Abstract
Bacteria are exposed to and tolerate diverse and potentially toxic compounds in the natural environment. While efflux transporters are generally thought to involve bacterial antibiotic resistance in vitro, their contributions to plant bacterial virulence have so far been poorly understood. Pseudomonas cannabina pv. alisalensis (Pcal) is a causal agent of bacterial blight of Brassicaceae. We here demonstrated that NU19, which is mutated in the resistance-nodulation-cell division (RND) transporter encoded gene, showed reduced virulence on cabbage compared to WT, indicating that the RND transporter contributes to Pcal virulence on cabbage. We also demonstrated that brassinin biosynthesis was induced after Pcal infection. Additionally, the RND transporter was involved in resistance to plant-derived antimicrobials and antibiotics, including the cabbage phytoalexin brassinin. These results suggest that the RND transporter extrudes plant-derived antimicrobials and contributes to Pcal virulence. We also found that the RND transporter contributes to Pcal virulence on Brassicaceae and tomato, but not on oat. These results suggest that the RND transporter contributes to Pcal virulence differentially depending on the host-plant species. Lastly, our expression-profile analysis indicated that the type-three secretion system (TTSS), which is essential for pathogenesis, is also involved in suppressing brassinin biosynthesis. Taken together, our results suggest that several Pcal virulence factors are involved in resistance to plant-derived antimicrobials and bacterial survival during infection.
1. Introduction
Plants produce diverse specialized secondary metabolites to protect against pathogens and pests [1]. Specialized metabolites differ between plant clades [2]. The simplest functional definitions recognize “phytoalexins” as metabolites that are synthesized de novo in response to a pathogen, and “phytoanticipins” as constitutively biosynthesized infection inhibitors [3]. Phytoanticipins and phytoalexins are structurally diverse and different in plant species. So far, at least 44 phytoalexins have been isolated from Brassicaceae, most of which are derived from the amino acid tryptophan [4]. These defense metabolites have inhibitory activity in vitro against various bacteria and fungi, and they confer disease resistance in plant–pathogen interactions [4,5,6,7].
The Brassicaceae family includes many economically important crops. More than 40 phytoalexins have been identified from cultivated and wild Brassicaceae. Brassica species produce indole sulfur phytoalexins, which are hallmarks of the Brassicaceae with different subsets produced by different edible crucifers [8,9]. The role of phytoalexins in pathogen resistance has been well-studied in the model plant Arabidopsis thaliana. Camalexin is a major phytoalexin of A. thaliana, and its production can be induced in A. thaliana leaves by a range of biotrophic and necrotrophic plant pathogens [4,10]. Camalexin antimicrobial activity was shown in vitro against bacteria, oomycetes, and fungi [11,12,13,14,15,16,17]. A mutation in the PHYTOALEXIN DEFICIENT 3 (PAD3) gene abolishes camalexin biosynthesis, resulting in enhanced susceptibility to necrotrophic pathogens, including Botrytis cinerea [16,18] and Alternaria brassicicola [19,20]. Several studies highlighted the importance of camalexin in response to hemibiotrophic pathogens [21,22], although camalexin accumulation was not always correlated with pathogen resistance. For instance, camalexin production was induced in response to various Pseudomonas syringae strains, but a pad3 mutant showed the same susceptibility to those strains [23,24,25].
Klein and Sattely (2017) identified the biosynthetic genes required to generate the cruciferous phytoalexin brassinin. Brassinin is a glucosinolate downstream product and is a starting point for the various other phytoalexins [26]. Brassinin is not present in A. thaliana but is produced by many cultivated Brassica species. The inability of A. thaliana to synthesize or tailor brassinin is associated with the absence of enzymes, including brassinin-associated β-glucosidase (BABG) and dithiocarvamate 5-methyltransferase (DTCMT) [8]. Brassinin antifungal activity in vitro has been reported [13]. Brassinin primarily targets mitochondrial functions in A. brassicicola, then induces secondary effects such as reactive oxygen species (ROS) production and changes in lipid homeostasis [27]. Camalexin contributes to plant resistance against various fungal and oomycete pathogens [11,18,21,28,29]. However, few studies have investigated the importance of brassinin in plant resistance, and especially focused on the importance of phytoalexin in resistance against bacterial pathogens.
Pseudomonas cannabina pv. alisalensis (Pcal) is a causal agent of bacterial blight of Brassicaceae [30]. Pcal has a wide plant-host range: the Brassicaceae family (including cabbage, broccoli, Japanese radish, Chinese cabbage), tomato, and portions of Poaceae families such as oat (Avena strigosa) and timothy (Phleum pratense) [30]. Currently, copper fungicides and antibiotics have mainly been used for bacterial disease control. However, bacterial strains (including a Pcal strain) have developed a resistance against these chemicals [31]. To develop new strategies for Pcal disease control, we need to identify Pcal infection mechanisms. We previously identified potential Pcal virulence factors [32]. Multiple virulence factors are needed for successful infection such as the type-three secretion system (TTSS), membrane transporters, transcriptional factors, and amino-acid metabolism [32]. Among these mutants, a NU19 mutant (where Tn5 is inserted in the resistance-nodulation-cell division (RND) transporter encoded gene (PMA4326_12408)), showed reduced virulence on cabbage [32]. However, the function of the RND transporter in Pcal virulence remains largely unclear.
For successful infection, plant pathogens need to eliminate the effects of host-derived antimicrobial compounds through extruding antimicrobials outside the cell, suppressing biosynthesis, and converting them to ineffective ones [7,33]. To extrude antimicrobials, bacteria have five structural groups of multidrug resistance (MDR) efflux-pump transporters: RND, small multidrug resistance, multiantimicrobial extrusion, the major facilitator superfamily, and ATP-binding cassette superfamilies. The RND efflux system functions to extrude various substrates, including antibiotics and host-derived molecules [34]. Fan et al. (2011) demonstrated that the sax (survival in Arabidopsis extracts) genes in P. syringae pv. tomato (Pto) DC3000 are required to overwhelm isothiocyanate-based defenses and facilitate a disease outcome. The sax genes form a subgroup of the RND efflux system [7]. In P. syringae, there are different operons for the RND efflux-pump transporter, mexAB-oprM and mexEF-oprN. mexAB-oprM deletion mutants in Pto DC3000, P. syringae pv. phaseolicola (Pph) 1448A, P. syringae pv. syringae (Psy) B728a, and P. amygdali pv. tabaci (formerly P. syringae pv. tabaci; Pta) 6605 exhibited increased antimicrobial susceptibility [35,36,37]. Helmann et al. (2022) demonstrated that Psy B728a MexB contributes to virulence differentially depending on the host-plant species. Therefore, although it is tempting to speculate that the Pcal RND transporter also contributes to virulence differentially depending on the host-plant species, few RND transporter studies focused on host-derived phytoalexin and its virulence contributions on different host plants.
We here investigated the importance of the RND transporter in Pcal virulence by inoculating the NU19 strain [32] on Brassicaceae crops. We demonstrated that brassinin accumulated in several brassica crops, and is induced by Pcal infection. We also showed that the RND transporter is involved in diverse antimicrobial sensitivity, including brassinin. Moreover, our results also indicated that the TTSS might be involved in suppressing brassinin biosynthesis. Together, our results suggest that several Pcal virulence factors are involved in resistance to plant-derived antimicrobials and bacterial survival during infection.
