Growing Maize Root: Lectins Involved in Consecutive Stages of Cell Development
Abstract
:1. Introduction
2. Results
2.1. Genes for Proteins with Lectin Domains in the Maize Genome
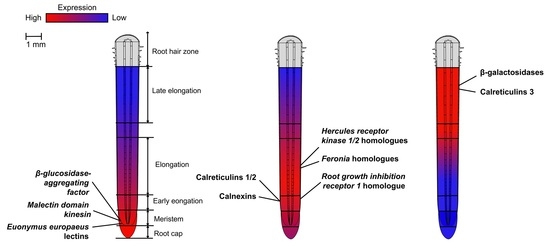
2.2. Expression of Genes of Proteins with Lectin Domains in Various Zones of Growing Maize Root
2.3. Phylogenetic Analysis of Gene Families Encoding Proteins with Lectin Domains
3. Discussion
3.1. Numerous Maize Lectins Have Access to Cell Wall Polysaccharides
3.2. Comparison of Lectin Sets in Monocots and Dicots
3.3. Lectin Genes with Zone-Specific Up-Regulation in Growing Maize Root
3.3.1. Meristematic Zone
3.3.2. The Early Elongation and Active Elongation
3.3.3. The Late Elongation
4. Materials and Methods
4.1. Identification of Genes for Proteins with Lectin Domains in the Maize Genome
4.2. Phylogenetic Analysis
4.3. Analysis of Transcriptomic Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hongqing, G.; Lei, L.; Huaxun, Y.; Xiaofei, Y.; Alexandria, A.; Yanhai, Y. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2009, 106, 7648–7653. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wu, H.-M.; Cheung, A.Y. FERONIA and her pals: Functions and mechanisms. Plant Physiol. 2016, 171, 2379–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Damme, E.J.M.; Peumans, W.J.; Barre, A.; Rougé, P. Plant lectins: A composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. CRC Crit. Rev. Plant Sci. 1998, 17, 575–692. [Google Scholar] [CrossRef]
- Van Holle, S.; De Schutter, K.; Eggermont, L.; Tsaneva, M.; Dang, L.; Van Damme, E.J.M. Comparative study of lectin domains in model species: New insights into evolutionary dynamics. Int. J. Mol. Sci. 2017, 18, 1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eggermont, L.; Verstraeten, B.; Van Damme, E.J.M. Genome-wide screening for lectin motifs in Arabidopsis thaliana. Plant Genome 2017, 10, plantgenome2017.02.0010. [Google Scholar] [CrossRef]
- Jiang, S.-Y.; Ma, Z.; Ramachandran, S. Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol. Biol. 2010, 10, 79. [Google Scholar] [CrossRef] [Green Version]
- Dang, L.; Van Damme, E.J.M. Genome-wide identification and domain organization of lectin domains in cucumber. Plant Physiol. Biochem. 2016, 108, 165–176. [Google Scholar] [CrossRef]
- Van Holle, S.; Van Damme, E.J.M. Distribution and evolution of the lectin family in soybean (Glycine max). Molecules 2015, 20, 2868–2891. [Google Scholar] [CrossRef] [Green Version]
- Saeed, B.; Baranwal, V.K.; Khurana, P. Identification and expression profiling of the lectin gene superfamily in mulberry. Plant Genome 2016, 9, plantgenome2015.10.0107. [Google Scholar] [CrossRef] [Green Version]
- Petrova, N.; Nazipova, A.; Gorshkov, O.; Mokshina, N.; Patova, O.; Gorshkova, T. Gene expression patterns for proteins with lectin domains in flax stem tissues are related to deposition of distinct cell wall types. Front. Plant Sci. 2021, 12, 634594. [Google Scholar] [CrossRef]
