Quantitative Proteomics of Chromochloris zofingiensis Reveals the Key Proteins Involved in Cell Growth and Bioactive Compound Biosynthesis
Abstract
:1. Introduction
2. Results
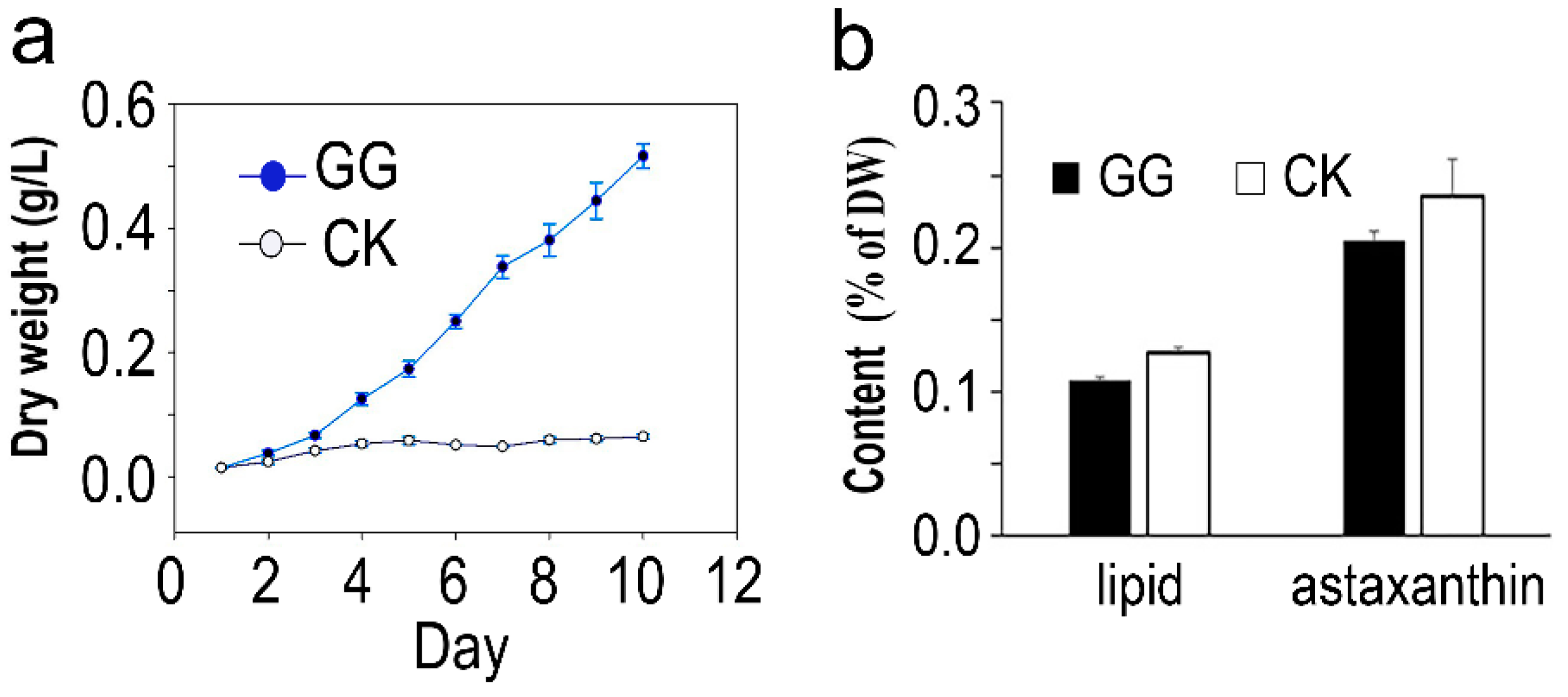
2.1. Cell Growth, Biomass, and Bioproduct Production of C. zofingiensis
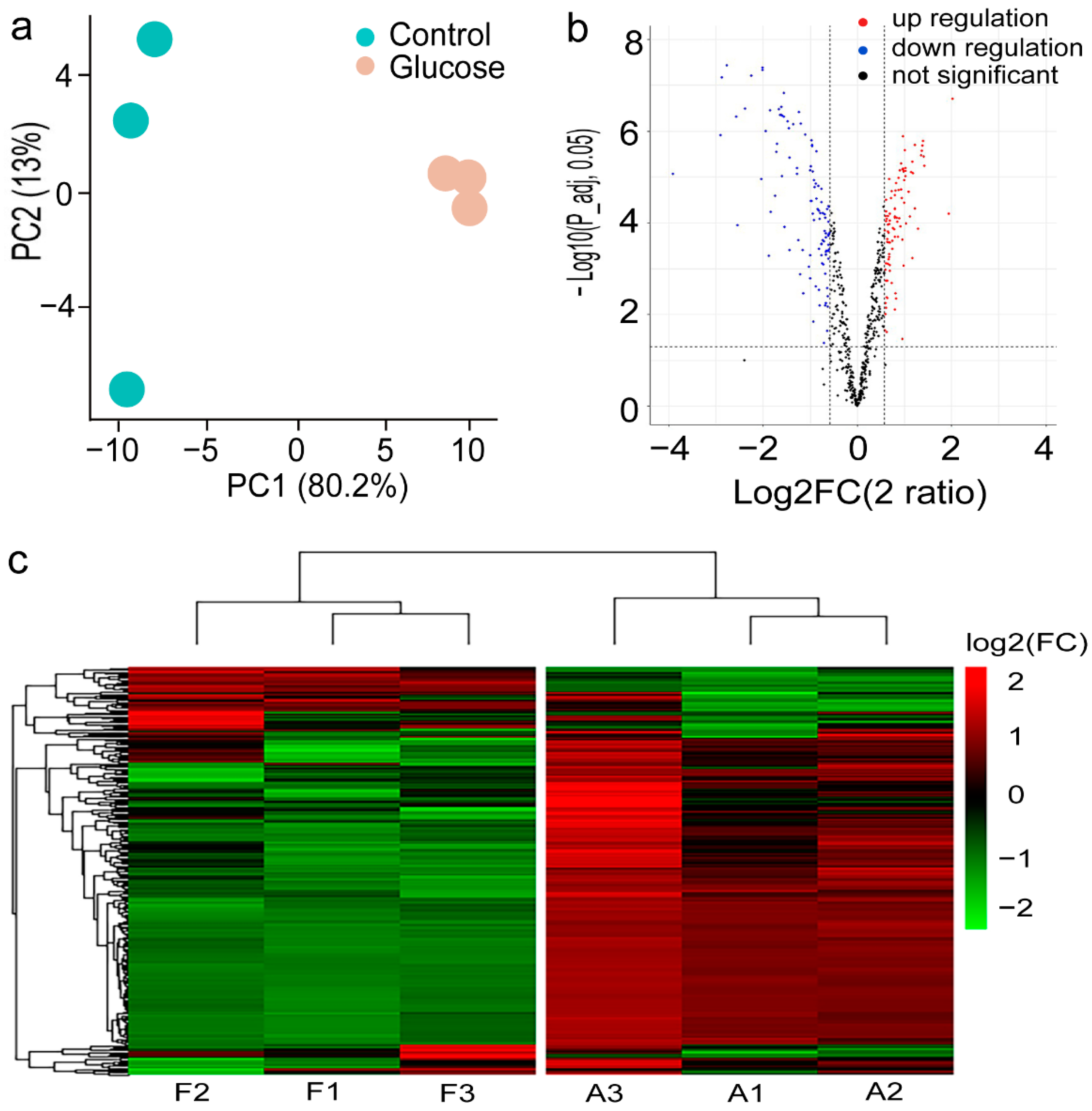
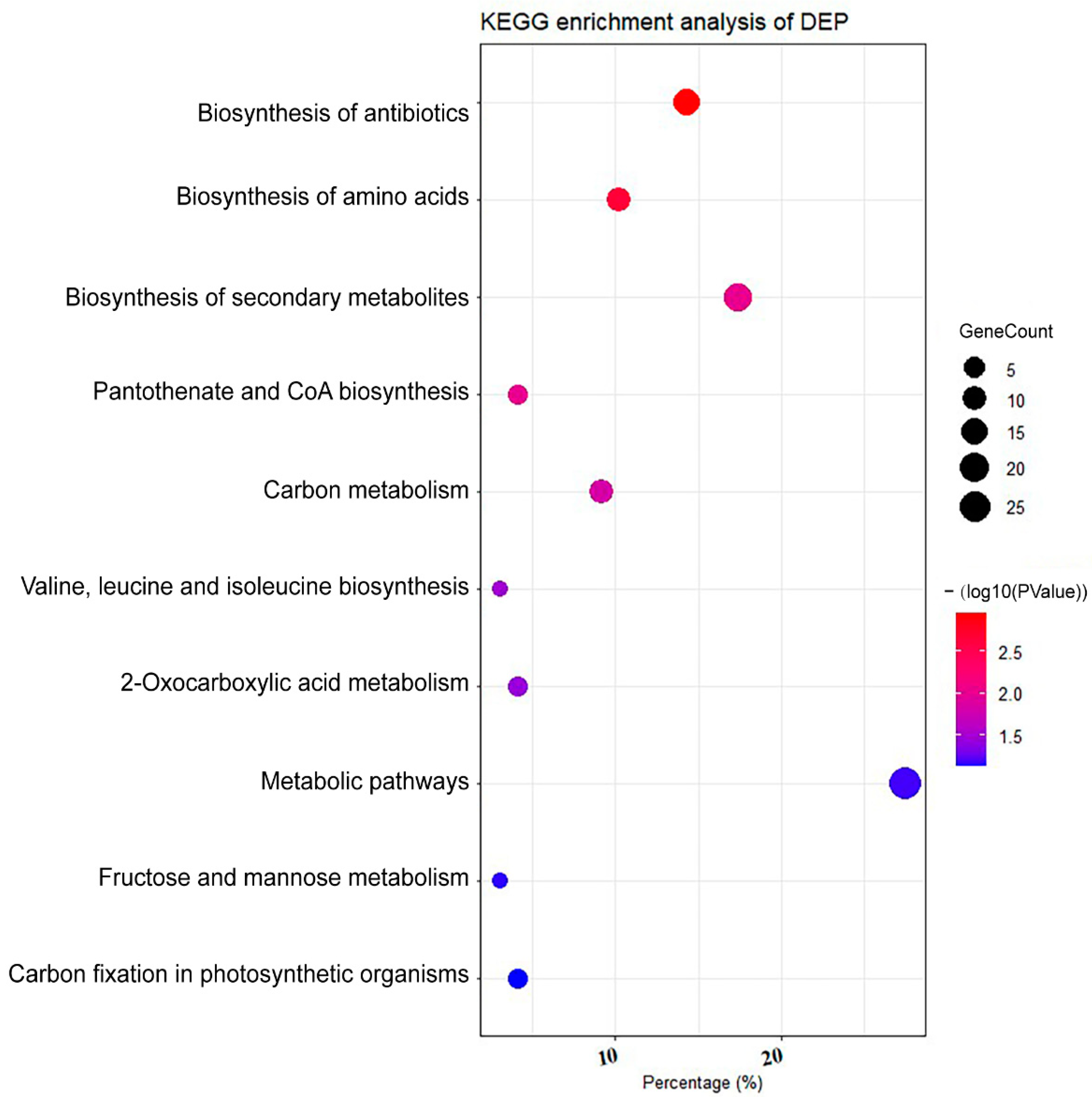
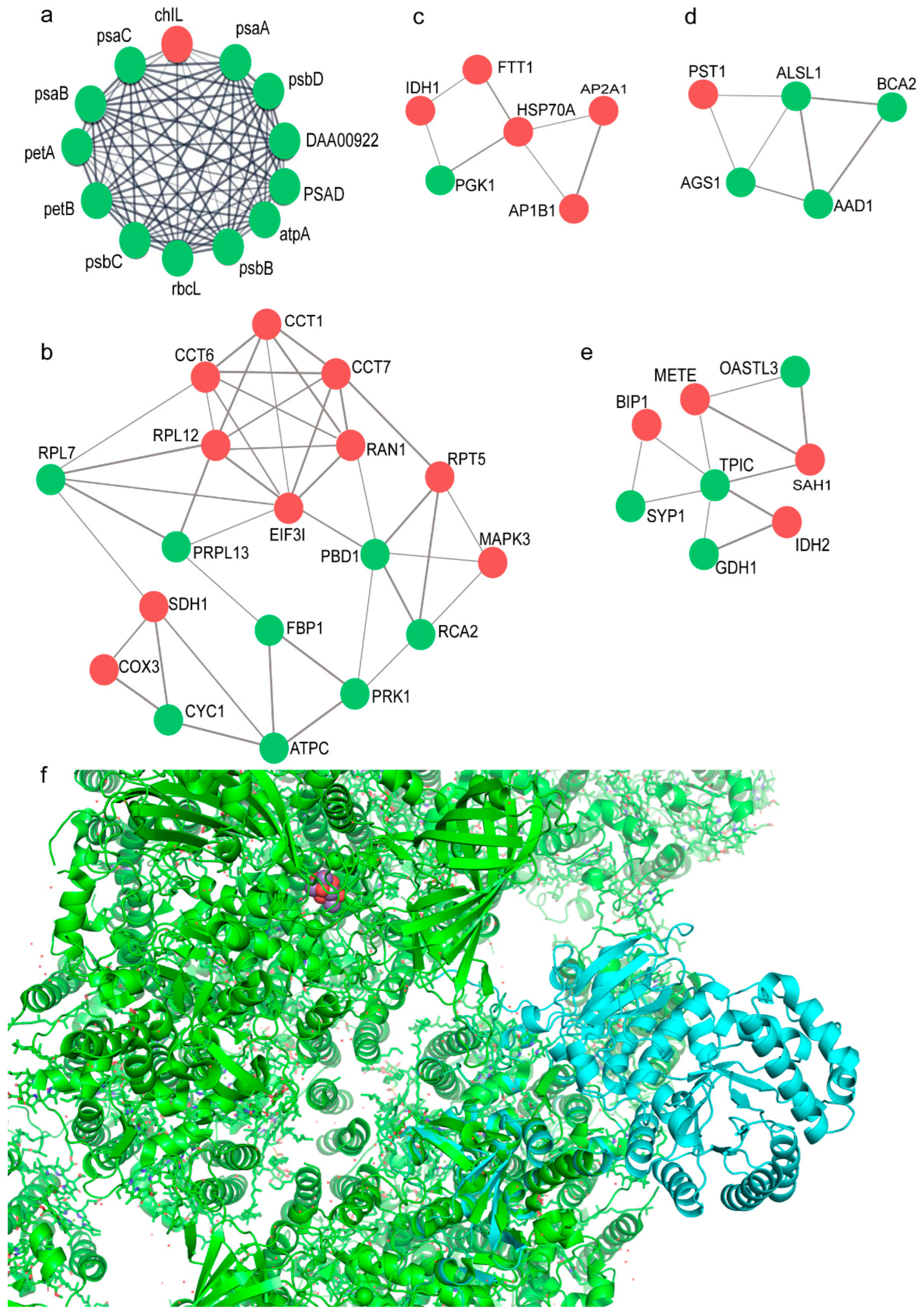
2.2. Proteomic Profile of C. zofingiensis Grown with/without Glucose
3. Discussion
3.1. Glucose Promote Rapid Growth of C. zofingiensis
3.2. Glucose Nutrient Induce Upregulation of Growth-Associated Protein of C. zofingiensis
3.3. Glucose Supplementation Decreases the Abundance of Proteins Involved in Amino Acid Metabolism
3.4. Glucose Triggers the Downregulation of Proteins in Photosynthesis-Associated Processes
4. Material and Methods
4.1. Microalgal Species, Growth Media, and Culture Conditions
4.2. Measurement of Dry Weight, Total Lipid Content, and Astaxanthin Content
4.3. Protein Extraction and Quantification
4.4. Protein Digestion, TMT Labeling, and Fractionation
4.5. Nano UHPLC–MS/MS-Based Protein Identification and Quantitation
4.6. Database Search
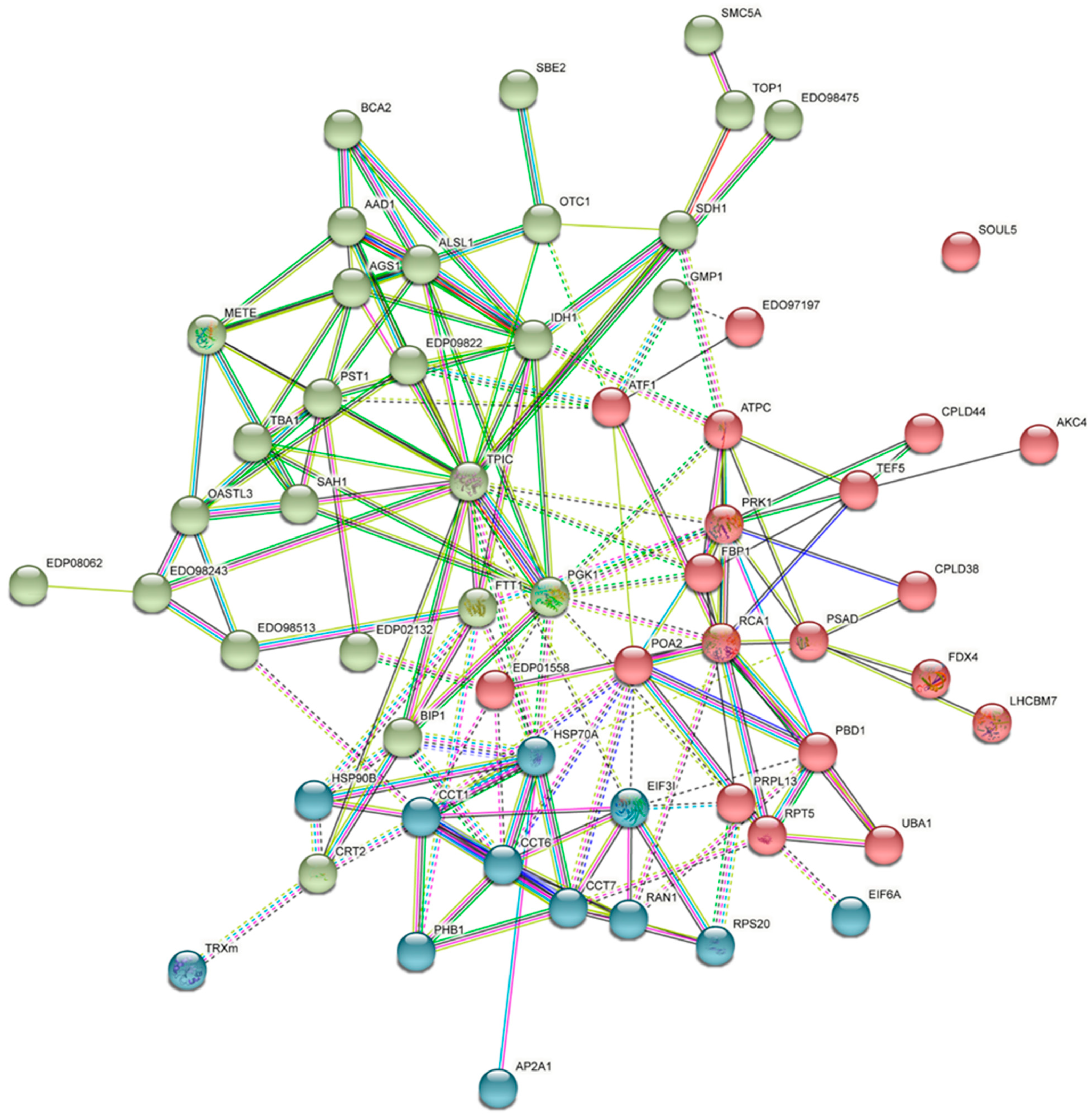
4.7. Protein Networks and Function Analysis
4.8. Protein–Protein Interaction Docking by ZDOCK
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kim, S.K. Handbook of Anticancer Drugs from Marine Origin; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Zhang, J.; Sun, Z.; Sun, P.; Chen, T.; Chen, F. Microalgal carotenoids: Beneficial effects and potential in human health. Food Funct. 2014, 5, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, Z.; Gerken, H.; Liu, Z.; Jiang, Y.; Chen, F. Chlorella zofingiensis as an alternative microalgal producer of astaxanthin: Biology and industrial potential. Mar. Drugs. 2014, 12, 3487–3515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solovchenko, A.E. Recent breakthroughs in the biology of astaxanthin accumulation by microalgal cell. Photosynth. Res. 2015, 125, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sommerfeld, M.; Chen, F.; Hu, Q. Effect of photon flux densities on regulation of carotenogenesis and cell viability of Haematococcus pluvialis (Chlorophyceae). J. Appl. Phycol. 2010, 22, 253–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Huang, J.J.; Sun, D.; Lee, Y.; Chen, F. Two-step cultivation for production of astaxanthin in Chlorella zofingiensis using a patented energy-free rotating floating photobioreactor (RFP). Bioresour. Technol. 2017, 224, 515–522. [Google Scholar] [CrossRef]