2. Results
2.1. RND Transporter Contributes to Pcal Virulence
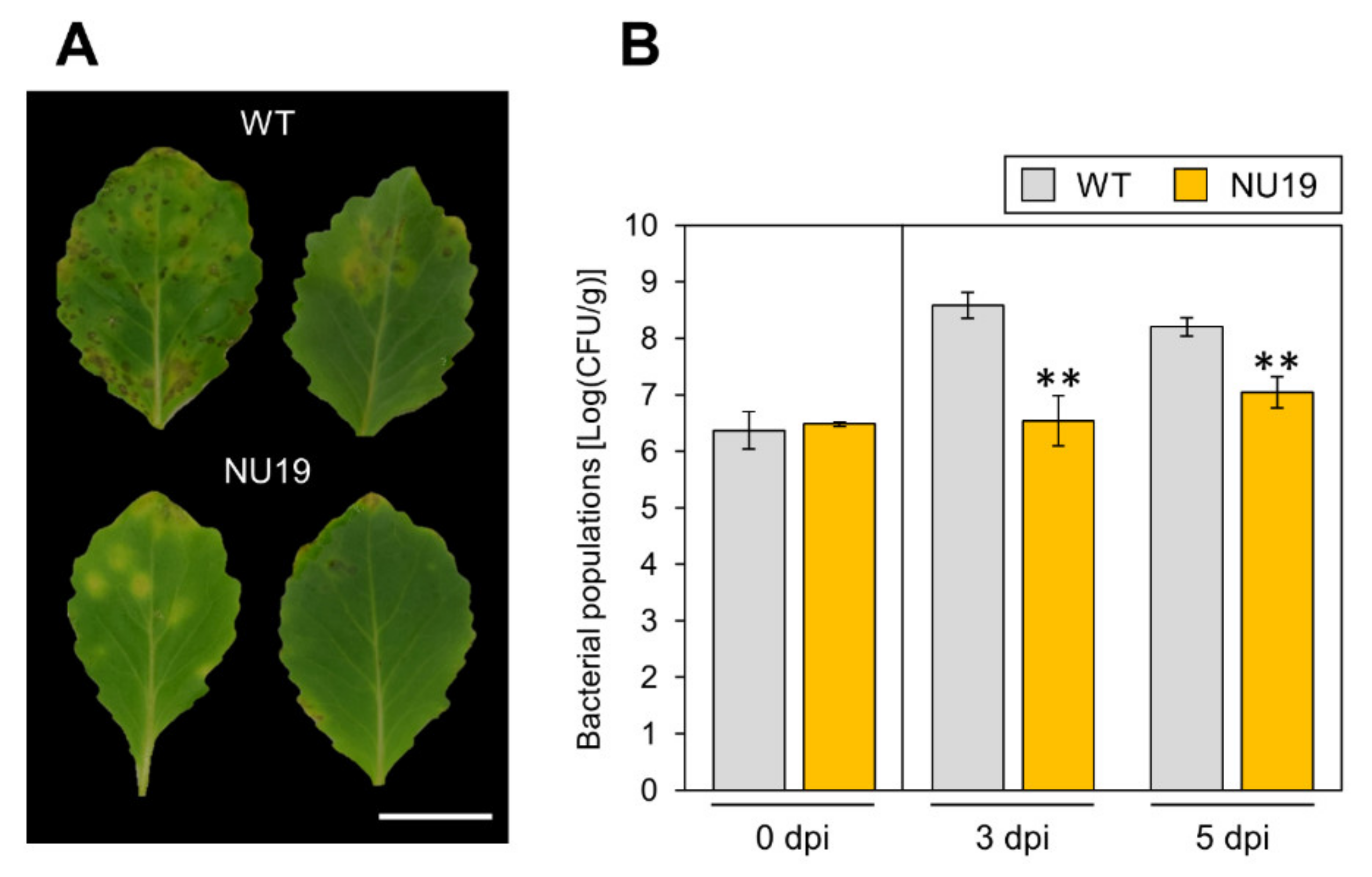
To investigate the RND transporter contributions to Pcal virulence, we conducted an inoculation assay with the RND transporter mutant NU19, which was isolated as a reduced virulence strain in a previous screening [32]. We firstly confirmed no significant differences in bacterial growth in KB medium between WT and NU19 after 12 h and 24 h incubation (Supplementary Figure S1). When we dip-inoculated plants with WT, cabbage showed chlorosis and necrosis (Figure 1A). However, cabbage inoculated with NU19 showed reduced symptoms (Figure 1A). Bacterial populations were also significantly reduced in plants inoculated with NU19 (Figure 1B). These results indicate that the RND transporter contributes to Pcal virulence on cabbage.
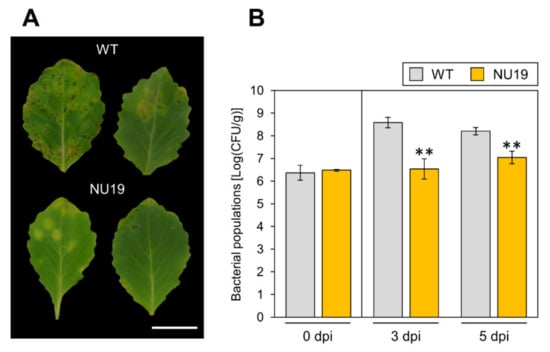
Figure 1.
Disease symptoms (A) and bacterial populations (B) on cabbage leaves dip-inoculated with Pseudomonas cannabina pv. alisalensis KB211 WT and NU19. Cabbage plants were dip-inoculated with 5 × 10 7 CFU/mL of inoculum containing 0.025% Silwet L-77. The bacterial populations in the plant were evaluated at 0, 3, and 5 dpi. The leaves were photographed at 5 dpi. Scale bar shows 2 cm. Vertical bars indicate the standard error for at least three independent experiments. Asterisks indicate a significant difference from the Pcal WT in a t-test (** p < 0.01).
2.2. Brassinin Biosynthesis Is Induced after Pcal Infection
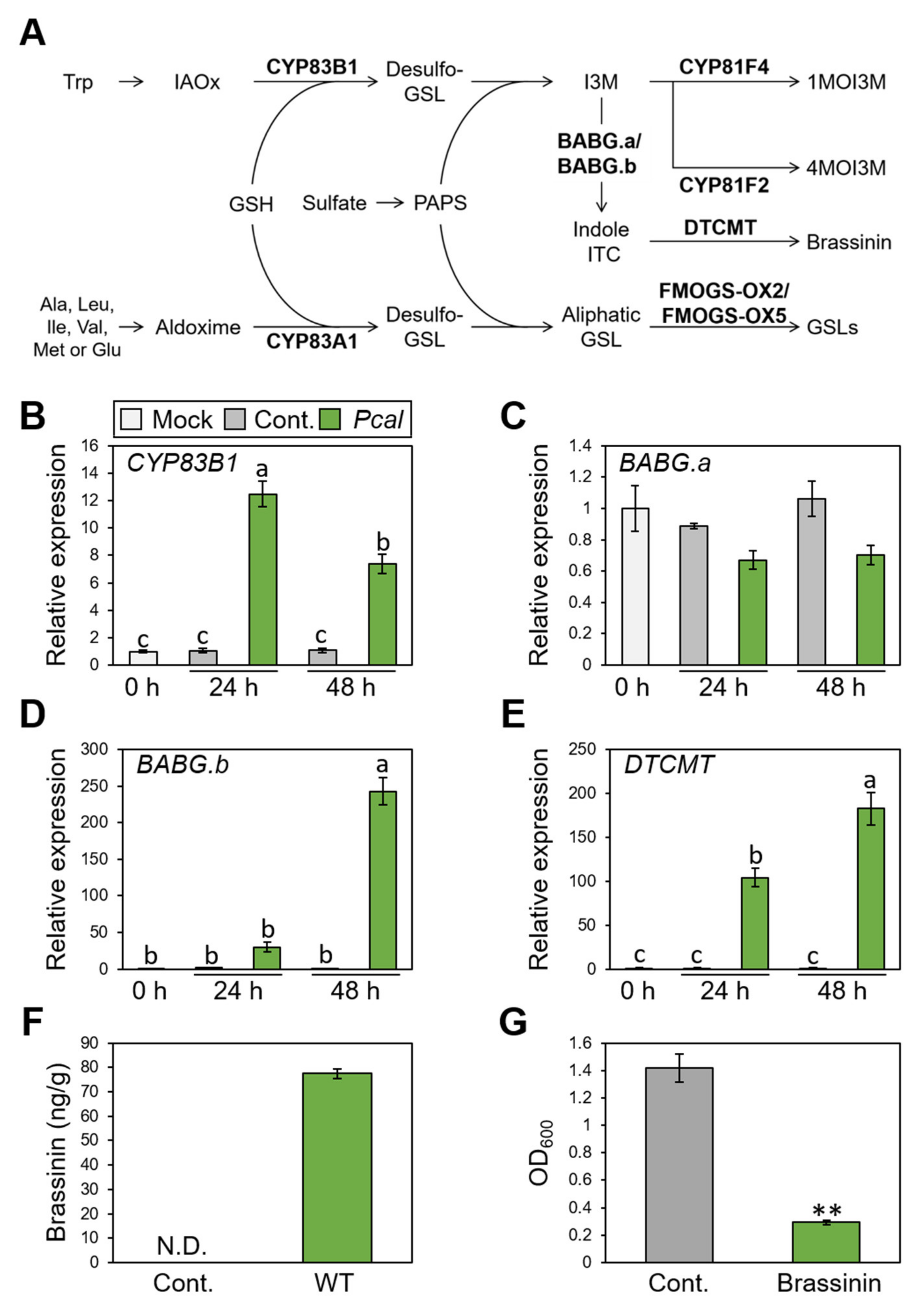
The RND transporter functions to extrude a wide range of substrates, including antibiotics and host-derived molecules [34]. We then hypothesized that NU19 exhibited reduced virulence by impairment in cabbage-derived antimicrobial efflux. Therefore, we firstly examined whether cabbage secondary metabolites, glucosinolate biosynthesis, are induced after Pcal infection. We investigated expression profiles of brassinin biosynthesis-related genes (CYP83B1, BABG.a, BABG.b, and DTCMT), indole glucosinolate biosynthesis-related genes (CYP81F2 and CYP81F4), and aliphatic glucosinolates biosynthesis-related genes (CYP83A1, FMOGS-OX2, and FMOG-OX5) (Figure 2A). Brassinin biosynthesis-related genes, except BABG.a, showed greater expression after Pcal infection (Figure 2B–E). However, aliphatic glucosinolate biosynthesis-related genes, including CYP83A1, FMOGS-OX2, and FMOGS-OX5, showed less expression after infection (Supplementary Figure S2A–C). Moreover, the indole glucosinolate pathway, CYP81F2 and CYP81F4, also showed greater expression after infection, same as the brassinin biosynthesis pathways (Supplementary Figure S2D,E).
