Understanding and Comprehensive Evaluation of Cold Resistance in the Seedlings of Multiple Maize Genotypes
Abstract
:1. Introduction
2. Results
2.1. Combined Analysis of Temperature Treatments and Maize Genotypes on All Traits
2.2. Maize Genetic Variation of All Traits
2.3. Maize Phenotypic and Physiological Variations of All Traits in Response to Low-Temperature Stress
2.4. Framework for Relationships among All Traits
2.5. Comprehensive Evaluation of Cold Resistance among All Maize Genotypes Seedlings
2.6. Differential Expression of Six Candidate Genes Involved in ROS Scavenging and PAs Metabolisms
3. Discussion
4. Materials and Methods
4.1. Maize Genotypes and Different Temperature Treatments
4.2. Phenotypic Observations
4.3. MSI and MDA Content
4.4. ROS Level
4.5. Pro Content
4.6. Antioxidant Enzyme Activity
4.7. PAs Concentration
4.8. Chlorophyll Content and Photosynthetic Parameters
4.9. RT-qPCR Analysis
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.H.; Hu, H.R.; Hu, X.M.; Wang, G.H.; Du, X.M.; Li, L.; Wang, F.; Fu, J.J.; Wang, G.Y.; Wang, J.H.; et al. Transcriptome analysis of near-isogenic lines provided novel insights into genes associated with seed low-temperature germination ability in maize (Zea mays L.). Plants 2022, 11, 887. [Google Scholar] [CrossRef] [PubMed]
- Sobkowiak, A.; Jończyk, M.; Jarochowska, E.; Biecek, P.; Trzcinska-Danielewicz, J.; Leipner, J.; Fronk, J.; Sowiński, P. Genome-wide transcriptiomic analysis of response to low temperature reveals camdidate genes determining divergent cold-sensitivity of maize inbred lines. Plant Mol. Biol. 2014, 85, 317–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avila, L.M.; Obeidat, W.; Earl, H.; Niu, X.M.; Hargreaves, W.; Lukens, L. Shared and genetically distinct Zea mays transcriptome responses to ongoing and past low temperature exposure. BMC Genom. 2018, 19, 761. [Google Scholar] [CrossRef] [Green Version]
- Meng, A.J.; Wen, D.X.; Zhang, C.Q. Maize seed germination under low-temperature stress impacts seedling growth under normal temperature by modulating photosynthesis and antioxidant metabolism. Front. Plant Sci. 2022, 13, 843033. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, V.M.; Butrón, A.; Rady, M.O.A.; Soengas, P.; Revilla, P. Identification of quantitative trait loci involved in the response to cold stress in maize (Zea mays L.). Mol. Breed. 2014, 33, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Hu, G.H.; Liu, X.F.; Zhou, Y.; Li, Y.; Zhang, X.; Yuan, X.H.; Zhang, Q.; Yang, D.G. Transcriptome sequencing identified genes and gene ontologies associated with early freezing tolerance in maize. Front. Plant Sci. 2016, 17, 1477. [Google Scholar] [CrossRef] [Green Version]
- Holubová, L.; Švubová, R.; Slováková, L.; Bokor, B.; Kročková, V.C.; Renčko, J.; Uhrin, F.; Medvecká, V.; Zahoranová, A.; Gálová, E. Cold atmospheric pressure plasma treatment of maize grains-induction of growth, enzyme activities and heat shock proteins. Int. J. Mol. Sci. 2021, 22, 8509. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, J.S.; Wang, G.F.; Fan, X.Y.; Sun, X.; Qin, H.L.; Xu, N.; Zhong, M.Y.; Qiao, Z.Y.; Tang, Y.P.; et al. Proline responding1 plays a critical role in regulating general protein synthesis and the cell cycle in maize. Plant Cell 2014, 26, 2582–2600. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.H.; Xiong, F.; Nong, S.H.; Liao, J.R.; Xing, A.Q.; Shen, Q.; Ma, Y.C.; Fang, W.P.; Zhu, X.J. Effects of nitric oxide on the GABA, polyamines, and proline in tea (Camellia sinensis) root under cold stress. Sci. Rep. 2020, 10, 12240. [Google Scholar] [CrossRef]
- Takahashi, T.; Kakehi, J.I. Polyamines: Ubiquitous polycations with unique roles in growth and stress responses. Ann. Bot. 2010, 105, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Igarashi, K.; Kashiwagi, K. Polyamines: Mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 2000, 271, 559–564. [Google Scholar] [CrossRef]
- Gao, C.H.; Sheteiwy, M.S.; Lin, C.; Guan, Y.J.; Ulhassan, Z.; Hu, J. Spermidine suppressed the inhibitory effects of polyamines inhibitors combination in maize (Zea mays) seedlings under chilling stress. Plants 2021, 10, 2421. [Google Scholar] [CrossRef]
