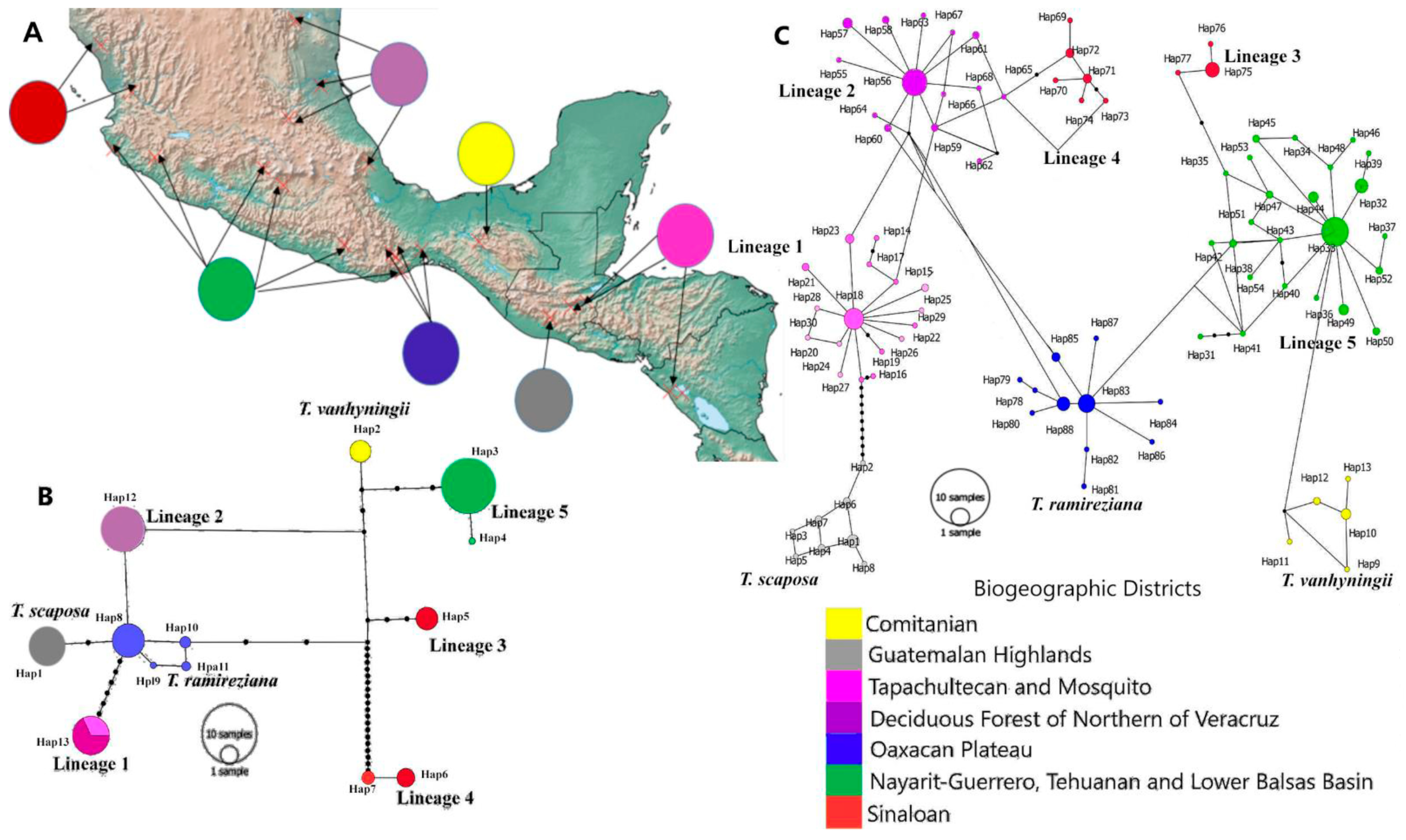
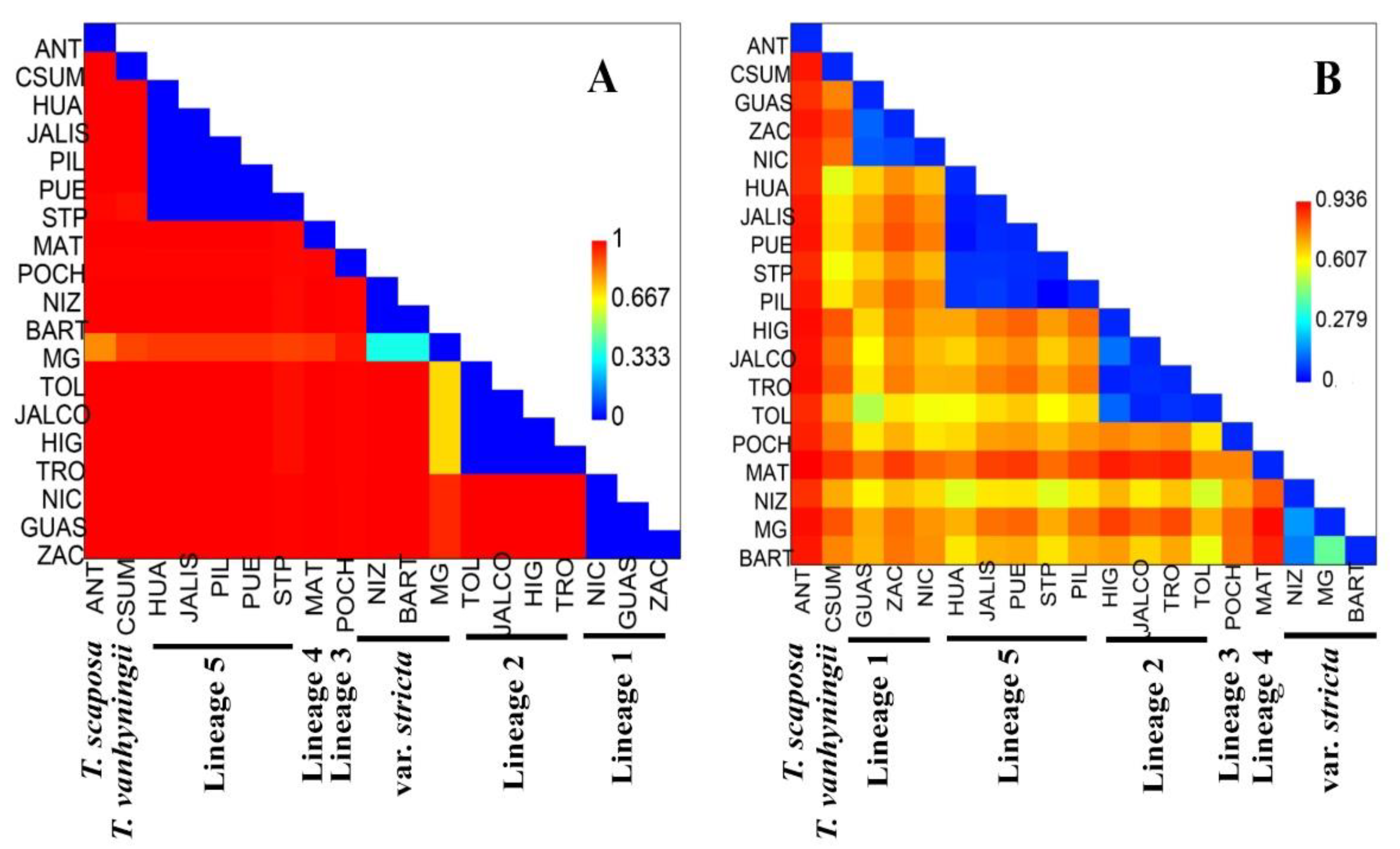
2.2. Genetic Diversity and Structure of cpDNA
2.3. Genetic Diversity and Structure of nDNA
2.4. Multispecies Coalescent
3. Discussion
4. Materials and Methods
4.1. Sampling Strategy
4.2. DNA Isolation, Amplification, and Sequencing
4.3. Phylogenetic Analyses
4.4. Genetic Diversity and Structure Analyzes
4.5. Species Delimitation Using STACEY
5. Conclusions
6. Taxonomy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef]
- Shneyer, V.S.; Kotseruba, V.V. Cryptic species in plants and their detection by genetic differentiation between populations. Russ. J. Genet. Appl. Res. 2015, 5, 528–541. [Google Scholar] [CrossRef]
- Struck, T.H.; Feder, J.L.; Bendiksby, M.; Birkeland, S.; Cerca, J.; Gusarov, V.I.; Kistenich, S.; Larsson, K.H.; Liow, L.H.; Nowak, M.D.; et al. Finding evolutionary processes hidden in cryptic species. Trends Ecol. Evol. 2018, 33, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Harper, J.T.; Gile, G.H.; James, E.R.; Kevin, J.; Carpenter, J.; Keeling, P.J. The inadequacy of morphology for species and genus delineation in microbial eukaryotes: An example from the parabasalian termite symbiont Coronympha. PLoS ONE 2009, 4, e6577. [Google Scholar] [CrossRef] [Green Version]
- Padial, J.M.; de la Riva, I. Taxonomic inflation and the stability of species lists: The perils of ostrich’s behavior. Syst. Biol. 2006, 55, 859–867. [Google Scholar] [CrossRef] [Green Version]
- Padial, J.M.; Miralles, A.; De la Riva, I.; Vences, M. The integrative future of taxonomy. Front. Zool. 2010, 7, 16. [Google Scholar] [CrossRef]
- Schlick-Steiner, B.C.; Steiner, F.M.; Seifert, B.; Stauffer, C.; Christian, E.; Crozier, R.H. Integrative taxonomy: A multisource approach to exploring biodiversity. Annu. Rev. Entomol. 2010, 55, 421–438. [Google Scholar] [CrossRef]
- de Queiroz, K. A unified concept of species and its consequences for the future of taxonomy. Proc. Calif. Acad. Sci. 2005, 56, 196–215. [Google Scholar]
- de Queiroz, K. Species concepts and species delimitation. Syst. Biol. 2007, 56, 879–886. [Google Scholar] [CrossRef] [Green Version]
- Fujita, M.K.; Leaché, A.D.; Burbrink, F.T.; McGuire, J.A.; Moritz, C. Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol. Evol. 2012, 27, 480–488. [Google Scholar] [CrossRef]