- Liu, J.; Sun, Z.; Zhong, Y.J.; Gerken, H.; Huang, J.C.; Chen, F. Utilization of cane molasses towards cost-saving astaxanthin production by a Chlorella zofingiensis mutant. J. Appl. Phycol. 2013, 25, 1447–1456. [Google Scholar] [CrossRef]
- Doucha, J.; Lívanský, K. Production of high-density Chlorella culture grown in fermenters. J. Appl. Phycol. 2012, 24, 35–43. [Google Scholar] [CrossRef]
- Huang, W.; Ye, J.; Zhang, J.; Lin, Y.; He, M.; Huang, J. Transcriptome analysis of Chlorella zofingiensis to identify genes and their expressions involved in astaxanthin and triacylglycerol biosynthesis. Algal. Res. 2016, 17, 236–243. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, D.; Zhang, Y.; Chen, F. Glucose triggers cell structure changes and regulates astaxanthin biosynthesis in Chromochloris zofingiensis. Algal. Res. 2019, 39, 101455. [Google Scholar] [CrossRef]
- Marudhupandi, T.; Sathishkumar, R.; Kumar, T.T. Heterotrophic cultivation of Nannochloropsis salina for enhancing biomass and lipid production. Biotechnol. Rep. 2016, 10, 8–16. [Google Scholar] [CrossRef] [Green Version]
- Fan, M.; Sun, X.; Liao, Z.; Wang, J.; Li, Y.; Xu, N. Comparative proteomic analysis of Ulva prolifera response to high temperature stress. Proteome Sci. 2018, 16, 17. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Chen, J.; Deschênes, J.S.; Tremblay, R.; Jolicoeur, M. Glucose feeding recalibrates carbon flux distribution and favours lipid accumulation in Chlorella protothecoides through cell energetic management. Algal. Res. 2016, 14, 83–91. [Google Scholar] [CrossRef]
- Puzanskiy, R.; Shavarda, A.; Tarakhovskaya, E.; Shishova, M. Analysis of metabolic profile of Chlamydomonas reinhardtii cultivated under autotrophic conditions. Appl. Biochem. Microbiol. 2014, 51, 83–94. [Google Scholar] [CrossRef]
- Yamane, Y.; Higashida, K.; Nakashimada, Y.; Kakizono, T.; Nishio, N. Influence of Oxygen and Glucose on Primary Metabolism and Astaxanthin Production by Phaffia rhodozyma in Batch and Fed-Batch Cultures: Kinetic and Stoichiometric Analysis. Appl. Environ. Microbiol. 1997, 63, 4471–4478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, L.E.; Lemoine, R.; Sauer, N. Sugar transporters in higher plants--a diversity of roles and complex regulation. Trends Plant Sci. 2000, 5, 283–290. [Google Scholar] [CrossRef]