- Chrispeels, M.J.; Raikhel, N.V. Lectins, lectin genes, and their role in plant defense. Plant Cell 1991, 3, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lannoo, N.; Van Damme, E.J.M. Nucleocytoplasmic plant lectins. Biochim. Biophys. Acta Gen. Subj. 2010, 1800, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Tsaneva, M.; Van Damme, E.J.M. 130 years of plant lectin research. Glycoconj. J. 2020, 37, 533–551. [Google Scholar] [CrossRef] [PubMed]
- De Coninck, T.; Van Damme, E.J.M. Review: The multiple roles of plant lectins. Plant Sci. 2021, 313, 111096. [Google Scholar] [CrossRef]
- Ivanov, V.B.; Dubrovsky, J.G. Longitudinal zonation pattern in plant roots: Conflicts and solutions. Trends Plant Sci. 2013, 18, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-K.; Han, C.-L.; Lin, S.-I.; Chen, Y.-J.; Tsai, Y.-C.; Chen, Y.-R.; Chen, J.-W.; Lin, W.-Y.; Chen, P.-M.; Liu, T.-Y.; et al. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 2013, 25, 4044–4060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcon, C.; Malik, W.A.; Walley, J.W.; Shen, Z.; Paschold, A.; Smith, L.G.; Piepho, H.-P.; Briggs, S.P.; Hochholdinger, F. A high-resolution tissue-specific proteome and phosphoproteome atlas of maize primary roots reveals functional gradients along the root axes. Plant Physiol. 2015, 168, 233–246. [Google Scholar] [CrossRef]
- Kozlova, L.V.; Nazipova, A.R.; Gorshkov, O.V.; Petrova, A.A.; Gorshkova, T.A. Elongating maize root: Zone-specific combinations of polysaccharides from type I and type II primary cell walls. Sci. Rep. 2020, 10, 10956. [Google Scholar] [CrossRef]
- Nazipova, A.; Gorshkov, O.; Eneyskaya, E.; Petrova, N.; Kulminskaya, A.; Gorshkova, T.; Kozlova, L. Forgotten actors: Glycoside hydrolases during elongation growth of maize primary root. Front. Plant Sci. 2022, 12, 802424. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Kumar, V.; Donev, E.N.; Barbut, F.R.; Kushwah, S.; Mannapperuma, C.; Urbancsok, J.; Mellerowicz, E.J. Genome-wide identification of Populus malectin/malectin-like domain-containing proteins and expression analyses reveal novel candidates for signaling and regulation of wood development. Front. Plant Sci. 2020, 11, 588846. [Google Scholar] [CrossRef]
- Van Holle, S.; Van Damme, E.J.M. Messages from the past: New insights in plant lectin evolution. Front. Plant Sci. 2019, 10, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambin, J.; Demirel Asci, S.; Dubiel, M.; Tsaneva, M.; Verbeke, I.; Wytynck, P.; De Zaeytijd, J.; Smagghe, G.; Subramanyam, K.; Van Damme, E.J.M. OsEUL lectin gene expression in rice: Stress regulation, subcellular localization and tissue specificity. Front. Plant Sci. 2020, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Wasąg, P.; Grajkowski, T.; Suwińska, A.; Lenartowska, M.; Lenartowski, R. Phylogenetic analysis of plant calreticulin homologs. Mol. Phylogenet. Evol. 2019, 134, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.-Y.; He, L.-H.; Jing, R.-L.; Li, R.-Z. Calreticulin: Conserved protein and diverse functions in plants. Physiol. Plant. 2009, 136, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, D.; Guo, L.; Pan, H.; Yvon, R.; Garman, S.; Wu, H.-M.; Cheung, A.Y. Malectin/malectin-like domain-containing proteins: A repertoire of cell surface molecules with broad functional potential. Cell Surf. 2021, 7, 100056. [Google Scholar] [CrossRef]