- Sites, J.W.; Marshall, J.C. Delimiting species: A Renaissance issue in systematic biology. Trends Ecol. Evol. 2003, 18, 462–470. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Rannala, B. Bayesian species delimitation using multilocus sequence data. Proc. Natl. Acad. Sci. USA 2010, 107, 9264–9269. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Rannala, B. Unguided species delimitation using DNA sequence data from multiple loci. Mol. Biol. Evol. 2014, 31, 3125–3135. [Google Scholar] [CrossRef] [Green Version]
- Rannala, B. The art and science of species delimitation. Curr. Zool. 2015, 61, 846–853. [Google Scholar] [CrossRef] [Green Version]
- Edwards, S. Is a new and general theory of molecular systematics emerging? Evolution 2009, 63, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Ancona, J.J.; Pinzón, J.P.; Ortiz-Díaz, J.J.; Ramírez-Morillo, I.; Tun-Garrido, J.; Palma-Silva, C.; Till, W. Botanical history and typification in the Tillandsia ionantha comaplex. Taxon 2021, 70, 1317–1326. [Google Scholar] [CrossRef]
- Ehlers, R. Tillandsia ionantha und ihre Verwandten. Die Bromel. 2000, 1, 6–9. [Google Scholar]
- Ehlers, R. Tillandsia delicata R. Ehlers sp. nov., eine besonders hübsche Neuentdeckung. Bromelie 1999, 3, 64–70. [Google Scholar]
- Foster, M.B. A new variety of Tilandsia. Bull. Bromel. Soc. 1957, 7, 71. [Google Scholar]
- Beutelspacher, C.R.; García-Martínez, R. Descripción de nuevos híbridos y una nueva combinación en Tillandsia (Bromeliaceae) para Chiapas, México. Lacandonia 2021, 15, 19–28. [Google Scholar]
- Várguez-Zapata, K. Variación Morfométrica del Complejo Tillandsia ionantha Planch. (Bromeliaceae). Master’s Thesis, Universidad Autónoma de Yucatán, Merida, Mexico, 2021. [Google Scholar]
- Ródríguez-Figueroa, S. Filogenia Molecular del Complejo Tillandsia ionantha (Bromeliaceae) y Especies Afines. Master’s Thesis, Universidad Autónoma de Yucatán, Merida, Mexico, 2021. [Google Scholar]
- Cronn, R.; Wendel, J.F. Cryptic trysts, genomic mergers, and plant speciation. New Phytol. 2004, 161, 133–142. [Google Scholar] [CrossRef] [Green Version]
- Linder, C.R.; Rieseberg, L.H. Reconstructing patterns of reticulate evolution in plants. Am. J. Bot. 2004, 91, 1700–1708. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.-B.; Huang, P.-H.; Li, D.-Z.; Wang, H. Incongruence between Nuclear and Chloroplast DNA Phylogenies in Pedicularis Section Cyathophora (Orobanchaceae). PLoS ONE 2013, 8, e74828. [Google Scholar] [CrossRef]