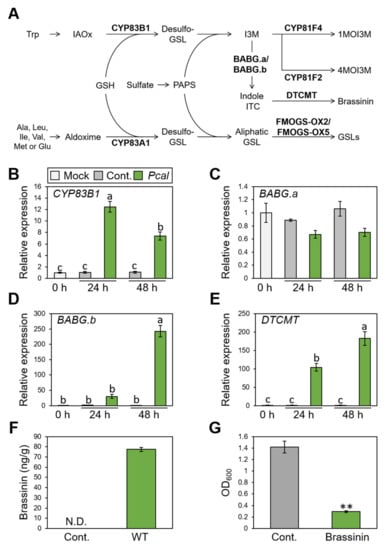
Figure 2.
Expression profiles of brassinin-related genes and brassinin production during Pseudomonas cannabina pv. alisalensis KB211 WT infection, and antimicrobial activity of brassinin. (A) The aliphatic and indolic glucosinolate biosynthesis pathways in cabbage. Schemic biosynthetic pathways with the specific biosynthetic enzyme locations used in this study are shown in bold. GSH, glutathione; GSL, glucosinolate; IAOx, indole-3-acetaldoxime; ITC, isothiocyanate; I3M, indole glucosinolates; PAPS, 3′-phosphoadenosine-5′-phosphosulfate; 1MOI3M, 1-Methoxyindole-3-yl methyl glucosinolate; 4MOI3M, 4-Methoxyindole-3-yl methyl glucosinolate. Brassinin biosynthesis gene expression profiles after syringe-inoculation with water (mock), or Pseudomonas cannabina pv. alisalensis KB211 WT. Expression profiles of CYP83B1 (B), BABG.a (C), BABG.b (D), and DTCMT (E) were determined 24 and 48 h after inoculation with 5 × 105 CFU/mL of WT or mock water-inoculated control, using real-time quantitative reverse-transcription PCR with gene-specific primer sets. Expression in cabbage was normalized using BoUBQ1. Vertical bars indicate the standard error for three biological replicates. Different letters indicate a significant difference among treatments based on a Tukey’s honestly significant different test (p < 0.05). (F) Total brassinin production in cabbage after syringe inoculation with Pcal WT or with water as a control. Cabbage leaves were collected at 48 hpi and were extracted with 80% methanol. Then, total brassinin were quantified by RP-LC-ESI-MS/MS. Vertical bars indicate the standard error for at least three independent experiments. N.D. indicates not detected. (G) Bacterial growth in LB medium after 24 h incubation with or without brassinin. The bacterial suspension was standardized to an OD600 of 0.01 in LB and coincubated with or without 200 µM brassinin. After 24 h, bacterial growth was measured at OD600. Asterisks indicate a significant difference from the water-treatment control in a t-test (** p < 0.01).
We next examined brassinin quantification using LC-MS/MS. The brassinin amount reached around 80 ng/g after Pcal infection at 48 h post inoculation (hpi) (Figure 2F). Moreover, brassinin has antimicrobial activity against Pcal (Figure 2G). Taken together, these results indicate that brassinin functions as a phytoalexin against Pcal infection.
2.3. RND Transporter Contributes to Resistance to Diverse Toxicants
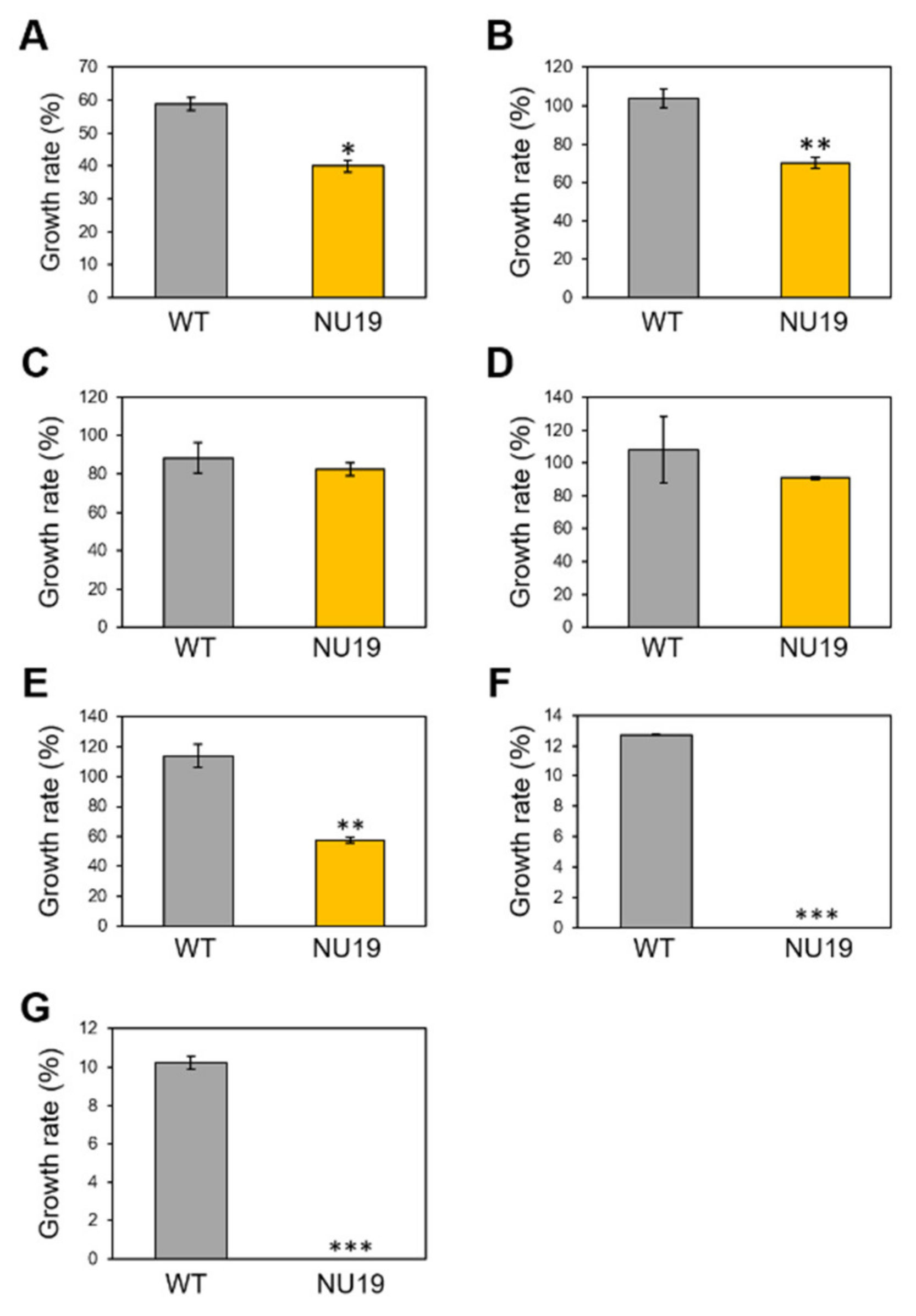
To investigate whether the RND transporter is involved in resistance to various toxicants, we firstly examined the brassinin sensitivity of WT and NU19. The NU19 growth rate was reduced compared to WT (Figure 3A), suggesting that the RND transporter contributes to brassinin resistance. We next examined the sensitivity to other plant-derived metabolites. NU19 was significantly susceptible to sulforaphane, genistein, indole, and phloretin in these experimental conditions (Figure 3B–G). Furthermore, NU19 was more sensitive to spectinomycin and streptomycin than WT (Table 1). Taken together, these results indicate that the RND transporter contributes to resistance to several plant-derived antimicrobials and antibiotics.