- Lee, K.H.; Minami, A.; Marshall, R.S.; Book, A.J.; Farmer, L.M.; Walker, J.M.; Vierstra, R.D. The RPT2 subunit of the 26S proteasome directs complex assembly, histone dynamics, and gametophyte and sporophyte development in Arabidopsis. Plant Cell. 2011, 23, 4298–4317. [Google Scholar] [CrossRef] [Green Version]
- Kurepa, J.; Wang, S.; Li, Y.; Zaitlin, D.; Pierce, A.J.; Smalle, J.A. Loss of 26S proteasome function leads to increased cell size and decreased cell number in Arabidopsis shoot organs. Plant Physiol. 2009, 150, 178–189. [Google Scholar] [CrossRef] [Green Version]
- Brosnan, J.T.; Brosnan, M.E. Branched-chain amino acids: Enzyme and substrate regulation. J. Nutr. 2006, 136, 207S–211S. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, H.D. Response of a wild type and a non-nitrogen-fixing mutant of Anabaena doliolum towards different amino acids. Z. Allg. Mikrobiol. 1981, 21, 353–359. [Google Scholar]
- Roth, M.S.; Gallaher, S.D.; Westcott, D.J.; Iwai, M.; Louie, K.B.; Mueller, M.; Walter, A.; Foflonker, F.; Bowen, B.P.; Ataii, N.N.; et al. Regulation of oxygenic photosynthesis during trophic transitions in the green alga Chromochloris zofingiensis. Plant Cell. 2019, 31, 579–601. [Google Scholar] [CrossRef] [Green Version]
- Roth, M.S.; Westcott, D.J.; Iwai, M.; Niyogi, K.K. Hexokinase is necessary for glucose-mediated photosynthesis repression and lipid accumulation in a green alga. Commun. Biol. 2019, 2, 347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, X.; Focke, M.; Pollard, M.; Ohlrogge, J. Understanding in vivo carbon precursor supply for fatty acid synthesis in leaf tissue. Plant. J. 2000, 22, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Tamoi, M.; Nagaoka, M.; Miyagawa, Y.; Shigeoka, S. Contribution of fructose-1,6-bisphosphatase and sedoheptulose-1,7-bisphosphatase to the photosynthetic rate and carbon flow in the Calvin cycle in transgenic plants. Plant Cell Physiol. 2006, 47, 380–390. [Google Scholar] [CrossRef] [Green Version]
- Funke, R.P.; Kovar, J.L.; Logsdon, J.M., Jr.; Corrette-Bennett, J.C.; Straus, D.R.; Weeks, D.P. Nucleus-encoded, plastid-targeted acetolactate synthase genes in two closely related chlorophytes, Chlamydomonas reihardtii and Volvox carteri: Phylogenetic origins and recent insertion of introns. Mol. Gen. Genet. 1999, 262, 12–21. [Google Scholar] [CrossRef]