- Nguyen, Q.-N.; Lee, Y.-S.; Cho, L.-H.; Jeong, H.-J.; An, G.; Jung, K.-H. Genome-wide identification and analysis of Catharanthus roseus RLK1-like kinases in rice. Planta 2015, 241, 603–613. [Google Scholar] [CrossRef]
- Davin, L.B.; Jourdes, M.; Patten, A.M.; Kim, K.-W.; Vassao, D.G.; Lewis, N.G. Dissection of lignin macromolecular configuration and assembly: Comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat. Prod. Rep. 2008, 25, 1015–1090. [Google Scholar] [CrossRef]
- Ma, Q.-H.; Liu, Y.-C. TaDIR13, a dirigent protein from wheat, promotes lignan biosynthesis and enhances pathogen resistance. Plant Mol. Biol. Rep. 2015, 33, 143–152. [Google Scholar] [CrossRef]
- Peumans, W.J.; Hause, B.; Van Damme, E.J.M. The galactose-binding and mannose-binding jacalin-related lectins are located in different sub-cellular compartments. FEBS Lett. 2000, 477, 186–192. [Google Scholar] [CrossRef]
- Grunwald, I.; Heinig, I.; Thole, H.H.; Neumann, D.; Kahmann, U.; Kloppstech, K.; Gau, A.E. Purification and characterisation of a jacalin-related, coleoptile specific lectin from Hordeum vulgare. Planta 2007, 226, 225. [Google Scholar] [CrossRef] [PubMed]
- Pinedo, M.; Orts, F.; de Oliveira Carvalho, A.; Regente, M.; Soares, J.R.; Gomes, V.M.; de la Canal, L. Molecular characterization of Helja, an extracellular jacalin-related protein from Helianthus annuus: Insights into the relationship of this protein with unconventionally secreted lectins. J. Plant Physiol. 2015, 183, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.A.; Rajewski, A.; He, J.; Castaneda, O.G.; Litt, A.; Kaloshian, I. Classification and phylogenetic analyses of the arabidopsis and tomato G-type lectin receptor kinases. BMC Genom. 2018, 19, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larroque, M.; Barriot, R.; Bottin, A.; Barre, A.; Rougé, P.; Dumas, B.; Gaulin, E. The unique architecture and function of cellulose-interacting proteins in oomycetes revealed by genomic and structural analyses. BMC Genom. 2012, 13, 605. [Google Scholar] [CrossRef] [Green Version]
- Esch, L.; Schaffrath, U. An update on jacalin-like lectins and their role in plant defense. Int. J. Mol. Sci. 2017, 18, 1592. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Kita, D.; Peaucelle, A.; Cartwright, H.N.; Doan, V.; Duan, Q.; Liu, M.-C.; Maman, J.; Steinhorst, L.; Schmitz-Thom, I.; et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 2018, 28, 666–675.e5. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Tang, W.; Pan, X.; Huang, A.; Gao, X.; Anderson, C.T.; Yang, Z. Arabidopsis pavement cell morphogenesis requires FERONIA binding to pectin for activation of ROP GTPase signaling. Curr. Biol. 2022, 32, 497–507.e4. [Google Scholar] [CrossRef]
- Moussu, S.; Augustin, S.; Roman, A.-O.; Broyart, C.; Santiago, J. Crystal structures of two tandem malectin-like receptor kinases involved in plant reproduction. Acta Crystallogr. Sect. D 2018, 74, 671–680. [Google Scholar] [CrossRef]