- Wang, W.; Li, H.; Chen, Z. Analysis of plastid and nuclear DNA data in plant phylogenetics evaluation and improvement. Sci. China Life Sci. 2014, 57, 280–286. [Google Scholar] [CrossRef] [Green Version]
- Harrison, R.G. Linking evolutionary pattern and process: The relevance of species concepts for the study of speciation. In Endless Forms: Species and Speciation; Howard, D.J., Berlocher, S.H., Eds.; Oxford University Press: New York, NY, USA, 1998; pp. 19–31. [Google Scholar]
- Morrone, J.J. Hacia una síntesis biogeográfica de México. Rev. Mex. Biodiv. 2005, 76, 207–252. [Google Scholar] [CrossRef]
- Morrone, J.J. Biogeographical regionalisation of the Neotropical region. Zootaxa 2014, 3782, 1–110. [Google Scholar] [CrossRef] [Green Version]
- Morrone, J.J. Regionalización biogeográfica y evolución biótica de México: Encrucijada de la biodiversidad del Nuevo Mundo. Rev. Mex. Biodiv. 2019, 90, e902980. [Google Scholar] [CrossRef]
- Rzedowski, J. Vegetación de México, 1ra. Edición Digital; CONABIO: Mexcio City, México, 1978; 504p. [Google Scholar]
- González-Medrano, F. Las Comunidades Vegetales de México. Segunda Edición; INE-SEMARNAT: Distrito Federal, México, 2004; 88p. [Google Scholar]
- Uribe-Salas, D.; España-Boquera, M.L.; Torres-Miranda, A. Aspectos biogeográficos y ecológicos del género Quercus (Fagaceae) en Michoacán, México. Act. Bot. Mex. 2019, 126. [Google Scholar] [CrossRef] [Green Version]
- Good-Avila, S.V.; Souza, V.; Gaut, B.S.; Eguiarte, L.E. Timing and rate of speciation in Agave (Agavaceae). Proc. Natl. Acad. Sci. USA 2006, 103, 9124–9129. [Google Scholar] [CrossRef] [Green Version]
- De Nova, J.A.; Medina, R.; Montero, J.C.; Weeks, A.; Rosell, J.A.; Olson, M.E.; Eguiarte, L.E.; Magallón, S. Insights into the historical construction of species-rich Mesoamerican seasonally dry tropical forests: The diversification of Bursera (Burseraceae, Sapindales). New Phytol. 2012, 193, 276–287. [Google Scholar] [CrossRef]
- Romero-Soler, K.J.; Ramírez-Morillo, I.M.; Ruiz-Sanchez, E.; Hornung-Leoni, C.T.; Carnevali, G.; Raigoza, N. Phylogenetic relationships within the Mexican genus Bakerantha (Hechtioideae, Bromeliaceae) based on plastid and nuclear DNA: Implications for taxonomy. J. Syst. Evol. 2022, 60, 55–72. [Google Scholar] [CrossRef]