- Lächler, K.; Imhof, J.; Reichelt, M.; Gershenzon, J.; Binder, S. The cytosolic branched-chain aminotransferases of Arabidopsis thaliana influence methionine supply, salvage and glucosinolate metabolism. Plant. Mol. Biol. 2015, 88, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Wang, Y.; Shen, Y.; Yan, D.; He, X.; Dai, J.; Wu, Q. Oil accumulation mechanisms of the oleaginous microalga Chlorella protothecoides revealed through its genome, transcriptomes, and proteomes. BMC Genom. 2014, 15, 582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, L.; Lin, H.X.; Low, C.S.; Wu, M.H.; Chow, Y.; Lee, Y.K. Expression of the Chlamydomonas reinhardtii sedoheptulose-1,7-bisphosphatase in Dunaliella bardawil leads to enhanced photosynthesis and increased glycerol production. Plant Biotechnol. J. 2012, 10, 1129–1135. [Google Scholar] [CrossRef]
- Hammel, A.; Sommer, F.; Zimmer, D.; Stitt, M.; Mühlhaus, T.; Schroda, M. Overexpression of sedoheptulose-1,7-bisphosphatase enhances photosynthesis in Chlamydomonas reinhardtii and has no effect on the abundance of other Calvin-Benson cycle enzymes. Front. Plant Sci. 2020, 11, 868. [Google Scholar] [CrossRef]
- Li, Y.; Huang, J.; Sandmann, G.; Chen, F. Glucose sensing and the mitochondrial alternative pathway are involved in the regulation of astaxanthin biosynthesis in the dark-grown Chlorella zofingiensis (Chlorophyceae). Planta 2008, 228, 735–743. [Google Scholar] [CrossRef]
- Kuo, R.C.; Zhang, H.; Stuart, J.D.; Provatas, A.A.; Hannick, L.; Lin, S. Abundant synthesis of long-chain polyunsaturated fatty acids in Eutreptiella sp. (Euglenozoa) revealed by chromatographic and transcriptomic analyses. J. Phycol. 2021, 57, 577–591. [Google Scholar] [CrossRef]
- Tan, Y.Y.; Hsu, W.H.; Shih, T.W.; Lin, C.H.; Pan, T.M. Proteomic insight into the effect of ethanol on citrinin biosynthesis pathway in Monascus purpureus NTU 568. Food Res Int. 2014, 64, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, Z.; Mao, X.; Gerken, H.; Wang, X.; Yang, W. Multiomics analysis reveals a distinct mechanism of oleaginousness in the emerging model alga Chromochloris zofingiensis. Plant J. 2019, 98, 1060–1077. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Mukherjee, J.; Hawkes, J.J.; Wilkinson, S.J. Optimization of lipid production for algal biodiesel in nitrogen stressed cells of Dunaliella salina using FTIR analysis. J. Chem. Technol. Biotechnol. 2013, 88, 1807–1814. [Google Scholar] [CrossRef]
- Wang, S.; Meng, Y.; Liu, J.; Cao, X.; Xue, S. Accurate quantification of astaxanthin from Haematococcus pluvialis using DMSO extraction and lipase-catalyzed hydrolysis pretreatment. Algal. Res. 2018, 35, 427–431. [Google Scholar] [CrossRef]
- Casella, P.; Iovine, A.; Mehariya, S.; Marino, T.; Musmarra, D.; Molino, A. Smart Method for Carotenoids Characterization in Haematococcus pluvialis red phase and Evaluation of Astaxanthin Thermal Stability. Antioxidants 2020, 9, 422. [Google Scholar] [CrossRef]