- Gao, C.H.; Sheteiwy, M.S.; Han, J.J.; Dong, Z.R.; Pan, R.H.; Guan, Y.J.; Hamoud, Y.A.; Hu, J. Polyamine biosynthetic pathways and their relation with the cold tolerance of maize (Zea mays L.) seedlings. Plant Signal. Behav. 2020, 15, 1807722. [Google Scholar] [CrossRef]
- Gao, C.H.; Hu, J.; Zhang, S.; Zheng, Y.Y.; Knapp, A. Association of polyamines in governing the chilling sensitivity of maize genotypes. Plant Growth Regul. 2009, 57, 31–38. [Google Scholar] [CrossRef]
- Leipner, J. Chilling Stress in Maize: From Physiology to Genetics and Molecular Mechanisms. Ph.D. Thesis, Eidgenössische Technische Hochschule Zürich, Department of Agricultural and Food Science, Zürich, Switzerland, 2009. [Google Scholar]
- Sobkowiak, A.; Joczyk, M.; Adamczyk, J.; Szczepank, J.; Solecka, D.; Kuciara, I.; Hetmańczyk, K.; Trzcinska-Danielewicz, J.; Grzybowski, M.; Skoneczny, M.; et al. Molecular foundations of chilling–tolerance of modern maize. BMC Genom. 2016, 17, 125. [Google Scholar] [CrossRef] [Green Version]
- Fracheboud, Y.; Jompuk, C.; Ribaut, J.M.; Stamp, P.; Leipner, J. Genetic analysis of cold–tolerance of photosynthesis in maize. Plant Mol. Biol. 2004, 56, 241–253. [Google Scholar] [CrossRef]
- Wang, H.Y.; Li, Z.B.; Yuan, L.Y.; Zhou, H.F.; Hou, X.L.; Liu, T.K. Cold acclimation can specifically inhibit chlorophyll biosynthesis in young leaves of Pakchoi. BMC Plant Biol. 2021, 21, 172. [Google Scholar] [CrossRef]
- Wei, Y.L.; Chen, H.Z.; Wang, L.; Zhao, Q.; Wang, D.; Zhang, T.E. Cold acclimation alleviates cold stress–induced PSII inhibition and oxidative damage in tobacco leaves. Plant Signal. Behav. 2022, 17, 2013638. [Google Scholar] [CrossRef]
- Ahmad, A.; Aslam, Z.; Javed, T.; Hussain, S.; Raza, A.; Shabbir, R.; Mora-Poblete, F.; Saeed, T.; Zulfiqar, F.; Ali, M.M.; et al. Screening of wheat (Triticum aestivum L.) genotypes for drought tolerance through agronomic and physiological response. Agronomy 2022, 12, 287. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Shi, J.; Niu, Y.N.; Lu, P.N.; Chen, X.J.; Mao, T.T. 24–epibrassinolide alleviates aluminum toxicity by improving leaf chlorophyll fluorescence and photosynthetic performance and root antioxidant-oxidant balance and ascorbate-glutathione cycle in maize. Russ. J. Plant Physiol. 2022, 69, 1–10. [Google Scholar] [CrossRef]
- Cofer, T.M.; Engelberth, M.; Engelberth, J. Green leaf volatiles protect maize (Zea mays) seedlings against damage from cold stress. Plant Cell Environ. 2018, 41, 1673–1682. [Google Scholar] [CrossRef]
- Yan, C.J.; Song, S.H.; Wang, W.B.; Wang, C.L.; Li, H.B.; Wang, F.; Li, S.Y.; Sun, X.G. Screening diverse soybean genotypes for drought tolerance by membership function value based on multiple traits and drought–tolerant coefficient of yield. BMC Plant Biol. 2020, 20, 321. [Google Scholar] [CrossRef]
- Yi, Q.; Malvar, R.A.; Álvarez-Iglesias, l.; Ordás, B.; Revilla, P. Dissecting the genetics of cold tolerance in a multiparental maize population. Theor. Appl. Genet. 2020, 133, 503–516. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, J.H.; Zhao, R.L.; Xu, K.; Xiao, Y.R.; Zhang, S.H.; Tian, J.C.; Yang, X.J. Genome-wide association study reveals the genetic basis of cold tolerance in wheat. Mol. Breed. 2020, 40, 36. [Google Scholar] [CrossRef]
- Liu, H.L.; Xin, W.; Wang, Y.L.; Zhang, D.Z.; Wang, J.G.; Zheng, H.L.; Yang, L.M.; Nie, S.J.; Zou, D.T. An integrated analysis of the rice transcriptome and lipidome reveals lipid metabolism plays a central role in rice cold tolerance. BMC Plant Biol. 2022, 22, 91. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, D.M.; Wang, Q.; Song, X.Y.; Wang, Y.B.; Yang, X.L.; Qin, D.L.; Xie, T.L.; Yang, D.G. Exogenous salicylic acid improves chilling tolerance in maize seedlings by improving plant growth and physiological characteristics. Agronomy 2021, 11, 1341. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, M.; Gong, X.L.; Liu, C.; Hong, M.M.; Wang, L.; Hong, F.S. Influence of lanthanides on the antioxidative defense system in maize seedlings under cold stress. Bio. Trace Res. 2011, 142, 819–830. [Google Scholar] [CrossRef]