- Pinzón, J.P.; Ramírez-Morillo, I.M.; Carnevali, G.; Barfuss, M.H.J.; Till, W.; Tun, J.; Ortiz-Díaz, J.J. Phylogenetics and evolution of the Tillandsia utriculata complex (Bromeliaceae, Tillandsioideae) inferred from three plastid DNA markers and the ETS of the nuclear ribosomal DNA. Bot. J. Linn. Soc. 2016, 181, 362–390. [Google Scholar] [CrossRef] [Green Version]
- Kessler, M.; Krömer, T. Patterns and ecological correlates of pollination modes among bromeliad communities of Andean forests in Bolivia. Plant Biol. 2000, 2, 659–669. [Google Scholar] [CrossRef]
- Krömer, T.; Kessler, M.; Herzog, S.K. Distribution and Flowering Ecology of Bromeliads along Two Climatically Contrasting Elevational Transects in the Bolivian Andes 1. Biotropica 2006, 38, 183–195. [Google Scholar] [CrossRef]
- Aguilar-Rodríguez, P.A.; Krömer, T.; García-Franco, J.G.; MacSwiney, G.M.C. From dusk till dawn: Nocturnal and diurnal pollination in the epiphyte Tillandsia heterophylla (Bromeliaceae). Plant Biol. 2016, 18, 37–45. [Google Scholar] [CrossRef]
- Petit, J.R.; Duminil, J.; Fineschi, S.; Hampe, A.; Salvini, D.; Ven-dramin, G.G. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 2005, 14, 689–701. [Google Scholar] [CrossRef] [Green Version]
- Petit, R.J.; Excoffier, L. Gene flow and species delimitation. Trends Ecol. Evol. 2009, 24, 386–393. [Google Scholar] [CrossRef]
- Yang, L.; Kong, H.; Huang, J.P.; Kang, M. Different species or genetically divergent populations? Integrative species delimitation of the Primulina hochiensis complex from isolated karst habitats. Mol. Phylogenet. Evol. 2019, 132, 219–231. [Google Scholar] [CrossRef]
- Carstens, B.C.; Satler, J.D. The carnivorous plant described as Sarracenia alata contains two cryptic species. Biol. J. Linn. Soc. 2013, 109, 737–746. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Liu, H.; Ru, D.; Hu, H.; Hu, Q. Population genomic evidence for radiative divergence of four Orychophragmus (Brassicaceae) species in eastern Asia. Bot. J. Linn. Soc. 2019, 191, 18–29. [Google Scholar] [CrossRef]
- Saag, L.; Mark, K.; Saag, A.; Randlane, T. Species delimitation in the lichenized fungal genus Vulpicida (Parmeliaceae, Ascomycota) using gene concatenation and coalescent-based species tree approaches. Am. J. Bot. 2014, 101, 2169–2182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, G.; Dal Grande, F.; Divakar, P.K.; Otte, J.; Leavitt, S.D.; Szczepanska, K.; Crespo, A.; Rico, V.J.; Aptroot, A.; da Silva Cáceres, M.E.; et al. Coalescent-Based Species Delimitation Approach Uncovers High Cryptic Diversity in the Cosmopolitan Lichen-Forming Fungal Genus Protoparmelia (Lecanorales, Ascomycota). PLoS ONE 2015, 10, e0124625. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, M.R.; Catullo, R.A.; Ruibal, M.; Dixon, K.W.; Peakall, R.; Linde, C.C. Evaluating multilocus Bayesian species delimitation for discovery of cryptic mycorrhizal diversity. Fungal Ecol. 2017, 26, 74–84. [Google Scholar] [CrossRef]
- Leaché, A.D.; Fujita, M.K. Bayesian species delimitation in West African forest geckos (Hemidactylus fasciatus). Proc. R. Soc. B 2010, 277, 3071–3077. [Google Scholar] [CrossRef] [Green Version]
- Bittencourt-Silva, G.B.; Lawson, L.P.; Tolley, K.A.; Portik, D.M.; Barratt, C.D.; Nagel, P.; Loader, S.P. Impact of species delimitation and sampling on niche models and phylogeographical inference: A case study of the East African reed frog Hyperolius substriatus Ahl, 1931. Mol. Phylogenet. Evol. 2017, 114, 261–270. [Google Scholar] [CrossRef] [PubMed]
- French, C.M.; Deutsch, M.S.; Chávez, G.; Almora, C.E.; Brown, J.L. Speciation with introgression: Phylogeography and systematics of the Ameerega petersi group (Dendrobatidae). Mol. Phylogenet. Evol. 2019, 138, 31–42. [Google Scholar] [CrossRef]
- Castello, L.V.; Barfuss, M.H.J.; Till, W.; Galetto, L.; Chiapella, J. Disentanding the Tillandsia capillaris complex: Phylogenetic relationships and taxon boundaries in Andean populations. Bot. J. Linn. Soc. 2016, 181, 391–414. [Google Scholar] [CrossRef] [Green Version]
- Leal, B.S.S.; Graciano, V.A.; Chaves, C.J.N.; Huacre, L.A.P.; Heuertz, M.; Palma-Silva, C. Dispersal and local persistence shape the genetic structure of a widespread Neotropical plant species with a patchy distribution. Ann. Bot. 2019, 124, 499–512. [Google Scholar] [CrossRef]
- Gonçalves-Oliveira, R.C.; Wöhrmann, T.; Benko-Iseppon, A.M.; Krapp, F.; Alves, M.; Wanderley, M.D.G.L.; Weising, K. Population genetic structure of the rock outcrop species Encholirium spectabile (Bromeliaceae): The role of pollination vs. seed dispersal and evolutionary implications. Am. J. Bot. 2017, 104, 868–878. [Google Scholar] [CrossRef] [Green Version]
- Thiers, B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. New York Botanical Garden’s Virtual Herbarium. 2016. Available online: http://sweetgum.nybg.org/science/ih/ (accessed on 1 October 2021).