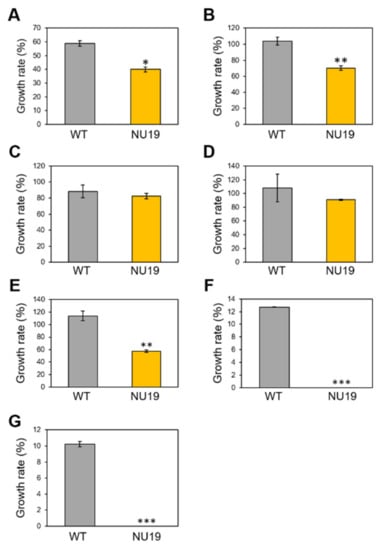
Figure 3.
Growth rate of Pseudomonas cannabina pv. alisalensis KB211 WT and NU19 in KB medium with or without plant-derived antimicrobials. The bacterial suspensions were standardized to an OD600 of 0.01 with KB, and after 6 h incubation, 200 µM brassinin (A), sulforaphane (B), camalexin (C), daidzein (D), genistein (E), indole (F), and phloretin (G) were added to each sample. Bacterial growth was measured at OD600 after 24 h incubation. Asterisks indicate a significant difference from the Pcal WT in a t-test (* p < 0.05, ** p < 0.01, *** p < 0.001).

Table 1.
Antimicrobial susceptibility of Pcal KB211 WT and NU19.
2.4. RND Transporter Contributes to Pcal Virulence on Multiple Host Plants
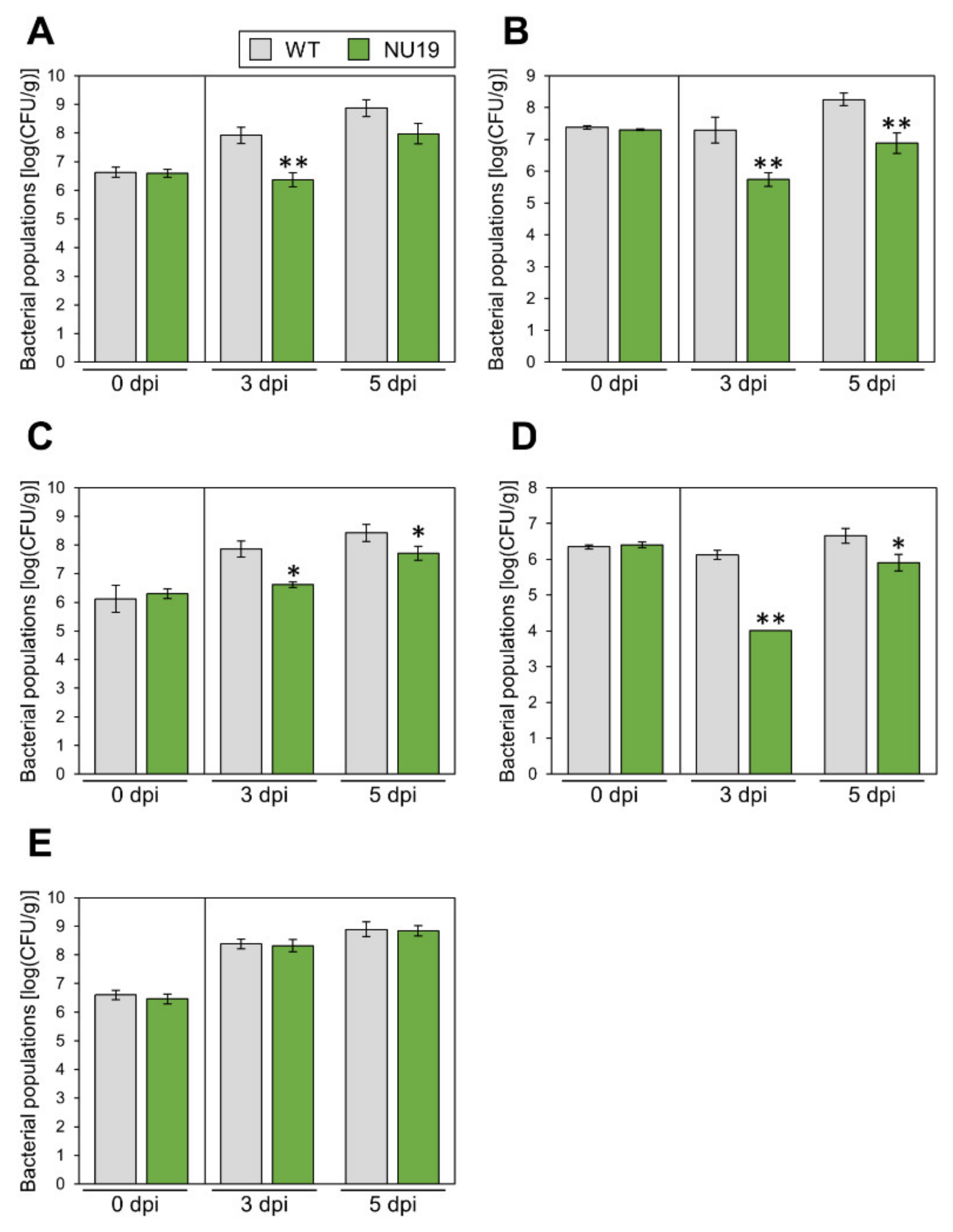
We next investigated whether the RND transporter contributes to Pcal virulence on multiple host plants. Disease symptoms and bacterial populations of NU19 were reduced in Brassica plants, including broccoli, Japanese radish, and Chinese cabbage (Figure 4A–C; Supplementary Figure S3A–C). These results indicate the RND transporter contributes to Pcal virulence on Brassica crops. Moreover, disease symptoms and bacterial populations of NU19 were reduced compared to WT in tomato (Figure 4D; Supplementary Figure S3D). Interestingly, however, disease symptoms and bacterial populations of NU19 and WT were almost the same in oat (Figure 4E; Supplementary Figure S4E). These results suggest that the RND transporter contributes less or does not contribute to disease on oat. Taken together, although the RND transporter contribution to Pcal virulence differed in infection on oat, the RND transporter contributes to disease on multiple host plants.
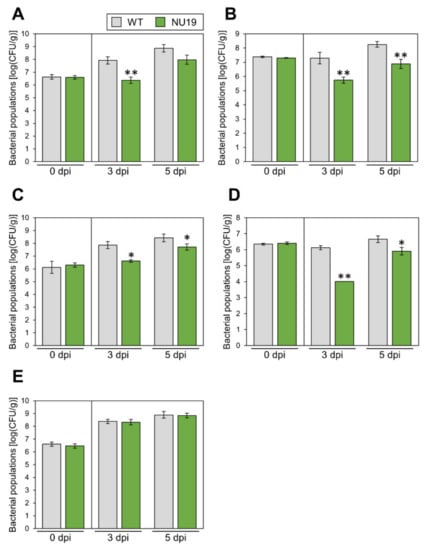
Figure 4.
Bacterial populations of Pseudomonas cannabina pv. alisalensis KB211 WT and NU19 in brocolli (A), Japanese radish (B), Chinese cabbage (C), tomato (D), and oat (E). All plants were dip-inoculated with 5 × 107 CFU/mL of inoculum containing 0.025% Silwet L-77. The bacterial populations in the plant were evaluated at 0, 3, and 5 dpi. Vertical bars indicate the standard error for at least three independent experiments. Asterisks indicate a significant difference from the Pcal WT in a t-test (* p < 0.05, ** p < 0.01).