- Schmelter, C.; Funke, S.; Treml, J.; Beschnitt, A.; Perumal, N.; Manicam, C.; Pfeiffer, N.; Grus, F.H. Comparison of two solid-phase extraction (SPE) methods for the identification and quantification of porcine retinal protein markers by LC-MS/MS. Int. J. Mol. Sci. 2018, 19, 3847. [Google Scholar] [CrossRef] [Green Version]
- Lyu, K.; Meng, Q.; Zhu, X.; Dai, D.; Zhang, L.; Huang, Y.; Yang, Z. Changes in iTRAQ-based proteomic profiling of the Cladoceran Daphnia magna exposed to microcystin-producing and microcystin-free Microcystis aeruginosa. Environ. Sci. Technol. 2016, 50, 4798–4807. [Google Scholar] [CrossRef]
- Gaspari, M.; Cuda, G. Nano LC-MS/MS: A robust setup for proteomic analysis. Methods Mol. Biol. 2011, 790, 115–126. [Google Scholar]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.H.; Vreven, T.; Weng, Z. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef]

| Gene Name | ID | Annotation | log2 (FC)for G+ vs. G− | Regulation | p-Value |
|---|---|---|---|---|---|
| OGD1 | A8IVG0 | 2-oxoglutarate dehydrogenase, E1 subunit | 0.27 | NS | 7.18 × 10−2 |
| CHLRE_17g713200v5 | A0A2K3CPR8 | oxoglutarate:malate antiporter | −0.73 | down | 3.91 × 10−5 |
| MNEG_0550 | A0A0D2LM39 | Putative 2-oxoglutarate/malatecarrier protein | 0.93 | up | 3.36 × 10−6 |
| MNEG_6327 | A0A0D2MM79 | 4-hydroxyphenylpyruvate dioxygenase | −0.40 | NS | 1.14 × 10−3 |
| MNEG_9313 | A0A0D2JH23 | Acyl-carrier-protein desaturase | 2.34 | up | 9.81 × 10−6 |
| DGAT1a | A0A411PNH6 | Diacylglycerol | 1.43 | up | 5.61 × 10−7 |
| CHLRE_03g158900v5 | A0A2K3DW88 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex | 0.03 | NS | 6.08 × 10−1 |
| MNEG_1234 | A0A0D2MW14 | Isocitrate dehydrogenase (NADP(+)) | −0.43 | NS | 5.66 × 10−4 |
| CHLRE_02g143250v5 | A0A2K3E3Z0 | Isocitrate dehydrogenase [NAD] subunit, mitochondrial | 0.72 | up | 7.06 × 10−6 |
| IDH3 | A8J9S7 | Isocitrate dehydrogenase [NADP] | −0.05 | NS | 5.90 × 10−1 |
| IDH1 | A8J6V1 | Isocitrate dehydrogenase, NAD-dependent | −0.66 | down | 5.74 × 10−5 |
| MNEG_16102 | A0A0D2LPD2 | Phosphoenolpyruvate carboxylase | −0.05 | NS | 5.09 × 10−1 |
| CHLRE_03g171950v5 | A0A2K3DX44 | Phosphoenolpyruvate carboxylase | 0.08 | NS | 3.91 × 10−1 |
| MNEG_10533 | A0A0D2MS98 | Phosphopyruvate hydratase | −0.63 | down | 1.28 × 10−2 |
| eno | Q946Z5 | Phosphopyruvate hydratase (Fragment) | −0.57 | NS | 1.65 × 10−4 |
| CHLRE_06g258700v5 | A0A2K3DMK8 | Pyruvate carboxylase | −0.74 | down | 3.39 × 10−5 |