- Barfuss, M.H.J.; Till, W.; Leme, E.M.C.; Pinzón, J.P.; Manzanares, J.M.; Halbritter, H.; Samuel, R.; Brown, G.K. Taxonomic revision of Bromeliaceae subfam. Tillandsioideae based on a multi-locus DNA sequence phylogeny and morphology. Phytotaxa 2016, 279, 1–97. [Google Scholar] [CrossRef]
- Barfuss, M.H.J. Molecular Studies in Bromeliaceae. Ph.D. Thesis, University Vienna, Vienna, Austria, 2012. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Rambaut, A. FigTree. Tree Figure Drawing Tool; Version 1.4.2; 2014. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-Del Barrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6. DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Met. Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Clement, M.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. In Proceedings of the Parallel and Distributed Processing Symposium, International, Fort Lauderdale, FL, USA, 15–19 April 2002; pp. 184–190. [Google Scholar]
- Pons, O.; Petit, R.J. Estimation, variance and optimal sampling of gene diversity. Theor. Appl. Genet. 1995, 90, 462–470. [Google Scholar] [CrossRef]
- Pons, O.; Petit, R.J. Measwring and Testing Genetic Differentiation with Ordered Versus Unordered Alleles. Genetics 1996, 144, 1237–1245. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Excoffier, L.; Smouse, P.E.; Quattro, J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, G.L. Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. J. Math. Biol. 2017, 74, 447–467. [Google Scholar] [CrossRef]
- Jones, G.; Aydin, Z.; Oxelman, B. DISSECT: An assignment-free Bayesian discovery method for species delimitation under the multispecies coalescent. Bioinformatics 2015, 31, 991–998. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.R.; Heled, J.; Kühnert, D.; Vaughan, T.G.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Comput. Biol. 2006, 4, e88. [Google Scholar] [CrossRef]
- Rambaut, A.; Suchard, M.A.; Xie, W.; Drummond, A.J. Tracer. ver. 1.6. 2014. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 1 January 2020).
- Koide, P. Tillandsia ionantha; Its Varieties, Forms, and Cultivars. J. Bromel. Soc. 1993, 43, 160–164. [Google Scholar]