2.5. TTSS Suppresses Brassinin Biosynthesis
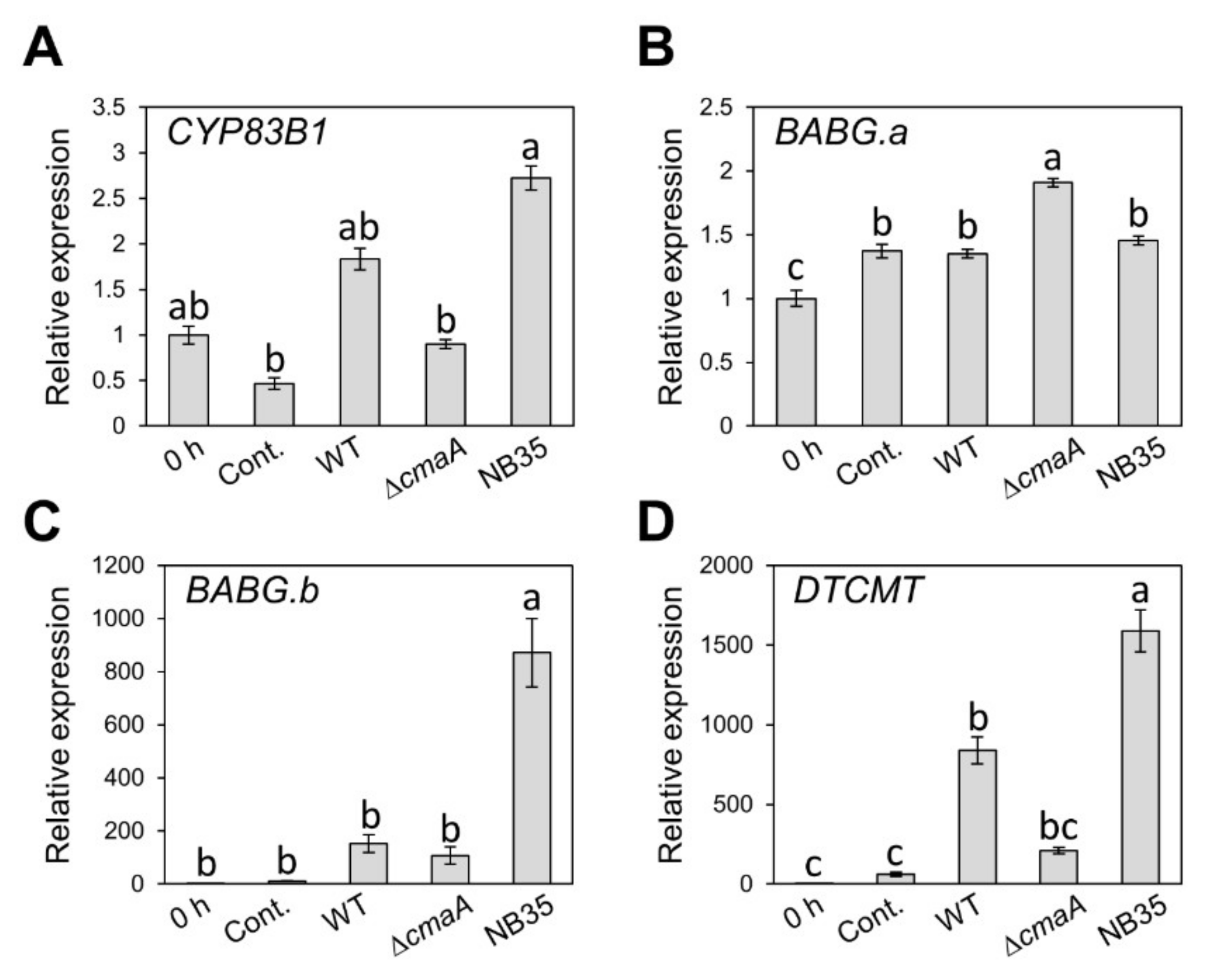
We demonstrated that brassinin biosynthesis is induced by Pcal infection. Therefore, it is tempting to speculate that efflux or detoxification of host-derived antimicrobials, including brassinin, is a critical step for successful Pcal infection. Thus, we assumed that the RND transporter is not the only virulence factor that suppresses brassinin accumulation. Therefore, to investigate whether the TTSS and coronatine (COR), which are important Pcal virulence factors [32,38], are involved in brassinin suppression, we examined the expression profiles of brassinin biosynthesis related-genes during infection with WT, NB35 (a TTSS mutant), and ΔcmaA. Since NB35 and ΔcmaA showed significantly reduced populations compared to WT [32,38,39], we first determined the time point at which these three strains have the same bacterial populations. Bacterial populations in cabbage inoculated with these strains were not significantly different at 6 hpi (Supplementary Figure S4). Therefore, we examined the expression profiles in plants inoculated with these Pcal strains at 6 hpi. All genes involved in brassinin biosynthesis, except BABG.a, showed significantly or tended to show greater expression during NB35 infection compared to WT and ΔcmaA (Figure 5A–D). These results indicate that the TTSS might be involved in brassinin biosynthesis suppression.

Figure 5.
Gene expression profiles of brassinin biosynthesis-related genes after syringe inoculation with water (mock), or Pseudomonas cannabina pv. alisalensis KB211 WT, ΔcmaA, and NB35 (TTSS mutant). Expression profiles of CYP83B1 (A), BABG.a (B), BABG.b (C), and DTCMT (D) were determined 6 h after inoculation with 5 × 107 CFU/mL of WT, ΔcmaA, NB35 or mock water-inoculated control, using real-time quantitative reverse transcription PCR with gene-specific primer sets. Expression in cabbage was normalized using BoUBQ1. Vertical bars indicate the standard error for three biological replicates. Different letters indicate a significant difference among treatments based on a Tukey’s honestly significant different test (p < 0.05).
3. Discussion
Bacteria are exposed to, and tolerate, diverse and potentially toxic compounds in the natural environment [40]. While efflux transporters are generally thought to involve bacterial antibiotic resistance in vitro, their contributions to plant bacterial virulence have so far been poorly understood. We here demonstrated that NU19, which is mutated in the RND transporter encoded gene, showed reduced virulence on cabbage compared to WT (Figure 1), indicating that the RND transporter contributes to Pcal virulence on cabbage. We also demonstrated that brassinin biosynthesis was induced after Pcal infection (Figure 2). Additionally, the RND transporter was involved in resistance to several plant-derived antimicrobials and antibiotics, including brassinin (Figure 3). These results suggests that the RND transporter contributes to Pcal virulence through extruding host-derived antimicrobials. The RND transporter also contributes to Pcal virulence on Brassicaceae plants and tomato, but not on oat (Figure 4), suggesting that RND transporter contributes to Pcal virulence differentially depending on the host plant species. Lastly, our expression-profile analysis indicates that the TTSS is also involved in brassinin biosynthesis suppression (Figure 5). Taken together, our results suggest that several Pcal virulence factors are involved in resistance to plant-derived antimicrobials and bacterial survival during infection.
Brassinin biosynthesis was induced in response to Pcal infection (Figure 2). Moreover, indole glucosinolates biosynthesis-related gene expression, including brassinin, were induced after Pcal infection (Figure 2; Supplementary Figure S2D,E). Conversely, aliphatic glucosinolate biosynthesis-related gene expression was downregulated in response to Pcal infection (Supplementary Figure S2A–C). Indeed, in A. thaliana, when the aliphatic glucosinolate pathway is blocked because of a cyp83a1 mutation, the pathways for indole glucosinolate and camalexin were enhanced [41]. One possible explanation for how the glucosinoalte synthetase CYP83A1 gene mutation affects camalexin accumulation is that it may cause crosstalk between the aliphatic glucosinolates and indole glucosinolates biosynthetic pathways [41]. Consistent with this, our data also indicated that indole glucosinolate pathways, including brassinin, were induced after Pcal infection, while alipathic glucosinolate pathways were downregulated (Figure 2; Supplementary Figure S2). Accumulation of indole indolic metabolites has been observed in response to P. syringae [23,24,42,43], and these indole metabolites, such as camalexin, 4-methoxyglucobrassicin, and 4-hydroxyindole-3-carbonyl nitrile, are important in A. thaliana basal defense [19,43,44,45,46]. Importantly, the increased biosynthesis of various glucosinolate classes depends on the type of challenging pathogens [26]. Together, the rapid and precise regulation of glucosinolate biosynthesis work in response to pathogen infection, and the downstream products of indole glucosinolates might function as the cabbage defense metabolites against bacterial pathogens.
The RND transporter contributes to Pcal virulence on Brassicaceae crops (Figure 1 and Figure 4A–C). We also demonstrated that the RND transporter contributes to susceptibility to the cabbage phytoalexin brassinin (Figure 3A), suggesting that RND transporters contribute to Pcal virulence by providing resistance to host-derived antimicrobials. The mexAB-oprM deletion mutants of Pto DC3000, Pph 1448A, Psy B728a, and Pta 6605 exhibited increased susceptibility to antimicrobials, and reduced disease-symptom development and bacterial populations [35,36,37]. In Pta, the RND transporter contributed not only directly to extrude antimicrobials, but also indirectly to regulate motility and N-acyl-homoserine lactone (AHL) production [36]. Thus, further investigation will lead to understanding the importance of the RND transporter in bacterial virulence.
Brassinin biosynthesis-related genes showed greater expression during the TTSS mutant infection compared to WT and the COR mutant (Figure 5), suggesting that the TTSS is involved in suppressing brassinin biosynthesis. Bais et al. (2005) also demonstrated that the TTSS and perhaps other virulence factors under HrpL control (but not COR) are required for blocking the synthesis or exudation of antimicrobial compounds in Pto DC3000 [47]. Moreover, P. syringae HopZ1 targeted a host enzyme to suppress isoflavone biosynthesis in soybean, which are important secondary metabolites during plant–microbe interactions in soybean [48]. These results suggest that the TTSS suppressed host-derived antimicrobial biosynthesis in addition to those emitted by the RND transporter. Conversely, a phytoanticipin, sulforaphane, inhibits P. syringae TTSS genes [49]. Chemoproteomics analyses showed that sulforaphane covalently modified the cysteine at position 209 of HrpS, a key transcriptional factor controlling TTSS gene expression [49]. This study indicated that sulforaphane inhibited virulence gene expression instead of targeting general bacterial activity. Taken together, although further analysis will be needed, there is a possibility that plant-derived antimicrobials and bacterial virulence factors target each other.