- Du, S.; Qu, L.-J.; Xiao, J. Crystal structures of the extracellular domains of the CrRLK1L receptor-like kinases ANXUR1 and ANXUR. Protein Sci. 2018, 27, 886–892. [Google Scholar] [CrossRef]
- Blixt, O.; Head, S.; Mondala, T.; Scanlan, C.; Huflejt, M.; Alvarez, R.; Bryan, M.; Fazio, F.; Calarese, D.; Stevens, D.; et al. Printed Covalent Glycan Array for Ligand Profiling of Diverse Glycan Binding Proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 17033–17038. [Google Scholar] [CrossRef] [Green Version]
- Van Damme, E.J.M.; Smith, D.F.; Cummings, R.; Peumans, W.J. Glycan Arrays to Decipher the Specificity of Plant Lectins. Adv. Exp. Med. Biol. 2011, 705, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Bojar, D.; Meche, L.; Meng, G.; Eng, W.; Smith, D.F.; Cummings, R.D.; Mahal, L.K. A Useful Guide to Lectin Binding: Machine-Learning Directed Annotation of 57 Unique Lectin Specificities. ACS Chem. Biol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Fouquaert, E.; Peumans, W.J.; Vandekerckhove, T.T.M.; Ongenaert, M.; Van Damme, E.J.M. Proteins with an euonymus lectin-like domain are ubiquitous in embryophyta. BMC Plant Biol. 2009, 9, 136. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Xu, W.; Xiang, Y.; Jia, H.; Zhang, L.; Ma, Z. Association of jacalin-related lectins with wheat responses to stresses revealed by transcriptional profiling. Plant Mol. Biol. 2014, 84, 95–110. [Google Scholar] [CrossRef]
- Laura, B.; Julia, S.; Timo, E.; Zdenka, B.; Lauri, V.; Guqi, Y.; Matthias, G.J.; Tereza, T.; Camilla, Ø.; Nora, G.-B.; et al. THESEUS1 modulates cell wall stiffness and abscisic acid production in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2022, 119, e2119258119. [Google Scholar] [CrossRef]
- Carpita, N.C. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Biol. 1996, 47, 445–476. [Google Scholar] [CrossRef]
- Van Hove, J.; Fouquaert, E.; Smith, D.F.; Proost, P.; Van Damme, E.J.M. Lectin activity of the nucleocytoplasmic EUL protein from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2011, 414, 101–105. [Google Scholar] [CrossRef] [Green Version]
- Al Atalah, B.; De Vleesschauwer, D.; Xu, J.; Fouquaert, E.; Höfte, M.; Van Damme, E.J.M. Transcriptional behavior of EUL-related rice lectins toward important abiotic and biotic stresses. J. Plant Physiol. 2014, 171, 986–992. [Google Scholar] [CrossRef]
- Van Hove, J.; De Jaeger, G.; De Winne, N.; Guisez, Y.; Van Damme, E.J.M. The Arabidopsis lectin EULS3 is involved in stomatal closure. Plant Sci. 2015, 238, 312–322. [Google Scholar] [CrossRef]
- Dubiel, M.; Beeckman, T.; Smagghe, G.; Van Damme, E.J.M. Arabidopsis lectin EULS3 is involved in ABA signaling in roots. Front. Plant Sci. 2020, 11, 437. [Google Scholar] [CrossRef]
- Galindo-Trigo, S.; Grand, T.M.; Voigt, C.A.; Smith, L.M. A malectin domain kinesin functions in pollen and seed development in Arabidopsis. J. Exp. Bot. 2020, 71, 1828–1841. [Google Scholar] [CrossRef] [PubMed]