- Oxelman, B.; Lidén, M.; Berglund, D. Chloroplastrps16 intron phylogeny of the tribeSileneae (Caryophyllaceae). Plant Syst. Evol. 1997, 206, 393–410. [Google Scholar] [CrossRef] [Green Version]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Model | Source Variation | D.F. | Variation (%) | F-Statistic | p Value |
|---|---|---|---|---|---|
| cpDNA | |||||
| All Populations | Among populations | 18 | 99.08 | FST = 0.99 | 1 × 10−5 |
| Within populations | 210 | 0.92 | |||
| Six Taxa * | Among groups | 5 | −9.46 | FCT = −0.09 | 0.44 |
| Among populations within groups | 13 | 100.58 | FST = 0.99 | 0.0001 | |
| Within groups | 210 | 0.96 | FSC = 0.99 | 1 × 10−5 | |
| Eigth Lineages | Among groups | 7 | 98.89 | FCT = 0.98 | 1 × 10−5 |
| Among populations within groups | 11 | 0.31 | FST = 0.99 | 1 × 10−6 | |
| Within groups | 210 | 0.8 | FSC = 0.28 | 1 × 10−6 | |
| nDNA | |||||
| All Populations | Among populations | 18 | 76.59 | FST = 0.765 | 0.0001 |
| Within populations | 208 | 23.42 | |||
| Six Taxa * | Among groups | 5 | 38.94 | FCT = 0.38 | 0.13 |
| Among populations within groups | 13 | 42.2 | FST = 0.81 | 1 × 10−5 | |
| Within groups | 208 | 18.887 | FSC = 0.69 | 1 × 10−5 | |
| Eigth Lineages | Among groups | 7 | 78 | FCT = 0.78 | 1 × 10−6 |
| Among populations within groups | 11 | 1.15 | FST = 0.79 | 1 × 10−5 | |
| Within groups | 208 | 20.86 | FSC = 0.052 | 1 × 10−6 |
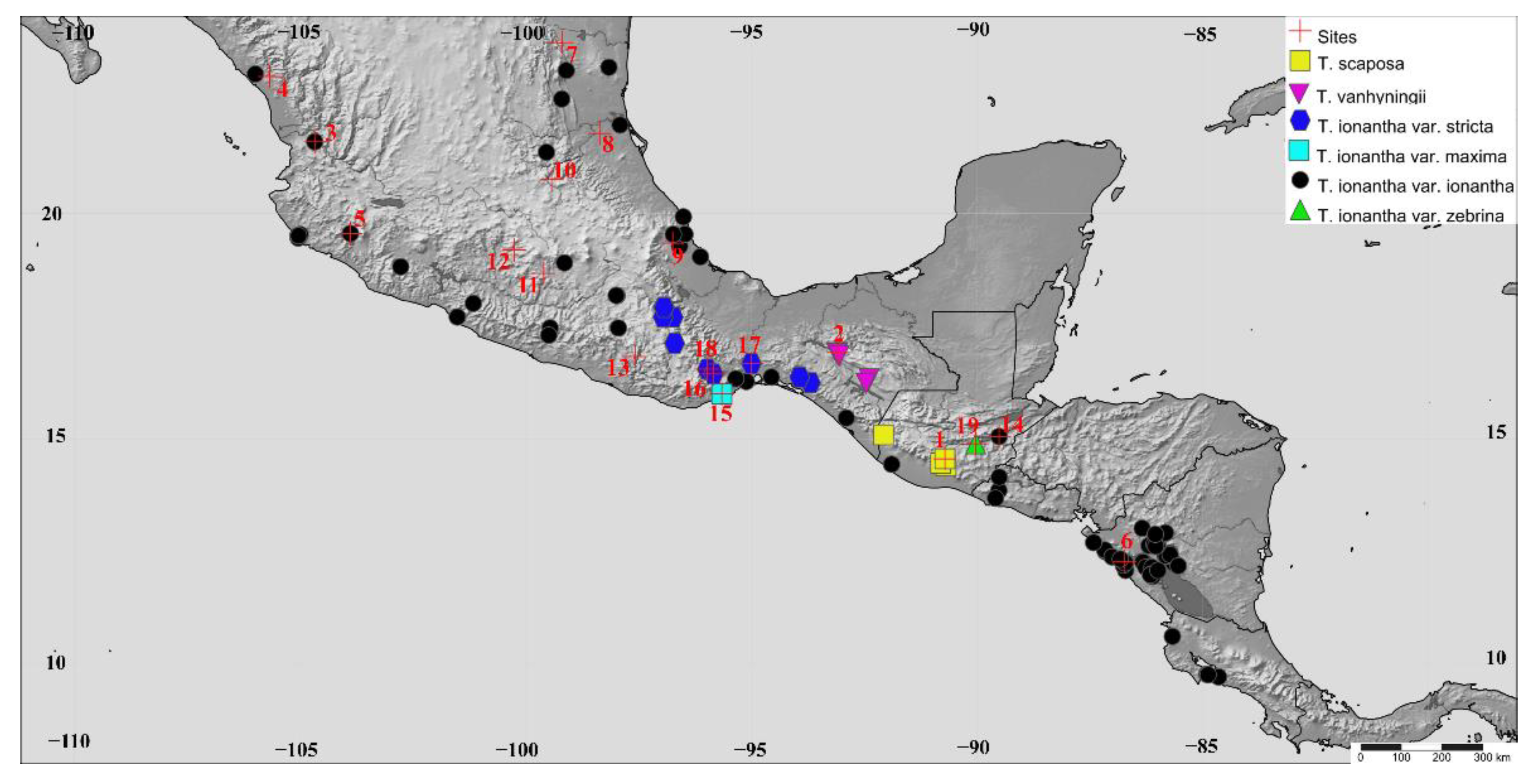
| Taxa | Population | Code | Biogeographical District | Geographic Location | Ele. | N |
|---|---|---|---|---|---|---|
| T. scaposa | 1. Antigua * | ANT | Guatemala Highlands | 14°33′17″ N, 90°43′14″ W | 1740 m | 16 |
| T. vanhyningii | 2. Cañón del Sumidero * | CSUM | Comitanian | 16°54′39″ N, 93°05′40″ W | 1100 m | 11 |
| T. ionantha var. ionantha | 3. Pochochitán | POCH | Sinaloan | 21°34′52″ N, 104°40′07” W | 700 m | 12 |
| 4. Matatán | MAT | Sinaloan | 23°1′42″ N, 105°40′45″ W | 131 m | 12 | |
| 5. Jalisco | JALIS | Nayarit-Gerrero | 19°48′44″ N, 105°16′27″ W | 49 m | 21 | |
| 6. Nicaragua | NIC | Tapachultecan | 12°16′17″ N, 86°44′07″ W | 24 m | 14 | |
| 7. El Troncón | TRO | Deciduous Forest of Northern of Veracruz | 23º46′42″ N, 99º12′22″ W | 700 m | 13 | |
| 8. El Higo | HIG | Deciduous Forest of Northern of Veracruz | 21°45′57″ N, 98°22′11″ W | 48 m | 12 | |
| 9. Jalcomulco * | JALCO | Deciduous Forest of Northern of Veracruz | 19°20′06″ N, 96°45′10″ W | 355 m | 12 | |
| 10. Barranca de Tolimán | TOL | Deciduous Forest of Northern of Veracruz | 20°44′38″ N, 99°26′09″ W | 1620 m | 10 | |