| PYC1 | A8HXT4 | Pyruvate carboxylase (Fragment) | −0.33 | NS | 9.08 × 10−3 |
| CHLRE_06g258733v5 | A0A2K3DMI3 | Pyruvate carboxyltransferase domain-containing protein | −0.12 | NS | 1.89 × 10−1 |
| MNEG_8717 | A0A0D2KV51 | Pyruvate dehydrogenase E1 component subunit alpha | −0.36 | NS | 1.37 × 10−3 |
| CHLRE_02g099850v5 | A0A2K3E272 | Pyruvate dehydrogenase E1 component subunit alpha | 0.02 | NS | 8.70 × 10−1 |
| MNEG_10719 | A0A0D2JC20 | Pyruvate dehydrogenase E1 component subunit alpha (Fragment) | −1.14 | down | 2.24 × 10−4 |
| MNEG_4864 | A0A0D2MRP4 | Pyruvate dehydrogenase E1 component subunit beta | −0.27 | NS | 9.03 × 10−2 |
| CHLRE_03g194200v5 | A0A2K3DYL5 | Pyruvate dehydrogenase E1 component subunit beta | 0.13 | NS | 1.43 × 10−1 |
| PDH1a|PDH1b | A8JBC6 | Pyruvate dehydrogenase E1 component subunit beta | 0.14 | NS | 6.99 × 10−2 |
| MNEG_8504 | A0A0D2MZ94 | Pyruvate dehydrogenase E2 component (Dihydrolipoamide acetyltransferase) | −0.20 | NS | 3.24 × 10−1 |
| MNEG_13760 | A0A0D2J2M4 | Pyruvate, phosphate dikinase | −0.73 | down | 6.85 × 10−5 |
| PPD1 | A8IC95 | Pyruvate, phosphate dikinase | 0.11 | NS | 2.71 × 10−1 |
| MNEG_3522 | A0A0D2LCG3 | 26S proteasome regulatory subunit T2 | 0.853 | up | 2.85 × 10−6 |
| MNEG_4662 | A0A0D2MJY5 | 20S proteasome subunit beta 6 | 0.620 | up | 7.11 × 10−5 |
| MNEG_6814 | A0A0D2MKU1 | 26S proteasome regulatory subunit T1 | 0.611 | up | 5.76 × 10−5 |
| MNEG_7470 | A0A0D2MIK8 | Putative 26S proteasome non-ATPase regulatory subunit 6 | 0.762 | up | 4.28 × 10−5 |
| MNEG_8800 | A0A0D2M741 | Proteasome subunit beta | 0.800 | up | 2.14 × 10−5 |
| RPT5 | A8IIP7 | 26S proteasome regulatory subunit | 0.785 | up | 1.19 × 10−5 |
| PBD1 | A8JAI8 | Proteasome subunit beta | −0.865 | down | 4.09 × 10−6 |
| POA2 | A8JEW4 | Proteasome subunit alpha type | −0.585 | down | 1.42 × 10−4 |
| Gene Name | ID | Annotation | log2 (FC) for G+ vs. G− | Regulation | p-Value |
|---|---|---|---|---|---|
| CHLREDRAFT_38643 | A8J0R6 | Alanine-tRNA ligase (Fragment) | 0.73 | up | 7.29 × 10−5 |
| MNEG_4651 | A0A0D2L8Z9 | Alanyl-tRNA synthetase | 0.64 | up | 3.26 × 10−5 |
| CHLRE_06g279150v5 | A8J1X8 | Aspartyl-tRNA synthetase | 0.62 | up | 3.96 × 10−5 |
| ATF1 | A8IZE7 | Glutamine-fructose−6-phosphate transaminase (isomerizing) | 0.59 | up | 1.99 × 10−3 |
| HemA | Q9FPR7 | Glutamyl-tRNA reductase | −0.73 | down | 3.98 × 10−4 |
| CHLRE_16g694850v5 | A0A2K3CSB8 | Arginine biosynthesis bifunctional protein ArgJ, chloroplastic | −1.06 | down | 2.48 × 10−6 |