- Petrova, N.; Mokshina, N. Using FIBexDB for in-depth analysis of flax lectin gene expression in response to Fusarium oxysporum infection. Plants 2022, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Kittur, F.S.; Lalgondar, M.; Yu, H.Y.; Bevan, D.R.; Esen, A. Maize β-glucosidase-aggregating factor is a polyspecific jacalin-related chimeric lectin, and its lectin domain is responsible for β-glucosidase aggregation. J. Biol. Chem. 2007, 282, 7299–7311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kittur, F.S.; Yu, H.Y.; Bevan, D.R.; Esen, A. Homolog of the maize β-glucosidase aggregating factor from sorghum is a jacalin-related GalNAc-specific lectin but lacks protein aggregating activity. Glycobiology 2009, 19, 277–287. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.-H.; Tian, B.; Li, Y.-L. Overexpression of a wheat jasmonate-regulated lectin increases pathogen resistance. Biochimie 2010, 92, 187–193. [Google Scholar] [CrossRef]
- Jiang, J.-F.; Xu, Y.-Y.; Chong, K. Overexpression of OsJAC1, a lectin gene, suppresses the coleoptile and stem elongation in rice. J. Integr. Plant Biol. 2007, 49, 230–237. [Google Scholar] [CrossRef]
- Weidenbach, D.; Esch, L.; Möller, C.; Hensel, G.; Kumlehn, J.; Höfle, C.; Hückelhoven, R.; Schaffrath, U. Polarized defense against fungal pathogens is mediated by the jacalin-related lectin domain of modular Poaceae-specific proteins. Mol. Plant 2016, 9, 514–527. [Google Scholar] [CrossRef] [Green Version]
- Jung, I.J.; Ahn, J.-W.; Jung, S.; Hwang, J.E.; Hong, M.J.; Choi, H.-I.; Kim, J.-B. Overexpression of rice jacalin-related mannose-binding lectin (OsJAC1) enhances resistance to ionizing radiation in Arabidopsis. BMC Plant Biol. 2019, 19, 561. [Google Scholar] [CrossRef] [Green Version]
- Del Bem, L.E.V. The evolutionary history of calreticulin and calnexin genes in green plants. Genetica 2011, 139, 255–259. [Google Scholar] [CrossRef]
- Persson, S.; Rosenquist, M.; Svensson, K.; Galvão, R.; Boss, W.F.; Sommarin, M. Phylogenetic analyses and expression studies reveal two distinct groups of calreticulin isoforms in higher plants. Plant Physiol. 2003, 133, 1385–1396. [Google Scholar] [CrossRef] [Green Version]
- Christensen, A.; Svensson, K.; Thelin, L.; Zhang, W.; Tintor, N.; Prins, D.; Funke, N.; Michalak, M.; Schulze-Lefert, P.; Saijo, Y.; et al. Higher plant calreticulins have acquired specialized functions in arabidopsis. PLoS ONE 2010, 5, e11342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, N. Regulation of Arabidopsis Root Development by Receptor-Like Kinase RGIR1 and Abiotic Stress. Ph.D. Thesis, University of Groningen, Groningen, The Netherlands, 2017. [Google Scholar]
- Juan-Miguel, E.-R.; Norbert, H.; Sharon, K.; Valeria, G.; Jacqueline, G.; Wei-Cai, Y.; Ueli, G. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 2007, 317, 656–660. [Google Scholar] [CrossRef]
- Duan, Q.; Liu, M.-C.J.; Kita, D.; Jordan, S.S.; Yeh, F.-L.J.; Yvon, R.; Carpenter, H.; Federico, A.N.; Garcia-Valencia, L.E.; Eyles, S.J.; et al. FERONIA controls pectin- and nitric oxide-mediated male–female interaction. Nature 2020, 579, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [Green Version]