Moreover, although the RND transporter contributes to Pcal virulence on multiple host plants (Figure 4A–D), the NU19 multiplication defect was not observed on oat, indicating that the RND transporter has less or no contribution to disease on oat (Figure 4E). Psy B728a MexB contributes to virulence in common bean, but was not required for growth in lima bean, fava bean, pepper, Nicotiana benthamiana, sunflower, and tomato [50]. Additionally, Sclerotinia scleotiorum induced both camalexin and aliphatic glucosinolate biosynthesis genes, while B. cinerea did not induce aliphatic glucosinolate and induced camalexin biosynthesis genes [14]. Therefore, the plant–microbe interaction must be considered. Moreover, despite the high degree of primary homology between two RND transporters, AcrAB-TolG and MexAB-OprM, these pumps do not efflux all substrates with equal efficiency [51]. These results indicated that RND transporters have substrate specificity. Related plant families generally make use of related chemical structures for defense [2]. Indeed, oat major specialized metabolites are amphiphilic saponins [52], while brassicaceae major specialized metabolites are glucosinolates and indole alkanoids [53]. Given these very different polarities, it is unlikely that the same RND transporter would be suitable to efflux them out of the cells. However, the reason for the difference in RND transporter contribution depending on the host plant remains unclear. Further studies on oat secondary metabolites and RND transporter roles in Pcal virulence on different host plants are necessary.
We here demonstrated that the RND transporter contributes to Pcal virulence on Brassicaceae plants. The RND transporter plays an important role in resistance to plant-derived antimicrobials and antibiotics. Moreover, we revealed that the TTSS might be involved in suppressing brassinin biosynthesis. Our study shed light on the importance of efflux or suppressing host-derived antimicrobials for successful bacterial infection. Further study on plant–bacterial interactions over host-derived metabolites will be needed to understand Pcal host diversity and virulence mechanisms.
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are described in Supplementary Table S1. Pseudomonas cannabina pv. alisalensis strain KB211 (Pcal KB211) was used as the pathogenic strain to inoculate cabbage, broccoli, Japanese radish, Chinese cabbage, tomato, and oat. Pcal wild type (WT) and ΔcmaA were grown on King’s B (KB; [54]) medium at 28 °C NU19 and NB35 were grown on KB medium containing kanamycin (10 µg/mL) (Km) (Supplementary Table S1). Before Pcal inoculation, bacteria were suspended in sterile distilled H2O, and the bacterial cell densities at 600 nm (OD600) were measured using a Biowave CO8000 Cell Density Meter (Funakoshi, Tokyo, Japan).
4.2. Bacterial In Vitro Growth Measurements
WT and NU19 were grown at 28 °C on Luria–Bertani (LB; [55]) medium. The bacterial suspensions were standardized to an OD600 of 0.01 with LB, and bacterial growth was measured at OD600 for 6, 9, 12, and 24 h.
4.3. Plant Materials
Plants used for Pcal virulence assays include cabbage (Brassica oleracea var. capitate) cv. Kinkei 201, broccoli (Brassica oleracea var. italica) cv. Midoribue, Japanese radish (Raphanus sativus var. longipinnatus) cv. Natsutsukasa, Chinese cabbage (Brassica rapa var. pekinensis) cv. Akimeki, tomato (Solanum lycopersicum) cv. Moneymaker, and oat (Avena strigosa) cv. Hayoat. All plants were grown from seed at 23–25 °C with a light intensity of 200 μEm−2s−1 and a 16 h light/8 h dark photoperiod. Seedlings were used for dip-inoculation assays around two weeks after germination.
4.4. Bacterial Inoculation
To assay for disease on cabbage, broccoli, Japanese radish, Chinese cabbage, tomato, and oat plants, dip inoculations were conducted by soaking seedlings in bacterial suspensions (5 × 107 CFU/mL) containing 0.025% Silwet L-77 (OSI Specialities, Danbury, CT, USA). The seedlings were then incubated in growth chambers at 85–95% RH for the first 24 h, then at 80–85% RH for the rest of the experimental period. Disease symptoms were photographed at 5 days postinoculation (dpi) for all plants. To assess bacterial growth in all plants, the internal bacterial populations were measured after dip inoculation. Inoculated seedlings were collected, and two inoculated leaves were measured. The leaves were surface-sterilized with 10% H2O2 for 3 min. After washing with sterile distilled water three times, the leaves were homogenized in sterile distilled water, and diluted samples were plated onto solid KB agar medium. Two or three days after dilution sample plating, the bacterial colony-forming units (CFUs) were counted and normalized as CFU per gram, using the total leaf weight. The bacterial populations at 0 dpi were estimated using leaves harvested at 1 hpi without surface sterilization. The bacterial populations were evaluated in at least three independent experiments.
Cabbage was syringe-inoculated with Pcal WT, ΔcmaA, and NB35 (5 × 107 CFU/mL) with a 1 mL blunt syringe. The plants were then incubated at 70–80% RH for the rest of the experimental period. To assess bacterial growth in cabbage, the internal bacterial population was measured at 6 hpi. Leaf disks were harvested using a 3.5 mm-diameter cork-borer from syringe-infiltrated zones. The bacterial populations were evaluated in at least three independent experiments.
4.5. Monitoring Gene Expression in Planta
To analyze plant gene expression profiles during infection, we syringe-inoculated cabbage plants with Pcal WT (5 × 105 CFU/mL), and sampled at 24 and 48 hpi. To compare gene expression profiles during infection, cabbage plants were syringe-inoculated with Pcal WT, ΔcmaA, and NB35 (5 × 107 CFU/mL), and sampled at 6 hpi, where the bacterial populations of even the virulence pathogen WT had not yet significantly increased. The total RNAs, including plant and bacterial RNAs, were extracted from infected leaves and purified. Total RNA extraction and real-time quantitative RT-PCR (RT-qPCR) were performed as described previously [56]. Two micrograms of total RNA were treated with gDNA Remover (Toyobo, Osaka, Japan) to eliminate genomic DNA, and the DNase-treated RNA was reverse-transcribed using the ReverTra Ace qPCR RT Master Mix (Toyobo). The cDNA (1:10) was then used for RT-qPCR using the primers shown in Table S2 with THUNDERBIRD SYBR qPCR Mix (Toyobo) on a Thermal Cycler Dice Real-Time System (Takara Bio, Kusatsu, Japan). Cabbage UBIQUITIN EXTENSION PROTEIN 1 (BoUBQ1) was used as an internal control to normalize gene expression. The reagent blank (no-template) controls were used to detect contamination. The expression profiles were evaluated in at least six independent samples.
4.6. Brassinin Quantification by RP-LC-ESI-MS/MS
Pcal WT bacterial suspension (5 × 105 CFU/mL), or water (mock) were infiltrated into three-week-old cabbage. Twenty leaf discs (3.5 mm diameter) from four cabbage leaves were collected 48 hpi, the weight was measured, and samples were frozen in liquid nitrogen and stored at −80 °C. Samples were extracted with 300 μL of 80% methanol.
Brassinin was measured by using the multiple reaction monitoring (MRM) mode on the LC-ESI-MS/MS (LCMS-8045; Shimadzu, Kyoto, Japan) under the following conditions: capillary voltage, 4.5 kV; desolvation line, 300 °C; heat block, 500 °C; nebulizer nitrogen gas 3 L/min; drying gas, 10 L/min. Ion-source polarity was set in the negative-ion mode. The separation was performed with the LC system equipped with a 150 × 2.1 mm ACQUITY UPLC CSH C18 Column (Waters Corp., Milford, MA, USA) with a particle and pore size of 1.7 μm and 130Å, respectively. The initial mobile phase was solvent A: solvent B = 95:5 (solvent A, 0.025% formic acid; solvent B, acetonitrile (LC/MS Grade, Merck KGaA, Darmstadt, Germany) and maintained for 4 min. The solvent B concentration was increased to 50% for 11 min and then maintained at that ratio for another 5 min. The column was re-equilibrated for 3 min. The 0.4 mL min−1 flow rate and the 40 °C column temperature were maintained throughout the analysis. The MRM-transition m/z 235→58 was used as a precursor and as productions, respectively. The dwell time, Q1 pre-bias, collision energy, and Q3 pre-bias were set at 100 ms, 26 V, 7 eV, 21 V, respectively. The brassinin ion peak was detected at the retention of 15.3 min, and the fragment ion peak area of m/z = 58 was used for the quantification.