| 11. Pilcaya | PIL | Lower Balsas Basin | 18°40′01″ N, 99°36′35″ W | 1300 m | 11 | |
| 12. Santo Tomás de los Plátanos | STP | Lower Balsas Basin | 19°10′59″ N, 100°15′37″ W | 1562 m | 12 | |
| 13.- El Puente | PUE | Lower Balsas Basin | 16°49′19″ N, 97°35′11″ W | 1250 m | 12 | |
| 14. Zacapa | ZAC | Mosquito | 15°02′58″ N, 89°31′14″ W | 200 m | 10 | |
| T. ionantha var. maxima | 15. San Pedro Huamelula * | HUA | Tehuanan | 15°59′40″ N, 95°39′54″ W | 70 m | 15 |
| T. ionantha var. stricta | 16. El Manguito * | MG | Oaxaca Plateau | 16°32′35″ N, 95°59′03″ W | 1120 m | 11 |
| 17. Nizanda | NIZ | Oaxaca Plateau | 16°39′58″ N, 95°00′28″ W | 180 m | 11 | |
| 18. San Bartolo | BART | Oaxaca Plateau | 15º26′05″ N, 95º50′58″ W | 1120 m | 11 | |
| T. ionantha var. zebrina | 19. Guastatoya * | GUAS | Mosquito | 14°53′16″ N, 90°02′23″ W | 460 m | 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ancona, J.J.; Pinzón-Esquivel, J.P.; Ruiz-Sánchez, E.; Palma-Silva, C.; Ortiz-Díaz, J.J.; Tun-Garrido, J.; Carnevali, G.; Raigoza, N.E. Multilocus Data Analysis Reveal the Diversity of Cryptic Species in the Tillandsia ionantha (Bromeliaceae: Tillansiodeae) Complex. Plants 2022, 11, 1706. https://doi.org/10.3390/plants11131706
Ancona JJ, Pinzón-Esquivel JP, Ruiz-Sánchez E, Palma-Silva C, Ortiz-Díaz JJ, Tun-Garrido J, Carnevali G, Raigoza NE. Multilocus Data Analysis Reveal the Diversity of Cryptic Species in the Tillandsia ionantha (Bromeliaceae: Tillansiodeae) Complex. Plants. 2022; 11(13):1706. https://doi.org/10.3390/plants11131706
Chicago/Turabian StyleAncona, Juan J., Juan P. Pinzón-Esquivel, Eduardo Ruiz-Sánchez, Clarisse Palma-Silva, Juan J. Ortiz-Díaz, Juan Tun-Garrido, Germán Carnevali, and Néstor E. Raigoza. 2022. "Multilocus Data Analysis Reveal the Diversity of Cryptic Species in the Tillandsia ionantha (Bromeliaceae: Tillansiodeae) Complex" Plants 11, no. 13: 1706. https://doi.org/10.3390/plants11131706
APA StyleAncona, J. J., Pinzón-Esquivel, J. P., Ruiz-Sánchez, E., Palma-Silva, C., Ortiz-Díaz, J. J., Tun-Garrido, J., Carnevali, G., & Raigoza, N. E. (2022). Multilocus Data Analysis Reveal the Diversity of Cryptic Species in the Tillandsia ionantha (Bromeliaceae: Tillansiodeae) Complex. Plants, 11(13), 1706. https://doi.org/10.3390/plants11131706