- Nissen, K.S.; Willats, W.G.T.; Malinovsky, F.G. Understanding CrRLK1L function: Cell walls and growth control. Trends Plant Sci. 2016, 21, 516–527. [Google Scholar] [CrossRef]
- Xu, E.; Vaahtera, L.; Brosché, M. Roles of defense hormones in the regulation of ozone-induced changes in gene expression and cell death. Mol. Plant 2015, 8, 1776–1794. [Google Scholar] [CrossRef] [Green Version]
- Liszkay, A.; van der Zalm, E.; Schopfer, P. Production of reactive oxygen intermediates (O2˙−, H2O2, and ˙OH) by maize roots and their role in wall loosening and elongation growth. Plant Physiol. 2004, 136, 3114–3123. [Google Scholar] [CrossRef] [Green Version]
- Petrova, A.; Gorshkova, T.; Kozlova, L. Gradients of cell wall nano-mechanical properties along and across elongating primary roots of maize. J. Exp. Bot. 2021, 72, 1764–1781. [Google Scholar] [CrossRef]
- Saijo, Y. ER quality control of immune receptors and regulators in plants. Cell. Microbiol. 2010, 12, 716–724. [Google Scholar] [CrossRef]
- Caño-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-García, S.; Cheng, J.-C.; Nam, K.H.; Li, J.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Keller, O.; Kollmar, M.; Stanke, M.; Waack, S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 2011, 27, 757–763. [Google Scholar] [CrossRef] [Green Version]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, S10. [Google Scholar] [CrossRef] [Green Version]
- Almagro Armenteros, J.J.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, T.; Hecht, M.; Hamp, T.; Karl, T.; Yachdav, G.; Ahmed, N.; Altermann, U.; Angerer, P.; Ansorge, S.; Balasz, K.; et al. LocTree3 prediction of localization. Nucleic Acids Res. 2014, 42, W350–W355. [Google Scholar] [CrossRef] [Green Version]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (ITOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, W.; Li, G.; Yu, Y.; Ouyang, Y. FunRiceGenes dataset for comprehensive understanding and application of rice functional genes. Gigascience 2018, 7, gix119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consortium, S.-I. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the sequencing quality control consortium. Nat. Biotechnol. 2014, 32, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]





| Lectin Families | ID (Pfam/cd/ PTHR) 1 | Number of Genes | Expressed (TGR > 16) 2 | Kinase Domain | Number of Expressed Genes in Each Cluster (Cutoff 100 TGR/16 TGR) | ||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | |||||
| GNA | PF01453 | 81 | 56 | 68 | 5/13 | 4/7 | 6/7 | 13/13 | 6/16 |
| Legume | PF00139 | 51 | 36 | 46 | 3/6 | 2/2 | 3/3 | 7/13 | 6/12 |
| Malectin-like | PF12819 | 32 | 21 | 22 | 2/2 | 3/3 | 6/6 | 6/6 | 2/4 |
| LysM | PF01476 | 29 | 22 | 10 | 3/3 | 2/2 | 4/4 | 8/8 | 4/5 |
| Jacalin-related | PF01419 | 20 | 13 | 3 | 1/2 | - | 6/7 | 3/3 | 1/1 |
| Nictaba | PF14299 | 18 | 18 | 0 | - | 1/2 | 5/7 | 6/8 | 2/2 |
| Galectin-like | PF00337 | 17 | 12 | 0 | 2/2 | 4/4 | 1/1 | 2/2 | 1/3 |