4.7. Antimicrobial-Activity Assay
To analyze brassinin antimicrobial activity against bacteria, the Pcal suspension was standardized to an OD600 of 0.01 in LB and coincubated with or without 200 µM brassinin (Merck KGaA). After 24 h, bacterial growth was measured at OD600.
4.8. Inhibition Assay
To analyze WT and NU19 susceptibility to plant-derived antimicrobials, WT and NU19 were grown at 28 °C on KB medium. The bacterial suspensions were standardized to an OD600 of 0.01 with KB, and after 6 h incubation, 200 µM antimicrobials, including brassinin (Merck KGaA), sulforaphane (Funakoshi), camalexin (Merck KGaA), daidzein (INDOFINE Chemical Company, Hillsborough, NJ, USA), genistein (Tokyo Chemical Industry, Tokyo, Japan), indole (Tokyo Chemical Industry), and phloretin (Funakoshi), was added to each sample. Bacterial growth was measured at OD600 after 24 h incubation.
4.9. Drug-Susceptibility Tests
The minimum inhibitory concentrations (MICs) of antibiotics for WT and NU19 were determined via cell growth in 2-fold dilutions of test compounds, including spectinomycin, streptomycin, nalidixic acid, cefotaxime, tetracycline, ampicillin, and carbenicillin (Merck KGaA), in 96-well plates containing KB medium to reach a total volume of 100 µL per well. The bacterial suspensions were standardized to an OD600 of 0.01 with KB, and bacterial growth was examined by visual inspection after 24 h of static incubation.
4.10. Statistical Analysis
All data are expressed as the mean with SE. All statistical analyses were performed using EZR (Saitama Medical Centre, Jichi Medical University, Saitama, Japan; [57]), a graphical user interface for R (version 3.6.3; R Foundation for statistical Computing, Vienna, Austria). Tukey’s honestly significant difference (HSD) test was used to analyze gene expression profiles. Differences of p < 0.05 were considered statistically significant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11131742/s1, Table S1: Bacterial strains and plasmids used in this study; Table S2: Primer sets used in this study; Figure S1: Pseudomonas cannabina pv. alisalensis KB211 WT and NU19 growth in KB medium; Figure S2: Expression profiles of CYP83A1 (A), FMOG-OX2 (B), FMOG-OX5 (C), CYP81F2 (D), CYP81F4 (E) were determined 24 and 48 h after inoculation with 5 × 105 CFU/mL of WT or mock water-inoculated control, using real-time quantitative reverse transcription PCR with gene-specific primer sets; Figure S3: Disease symptoms on broccoli (A), Japanese radish (B), Chinese cabbage (C), tomato (D), and oat (E) dip-inoculated with Pseudomonas cannabina pv. alisalensis KB211 WT and NU19; Figure S4: Bacterial populations in cabbage syringe-inoculated with Pseudomonas cannabina pv. alisalensis KB211 WT, ΔcmaA, and NB35 (TTSS mutant); Spreadsheets: The data presented in this study.
Author Contributions
N.S. and Y.I. designed the experiments; N.S., T.H., S.M., T.I. and Y.I. performed the experiments; N.S. and Y.I. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported: in part, by the Japan Society for the Promotion of Science (JSPS), Grant Number: 19K06045 (Y.I.), and by the JSPS, Grant Number: 21J10765 (N.S.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in Supplementary Materials here.
Acknowledgments
We thank Christina Baker for editing the manuscript. Pcal KB211 was kindly given from the Nagano vegetable and ornamental crops experiment station, Nagano, Japan.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Piasecka, A.; Jedrzejczak-Rey, N.; Bednarek, P. Secondary metabolites in plant innate immunity: Conserved function of divergent chemicals. New Phytol. 2015, 206, 948–964. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A. Natural products and plant disease resistance. Nature 2001, 411, 843–847. [Google Scholar] [CrossRef]
- VanEtten, H.D.; Mansfield, J.W.; Bailey, J.A.; Farmer, E.E. Two classes of plant antibiotics: Phytoalexins versus phytoanticipins. Plant Cell 1994, 6, 1191–1192. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- González-Lamothe, R.; Mitchell, G.; Gattuso, M.; Diarra, M.S.; Malouin, F.; Bouarab, K. Plant antimicrobial agents and their effects on plant and human pathogens. Int. J. Mol. Sci. 2009, 10, 3400–3419. [Google Scholar] [CrossRef]
- Pedras, M.S.C.; Abdoli, A. Pathogen inactivation of cruciferous phytoalexins: Detoxification reactions, enzymes and inhibitors. RSC Adv. 2017, 7, 23633–23646. [Google Scholar] [CrossRef]
- Fan, J.; Doerner, P.; Lamb, C. Pseudomonas sax genes overcome non-host resistance in Arabidopsis. Science 2011, 1185, 1185–1188. [Google Scholar] [CrossRef]
- Klein, A.P.; Sattely, E.S. Biosynthesis of cabbage phytoalexins from indole glucosinolate. Proc. Natl. Acad. Sci. USA 2017, 114, 1910–1915. [Google Scholar] [CrossRef]
- Pedras, M.S.C.; Yaya, E.E.; Glawischnig, E. The phytoalexins from cultivated and wild crucifers: Chemistry and biology. Nat. Prod. Rep. 2011, 28, 1381–1405. [Google Scholar] [CrossRef]
- Glawischnig, E. Camalexin. Phytochemistry 2007, 68, 401–406. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Abou-Mansour, E.; Buchala, A.; Mauch, F. Disease resistance of Arabidopsis to Phytophthora brassicae is established by the sequential action of indole glucosinolates and camalexin. Plant J. 2010, 62, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Rogers, E.E.; Glazebrook, J.; Ausubel, F.M. Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol. Plant. Microbe. Interact. 1996, 9, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Sellam, A.; Iacomi-Vasilescu, B.; Hudhomme, P.; Simoneau, P. In vitro antifungal activity of brassinin, camalexin and two isothiocyanates against the crucifer pathogens Alternaria brassicicola and Alternaria brassicae. Plant Pathol. 2007, 56, 296–301. [Google Scholar] [CrossRef]
- Stotz, H.U.; Sawada, Y.; Shimada, Y.; Hirai, M.Y.; Sasaki, E.; Krischke, M.; Brown, P.D.; Saito, K.; Kamiya, Y. Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J. 2011, 67, 81–93. [Google Scholar] [CrossRef]
- Sanchez-Vallet, A.; Ramos, B.; Bednarek, P.; López, G.; Piślewska-Bednarek, M.; Schulze-Lefert, P.; Molina, A. Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J. 2010, 63, 115–127. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Rowe, H.C.; Denby, K.J. Secondary metabolites influence Arabidopsis/Botrytis interactions: Variation in host production and pathogen sensitivity. Plant J. 2005, 44, 25–36. [Google Scholar] [CrossRef]
- Pedras, M.S.; Khan, A.Q. Biotransformation of the phytoalexin camalexin by the phytopathogen Rhizoctonia solani. Phytochemistry 2000, 53, 59–69. [Google Scholar] [CrossRef]
- Ferrari, S.; Plotnikova, J.M.; De Lorenzo, G.; Ausubel, F.M. Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 2003, 35, 193–205. [Google Scholar] [CrossRef]
- Thomma, B.P.; Nelissen, I.; Eggermont, K.; Broekaert, W.F. Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 1999, 19, 163–171. [Google Scholar] [CrossRef]
- Nafisi, M.; Goregaoker, S.; Botanga, C.J.; Glawischnig, E.; Olsen, C.E.; Halkier, B.A.; Glazebrook, J. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 2007, 19, 2039–2052. [Google Scholar] [CrossRef]