| MNEG_2778 | A0A0D2LET9 | Argininosuccinate lyase | −0.72 | down | 2.12 × 10−5 |
| MNEG_6059 | A0A0D2L3Z0 | Glutamate synthase (NADPH/NADH) | −0.84 | down | 5.27 × 10−5 |
| MNEG_3551 | A0A0D2LCD0 | Glutamate synthase (NADPH/NADH) | −0.65 | down | 8.17 × 10−4 |
| MNEG_0007 | A0A0D2LNS4 | 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase | 1.13 | up | 2.00 × 10−6 |
| METE | A8JH37 | 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase | 2.05 | up | 1.69 × 10−7 |
| OASTL3 | A8IEE5 | Cysteine synthase | −0.98 | down | 1.37 × 10−6 |
| MNEG_11543 | A0A0D2KKW6 | Cysteine synthase A | −0.76 | down | 1.62 × 10−5 |
| MNEG_10819 | A0A0D2M0L3 | Cysteine synthase A | −0.62 | down | 1.57 × 10−4 |
| MNEG_3728 | A0A0D2K0T8 | Glutamine synthetase | −0.64 | down | 2.26 × 10−4 |
| MNEG_7613 | A0A0D2MI17 | Glutamine synthetase (Fragment) | −0.77 | down | 9.52 × 10−5 |
| CHLRE_06g293950v5 | A0A2K3DQH9 | Serine hydroxymethyltransferase | 0.95 | up | 3.64 × 10−6 |
| PST1 | A8IH03 | Phosphoserine aminotransferase | 1.14 | up | 4.96 × 10−6 |
| MNEG_16458 | A0A0D2LNB1 | Serine/threonine-protein phosphatase | −1.24 | down | 6.35 × 10−7 |
| AAD1 | A8IX80 | Acetohydroxyacid dehydratase | −2.28 | down | 8.58 × 10−7 |
| ALSL1 | A8J1U3 | Acetolactate synthase, large subunit | −0.84 | down | 1.81 × 10−4 |
| MNEG_15388 | A0A0D2MB71 | Isoleucyl-tRNA synthetase | 0.65 | up | 2.83 × 10−5 |
| BCA2 | A8I5J8 | Branched-chain-amino-acid aminotransferase | −0.66 | down | 1.57 × 10−4 |
| Gene Name | ID | Annotation | log2 (FC) for G+ vs. G− | Regulation | p-Value |
|---|---|---|---|---|---|
| ycf3 | A0A140HA77 | Photosystem I assembly protein Ycf3 | −0.31 | NS | 3.37 × 10−3 |
| psaC | A0A140HA43 | Photosystem I iron-sulfur center | −1.36 | down | 2.14 × 10−6 |
| psaA | A0A140HA40 | Photosystem I P700 chlorophyll a apoprotein A1 | −2.29 | down | 4.13 × 10−8 |
| psaB | A0A140HA64 | Photosystem I P700 chlorophyll a apoprotein A2 | −1.82 | down | 1.41 × 10−7 |
| PsaD | Q5NKW4 | Photosystem I reaction center subunit II, 20 kDa | −1.44 | down | 2.86 × 10−4 |
| psaN | Q9AXJ2 | Photosystem I reaction center subunit N | −2.04 | down | 1.06 × 10−5 |
| psbC | A0A140HA26 | Photosystem II CP43 reaction center protein | −1.64 | down | 2.98 × 10−7 |
| psbB | A0A140HA55 | Photosystem II CP47 reaction center protein | −1.49 | down | 3.47 × 10−7 |
| psbD | A0A140HA41 | Photosystem II D2 protein | −1.58 | down | 3.30 × 10−7 |
| psbA | A0A140HA23 | Photosystem II protein D1 | −1.48 | down | 3.43 × 10−7 |
| MNEG_8562 | A0A0D2M7P8 | Photosystem II stability/assembly factor (Fragment) | −1.37 | down | 2.00 × 10−7 |