| Galactose-binding | PF02140 | 13 | 10 | 0 | 1/2 | 3/3 | 1/1 | 1/1 | 2/3 |
| Calreticulin | PF00262 | 11 | 7 | 0 | - | 4/5 | - | 2/2 | - |
| Hevein | PF00187 | 10 | 7 | 0 | 2/2 | 1/2 | - | - | 3/3 |
| Malectin | PF11721 | 9 | 9 | 6 | 2/3 | 1/2 | - | 2/2 | 1/2 |
| EUL | PF14200, PTHR31257 | 8 | 8 | 0 | 3/4 | - | 1/1 | - | 3/3 |
| Amaranthin | PF07468 | 4 | 0 | 0 | - | - | - | - | - |
| RicinB | PF00652 | 3 | 3 | 0 | 1/1 | - | - | 1/1 | 1/1 |
| C-type | PF00059 | 1 | 1 | 1 | 1/1 | - | - | - | - |
| ABA | PF07367 | 0 | 0 | 0 | - | - | - | - | - |
| CRA | cd02879 | 0 | 0 | 0 | - | - | - | - | - |
| CV-N | PF08881 | 0 | 0 | 0 | - | - | - | - | - |
| Total | 307 | 223 | 156 | 26/41 | 25/32 | 33/36 | 51/59 | 32/55 | |
| Cluster | Gene ID | Family | Cap | Mer | eElong | Elong | lateElong | Homolog | Name |
|---|---|---|---|---|---|---|---|---|---|
| I | Zm00001d019312 | Jacalin | 24,787 | 26,384 | 11,923 | 2309 | 5341 | TaJRL4, LOC_Os12g14440 | TaJA1, OsJAC1 |
| I | Zm00001d028998 | EUL | 14,636 | 7917 | 2056 | 2581 | 6146 | LOC_Os03g21040 | OsEULD1b |
| I | Zm00001d022421 | EUL | 13,268 | 15,200 | 4837 | 5271 | 15,761 | LOC_Os07g48490 | OsEULD1a |
| I | Zm00001d026303 | Malectin | 3199 | 1841 | 830 | 343 | 378 | At1g56120, At1g56130… | |
| I | Zm00001d021447 | LysM | 2833 | 1071 | 101 | 20 | 7 | At2g17120 | LYM2 |
| I | Zm00001d027803 | Gal-binding | 2718 | 2324 | 1955 | 1144 | 947 | At4g36360, At1g45130 | AtBGAL3/5 |
| I | Zm00001d027337 | Malectin | 1724 | 1626 | 433 | 60 | 150 | At2g22610 | MDKIN2 |
| I | Zm00001d024982 | EUL | 1648 | 704 | 178 | 21 | 1 | At2g39050 | ArathEULS3 |
| I | Zm00001d040724 | Galectin-like | 1241 | 1194 | 1022 | 666 | 547 | At3g06440 | GALT3 |
| I | Zm00001d036370 | Hevein | 1216 | 706 | 447 | 174 | 178 | At3g12500 | CHI-B |
| II | Zm00001d019283 | Calreticulin | 19,305 | 30,606 | 41,660 | 21,782 | 6699 | At1g56340, At1g09210 | CRT1/2 |
| II | Zm00001d003857 | Calreticulin | 12,625 | 20,599 | 30,997 | 19,097 | 5488 | At5g61790, At5g07340 | CNX1, CNX2 |
| II | Zm00001d025305 | Calreticulin | 9543 | 17,954 | 25,639 | 10,488 | 3021 | At5g61790, At5g07340 | CNX1, CNX2 |
| II | Zm00001d005460 | Calreticulin | 3190 | 6521 | 8847 | 3301 | 944 | At1g56340, At1g09210 | CRT1/2 |
| II | Zm00001d046838 | Malectin-like | 1217 | 1881 | 5314 | 9648 | 1813 | At3g46290, At2g39360 | HERK1, CVY1 |
| II | Zm00001d003190 | Hevein | 3935 | 3508 | 4029 | 3823 | 1784 | At3g54420 | EP3 |
| II | Zm00001d044290 | Gal-binding | 1334 | 1890 | 2760 | 1566 | 738 | At3g13750 | AtBGAL1 |
| II | Zm00001d048440 | Gal-binding | 1695 | 2283 | 2677 | 857 | 496 | At4g36360, At1g45130 | AtBGAL3/5 |
| II | Zm00001d049741 | Legume | 372 | 540 | 2019 | 2006 | 176 | ||
| II | Zm00001d030759 | Legume | 1311 | 751 | 1416 | 1657 | 952 | At3g55550 | LECRK-S.4 |
| III | Zm00001d040190 | EUL | 1653 | 4018 | 10,050 | 13,790 | 12,100 | LOC_Os01g01450 | OsEULS3 |
| III | Zm00001d028474 | Gal-binding | 905 | 3794 | 6665 | 2806 | 4568 | At2g28470 | AtBGAL8 |
| III | Zm00001d029047 | Malectin-like | 1084 | 1740 | 6279 | 11,398 | 5394 | At3g51550 | FER |