- Bohman, S.; Staal, J.; Thomma, B.P.H.J.; Wang, M.; Dixelius, C. Characterisation of an Arabidopsis-Leptosphaeria maculans pathosystem: Resistance partially requires camalexin biosynthesis and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J. 2004, 37, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Lemarié, S.; Robert-Seilaniantz, A.; Lariagon, C.; Lemoine, J.; Marnet, N.; Levrel, A.; Jubault, M.; Manzanares-Dauleux, M.J.; Gravot, A. Camalexin contributes to the partial resistance of Arabidopsis thaliana to the biotrophic soilborne protist Plasmodiophora brassicae. Front. Plant Sci. 2015, 6, 539. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J.; Ausubel, F.M. Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 1994, 91, 8955–8959. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Tootle, T.L.; Glazebrook, J. Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 1999, 11, 2419–2428. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Trotel-Aziz, P.; Villaume, S.; Rabenoelina, F.; Clément, C.; Baillieul, F.; Aziz, A. Priming of camalexin accumulation in induced systemic resistance by beneficial bacteria against Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000. J. Exp. Bot. 2022, 73, 3743–3757. [Google Scholar] [CrossRef]
- Plaszkó, T.; Szűcs, Z.; Vasas, G.; Gonda, S. Interactions of fungi with non-isothiocyanate products of the plant glucosinolate pathway: A review on product formation, antifungal activity, mode of action and biotransformation. Phytochemistry 2022, 200, 113245. [Google Scholar] [CrossRef]
- N’Guyen, G.Q.; Raulo, R.; Porquier, A.; Iacomi, B.; Pelletier, S.; Renou, J.-P.; Bataillé-Simoneau, N.; Campion, C.; Hamon, B.; Kwasiborski, A.; et al. Responses of the necrotrophic fungus Alternaria brassisicola to the indolic phytoalexin brassinin. Front. Plant Sci. 2021, 11, 2216. [Google Scholar] [CrossRef]
- Chassot, C.; Buchala, A.; Schoonbeek, H.-J.; Métraux, J.-P.; Lamotte, O. Wounding of Arabidopsis leaves causes a powerful but transient protection against Botrytis infection. Plant J. 2008, 55, 555–567. [Google Scholar] [CrossRef]
- Van Baarlen, P.; Woltering, E.J.; Staats, M.; VAN Kan, J.A.L. Histochemical and genetic analysis of host and non-host interactions of Arabidopsis with three Botrytis species: An important role for cell death control. Mol. Plant Pathol. 2007, 8, 41–54. [Google Scholar] [CrossRef]
- Takikawa, Y.; Takahashi, F. Bacterial leaf spot and blight of crucifer plants (Brassicaceae) caused by Pseudomonas syringae pv. maculicola and P. cannabina pv. alisalensis. J. Gen. Plant Pathol. 2014, 80, 466–474. [Google Scholar] [CrossRef]
- Takahashi, F.; Ochiai, M.; Ikeda, K.; Takikawa, Y. Streptomycin and copper resistance in Pseudomonas cannabina pv. alisalensis. Jpn. J. Phytopathol. 2013, 35. (abstract in Japanese). [Google Scholar]
- Sakata, N.; Ishiga, T.; Saito, H.; Nguyen, V.T.; Ishiga, Y. Transposon mutagenesis reveals Pseudomonas cannabina pv. alisalensis optimizes its virulence factors for pathogenicity on different hosts. PeerJ 2019, 7, e7698. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J.V. Multidrug-resistance efflux pumps - not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.A.; Piddock, L.J.V. Structure, function and inhibition of RND efflux pumps in gram-negative bacteria: An update. Curr. Opin. Microbiol. 2009, 12, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Stoitsova, S.O.; Braun, Y.; Ullrich, M.S.; Weingart, H. Characterization of the RND-type multidrug efflux pump MexAB-OprM of the plant pathogen Pseudomonas syringae. Appl. Environ. Microbiol. 2008, 74, 3387–3393. [Google Scholar] [CrossRef]
- Ichinose, Y.; Nishimura, T.; Harada, M.; Kashiwagi, R.; Yamamoto, M.; Noutoshi, Y.; Toyoda, K.; Taguchi, F.; Takemoto, D.; Matsui, H. Role of two sets of RND-type multidrug efflux pump transporter genes, MexAB-OprM and MexEF-OprN, in virulence of Pseudomonas syringae pv. tabaci 6605. Plant Pathol. J. 2020, 36, 148–156. [Google Scholar] [CrossRef]
- Helmann, T.C.; Ongsarte, C.L.; Lam, J.; Deutschbauer, A.M.; Lindow, S.E. Genome-wide transposon screen of a Pseudomonas syringae mexB mutant reveals the substrates of efflux transporters. MBio 2019, 10, e02614-19. [Google Scholar] [CrossRef]
- Sakata, N.; Ishiga, T.; Masuo, S.; Hashimoto, Y.; Ishiga, Y. Coronatine contributes to Pseudomonas cannabina pv. alisalensis virulence by overcoming both stomatal and apoplastic defenses in dicot and monocot plants. Mol. Plant. Microbe. Interact. 2021, 34, 746–757. [Google Scholar] [CrossRef]
- Sakata, N.; Ishiga, T.; Ishiga, Y. Pseudmonas cannabina pv. alisalensis TrpA is required for virulence in multiple host plants. Front. Microbiol. 2021, 12, 659734. [Google Scholar] [CrossRef]
- Mareri, L.; Parrotta, L.; Cai, G. Environmental stress and plants. Int. J. Mol. Sci. 2022, 23, 5416. [Google Scholar] [CrossRef]
- Liu, S.; Bartnikas, L.M.; Volko, S.M.; Ausubel, F.M.; Tang, D. Mutation of the glucosinolate biosynthesis enzyme cytochrome P450 83A1 monooxygenase increases camalexin accumulation and powdery mildew resistance. Front. Plant Sci. 2016, 7, 227. [Google Scholar] [CrossRef] [PubMed]
- Forcat, S.; Bennett, M.; Grant, M.; Mansfield, J.W. Rapid linkage of indole carboxylic acid to the plant cell wall identified as a component of basal defence in Arabidopsis against hrp mutant bacteria. Phytochemistry 2010, 71, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Rajniak, J.; Barco, B.; Clay, N.K.; Sattely, E.S. A new cyanogenic metabolite in Arabidopsis required for inducible pathogen defence. Nature 2015, 525, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P.; Pislewska-Bednarek, M.; Svatos, A.; Schneider, B.; Doubsky, J.; Mansurova, M.; Humphry, M.; Consonni, C.; Panstruga, R.; Sanchez-Vallet, A.; et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 2009, 323, 101–106. [Google Scholar] [CrossRef]
- Clay, N.K.; Adio, A.M.; Denoux, C.; Jander, G.; Ausubel, F.M. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 2009, 323, 95–101. [Google Scholar] [CrossRef]
- Millet, Y.A.; Danna, C.H.; Clay, N.K.; Songnuan, W.; Simon, M.D.; Werck-Reichhart, D.; Ausubel, F.M. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 2010, 22, 973–990. [Google Scholar] [CrossRef]
- Bais, H.P.; Prithiviraj, B.; Jha, A.K.; Ausubel, F.M.; Vivanco, J.M. Mediation of pathogen resistance by exudation of antimicrobials from roots. Nature 2005, 434, 217–221. [Google Scholar] [CrossRef]
- Zhou, H.; Lin, J.; Johnson, A.; Morgan, R.L.; Zhong, W.; Ma, W. Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host Microbe 2011, 9, 177–186. [Google Scholar] [CrossRef]
- Wang, W.; Yang, J.; Zhang, J.; Liu, Y.X.; Tian, C.; Qu, B.; Gao, C.; Xin, P.; Cheng, S.; Zhang, W.; et al. An Arabidopsis secondary metabolite directly targets expression of the bacterial type III secretion system to inhibit bacterial virulence. Cell Host Microbe 2020, 27, 601–613.e7. [Google Scholar] [CrossRef]
- Helmann, T.; King, D.; Lindow, S. Differential virulence contributions of the efflux transporter MexAB-OprM in Pseudomonas syringae infecting variety of host plants. Mol. Plant. Microbe. Interact. 2022, in press. [Google Scholar] [CrossRef]
- Tikhonova, E.B.; Wang, Q.; Zgurskaya, H.I. Chimeric Analysis of the Multicomponent Multidrug Efflux Transporters from Gram-Negative Bacteria. J. Bacteriol. 2002, 184, 6499–6507. [Google Scholar] [CrossRef] [PubMed]
- Raguindin, P.F.; Adam Itodo, O.; Stoyanov, J.; Dejanovic, G.M.; Gamba, M.; Asllanaj, E.; Minder, B.; Bussler, W.; Metzger, B.; Muka, T.; et al. A systematic review of phytochemicals in oat and buckwheat. Food Chem. 2021, 338, 127982. [Google Scholar] [CrossRef] [PubMed]
- Nafisi, M.; Sønderby, I.E.; Hansen, B.G.; Geu-Flores, F.; Nour-Eldin, H.H.; Nørholm, M.H.H.; Jensen, N.B.; Li, J.; Halkier, B.A. Cytochromes P450 in the biosynthesis of glucosinolates and indole alkaloids. Phytochem. Rev. 2006, 5, 331–346. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning: A laboratory manual, 2nd ed.; Cold Spring Harbor Laboratory: Huntington, NY, USA, 1989. [Google Scholar]
- Ishiga, Y.; Ichinose, Y. Pseudomonas syringae pv. tomato OxyR is required for virulence in tomato and Arabidopsis. Mol. Plant. Microbe. Interact. 2016, 29, 119–131. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).