| PsbP domain-containing protein | A0A2K3D661 | Photosystem II PsbP domain-containing protein | −1.55 | down | 1.00 × 10−4 |
| rbcL | A0A140HA49 | Ribulose bisphosphate carboxylase large chain | −2.05 | down | 4.65 × 10−8 |
| rbcL.1 | A0A218N8A3 | Ribulose bisphosphate carboxylase large chain | −0.23 | NS | 2.00 × 10−1 |
| rbcL.3 | A0A517BB24 | Ribulose bisphosphate carboxylase large chain (Fragment) | −1.36 | down | 4.31 × 10−4 |
| rbcL.4 | Q3HTJ4 | Ribulose bisphosphate carboxylase large chain (Fragment) | −2.29 | down | 3.03 × 10−4 |
| RBCS | M4QL06 | Ribulose bisphosphate carboxylase small chain | −0.97 | down | 7.98 × 10−4 |
| MNEG_12441 | A0A0D2KIA7 | Ribulose bisphosphate carboxylase/oxygenaseactivase | −1.04 | down | 3.04 × 10−3 |
| SDH1 | A8HP06 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | 0.83 | up | 5.93 × 10−6 |
| MNEG_7567 | A0A0D2N2H0 | Thylakoid formation protein 1 | −0.39 | NS | 1.03 × 10−1 |
| MNEG_2022 | A0A0D2MTM8 | Thylakoid lumenal 17.4 kDa protein, chloroplastic | −1.50 | down | 4.25 × 10−5 |
| CPLD44 | A8J6G0 | Thylakoid lumenal protein | −1.16 | down | 8.51 × 10−6 |
| MNEG_1868 | A0A0D2K714 | Pyruvate kinase | 0.19 | NS | 2.96 × 10−2 |
| MNEG_11538 | A0A0D2LYE6 | Pyruvate kinase | −0.19 | NS | 3.69 × 10−2 |
| PYK1 | A8IVR6 | Pyruvate kinase | 0.03 | NS | 8.10 × 10−1 |
| PYK2 | A8J214 | Pyruvate kinase | 0.56 | NS | 1.13 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, W.; Chen, R.; Wang, X.; Liu, J.; Lv, W. Quantitative Proteomics of Chromochloris zofingiensis Reveals the Key Proteins Involved in Cell Growth and Bioactive Compound Biosynthesis. Plants 2022, 11, 1851. https://doi.org/10.3390/plants11141851
Qiu W, Chen R, Wang X, Liu J, Lv W. Quantitative Proteomics of Chromochloris zofingiensis Reveals the Key Proteins Involved in Cell Growth and Bioactive Compound Biosynthesis. Plants. 2022; 11(14):1851. https://doi.org/10.3390/plants11141851
Chicago/Turabian StyleQiu, Wen, Rongfeng Chen, Xianxian Wang, Junying Liu, and Weiguang Lv. 2022. "Quantitative Proteomics of Chromochloris zofingiensis Reveals the Key Proteins Involved in Cell Growth and Bioactive Compound Biosynthesis" Plants 11, no. 14: 1851. https://doi.org/10.3390/plants11141851
APA StyleQiu, W., Chen, R., Wang, X., Liu, J., & Lv, W. (2022). Quantitative Proteomics of Chromochloris zofingiensis Reveals the Key Proteins Involved in Cell Growth and Bioactive Compound Biosynthesis. Plants, 11(14), 1851. https://doi.org/10.3390/plants11141851