| III | Zm00001d047533 | Malectin-like | 674 | 959 | 3336 | 5279 | 2055 | At3g51550 | FER |
| III | Zm00001d043252 | GNA | 63 | 437 | 3282 | 6739 | 4529 | ||
| III | Zm00001d020691 | LysM | 131 | 841 | 2942 | 2065 | 386 | At1g21880, At1g77630 | LYM1, LYM3 |
| III | Zm00001d018789 | Malectin-like | 628 | 1183 | 2500 | 2943 | 1197 | At1g30570 | HERK2 |
| III | Zm00001d029673 | Nictaba | 694 | 888 | 1929 | 3115 | 1876 | LOC_Os10g37830 | |
| III | Zm00001d010169 | Jacalin | 2 | 264 | 1667 | 2354 | 208 | ||
| III | Zm00001d052306 | Malectin-like | 1052 | 1127 | 1621 | 1366 | 1248 | At2g37050 | RGIR1/SIMP1 |
| IV | Zm00001d041880 | Gal-binding | 472 | 628 | 932 | 2616 | 8397 | At2g32810 | AtBGAL9 |
| IV | Zm00001d038648 | Calreticulin | 1986 | 1912 | 3037 | 4522 | 7938 | At1g08450 | CRT3 |
| IV | Zm00001d002313 | Malectin | 1197 | 1244 | 3005 | 9336 | 6326 | At1g56120, At1g56130… | |
| IV | Zm00001d021729 | GNA | 834 | 900 | 2303 | 5231 | 4371 | At4g21390, At1g91910 | B120, - |
| IV | Zm00001d007789 | GNA | 148 | 148 | 413 | 1077 | 4219 | ||
| IV | Zm00001d021224 | Nictaba | 848 | 666 | 1430 | 3516 | 3846 | At3g53000 | PP2-A15 |
| IV | Zm00001d012170 | Calreticulin | 1636 | 1723 | 2684 | 3413 | 3109 | At1g08450 | CRT3 |
| IV | Zm00001d026306 | Malectin | 540 | 378 | 1043 | 4421 | 3015 | At1g56120, At1g56130… | |
| IV | Zm00001d018452 | Nictaba | 889 | 954 | 1494 | 2257 | 2650 | LOC_Os02g56840 | |
| IV | Zm00001d027439 | Malectin-like | 336 | 431 | 1310 | 3297 | 2614 | At4g00300 | |
| V | Zm00001d042654 | Gal-binding | 1275 | 1364 | 1319 | 1977 | 3103 | At5g63810 | AtBGAL10 |
| V | Zm00001d009936 | Hevein | 447 | 290 | 233 | 620 | 1533 | At3g12500 | CHI-B |
| V | Zm00001d010971 | Legume | 486 | 400 | 602 | 718 | 1413 | At5g03140, At3g53380 | LECRK-VIII.2/VIII.1 |
| V | Zm00001d007187 | EUL | 385 | 160 | 68 | 220 | 1124 | LOC_Os07g48460 | OsEULD2 |
| V | Zm00001d025921 | Legume | 527 | 411 | 404 | 640 | 867 | - | |
| V | Zm00001d036366 | Hevein | 1089 | 65 | 150 | 198 | 853 | At3g12500 | CHI-B |
| V | Zm00001d053695 | LysM | 160 | 86 | 85 | 247 | 744 | At2g33580 | LYK5 |
| V | Zm00001d017152 | Hevein | 682 | 227 | 150 | 468 | 658 | At3g54420 | EP3 |
| V | Zm00001d038651 | Jacalin | 212 | 141 | 112 | 297 | 537 | LOC_Os05g43240 | |
| V | Zm00001d022420 | EUL | 21 | 22 | 14 | 67 | 520 | LOC_Os07g48500 | OsEULS2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aglyamova, A.; Petrova, N.; Gorshkov, O.; Kozlova, L.; Gorshkova, T. Growing Maize Root: Lectins Involved in Consecutive Stages of Cell Development. Plants 2022, 11, 1799. https://doi.org/10.3390/plants11141799
Aglyamova A, Petrova N, Gorshkov O, Kozlova L, Gorshkova T. Growing Maize Root: Lectins Involved in Consecutive Stages of Cell Development. Plants. 2022; 11(14):1799. https://doi.org/10.3390/plants11141799
Chicago/Turabian StyleAglyamova, Aliya, Natalia Petrova, Oleg Gorshkov, Liudmila Kozlova, and Tatyana Gorshkova. 2022. "Growing Maize Root: Lectins Involved in Consecutive Stages of Cell Development" Plants 11, no. 14: 1799. https://doi.org/10.3390/plants11141799