- Stewart, C.R.; Martin, B.A.; Reding, L.; Cerwick, S. Seedling growth, mitochondrial characteristics and alternative respiratory capacity of corn genotypes differing in cold tolerance. Plant Physiol. 1990, 92, 761–766. [Google Scholar] [CrossRef] [Green Version]
- Obeidat, W.; Avila, L.; Earl, H.; Lukens, L. Leaf spectral reflectance of maize seedlings and its relationship to cold tolerance. Crop Sci. 2018, 58, 2569–2580. [Google Scholar] [CrossRef]
- Miedema, P. The effects of low temperature on Zea mays L. Adv. Agron. 1982, 35, 93–128. [Google Scholar]
- Len, J.S.; Koh, W.S.D.K.; Tan, S.X. The roles of reactive oxygen species and antioxidants in cryopreservation. BioScience Rep. 2019, 39, BSR20191601. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.Q.; Niu, Y.N.; Bai, X.D.; Mao, T.T. Transcriptomic and metabolic profiling reveals a lignin metabolism network involved in mesocotyl elongation during maize seed germination. Plants 2022, 11, 1034. [Google Scholar] [CrossRef]
- Oracz, K.; Karpiński, S. Phytohormones signaling pathways and ROS involvement in seed germination. Front. Plant Sci. 2016, 7, 864. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.T.; Wang, W.; Mao, B.G.; Chu, C.C. Cold stress tolerance in rice: Physiological changes, molecular mechanism, and future prospects. Hereditas 2018, 40, 171–185. [Google Scholar]
- Melanie, M.; Sergi, M.B. Malondialdehyde: Facts and artifacts. Plant Physiol. 2019, 180, 1246–1250. [Google Scholar]
- Popov, V.N.; Naraikina, N.V. Change of antioxidant enzymes activity during low-temperature hardening of Nicotiana tabacum L. and Secale cereal L. Russ. J. Plant Physiol. 2020, 67, 898–905. [Google Scholar] [CrossRef]
- Ramazan, S.; Qazi, H.A.; Dar, Z.A.; John, R. Low temperature elicits differential biochemical and antioxidant responses in maize (Zea Mays) genotypes with different susceptibility to low temperature stress. Physiol. Mol. Biol. Plants 2021, 27, 1395–1412. [Google Scholar] [CrossRef]
- Dreyer, A.; Dietz, K.J. Reactive oxygen species and the redox–regulatory network in cold stress acclimation. Antioxidants 2018, 7, 169. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.Y.; Liu, Z.G.; Mi, W.B.; Xu, C.M.; Xu, M.X.; Zhou, Y.; Zhen, G.Q.; Cao, X.D.; Fang, X.L.; Mi, C. Overexpression of BrAFP1 gene from winter rapeseed (Brassica rapa) confers cold tolerance in Arabidopsis. Plant Physiol. Bioch. 2020, 155, 338–345. [Google Scholar] [CrossRef]
- Qin, Q.Y.; Liu, T.; Qu, J.X.; Liu, D.M.; Zhang, X.W.; Yan, J. Research progress of proline as cryoprotectant. Chin. J. Reprod. Contracept. 2022, 42, 213–217. [Google Scholar]
- Liu, Z.G.; Sun, W.C.; Zhao, Y.N.; Li, X.C.; Fang, Y.; Wu, J.Y.; Zeng, X.C.; Yang, N.N.; Wang, Y.; He, L. Effects of low nocturnal temperature on photosynthetic characteristics and chloroplast ultrastructure of winter rapeseed. Russ. J. Plant Physiol. 2016, 63, 451–460. [Google Scholar] [CrossRef]
- Gilmore, A.M. 1997. Mechanistic aspects of xanthophyll cycledependent photoprotection in higher plant chloroplasts and leaves. Physiol. Plant 1997, 99, 197–209. [Google Scholar] [CrossRef]
- Pfündel, E.; Bilger, W. Regulation and possible function of the violaxanthin cycle. Photosynth. Res. 1994, 42, 89–109. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.G.; Liu, P.P.; Jiang, Z.S.; Ai, X.Z. Overexpression of the rubisco activase gene improves growth and low temperature and weak light tolerance in Cucumix sativus. Physiol. Plant. 2017, 161, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Bragina, T.V.; Ponomareva, Y.V.; Drozdova, I.S.; Grinieva, G.M. Photosynthesis and dark respiration in leaves of different ages of partly flooded maize seedlings. Russ. J. Plant Physiol. 2004, 51, 383–389. [Google Scholar] [CrossRef]
- Garcia, M.J.; Littler, A.S.; Sriram, A.; Teets, N.M. Distinct cold tolerance traits in independently across genotypes in drosophila melanogaster. Evolution 2020, 74, 1437–1450. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Zhong, Y. Genetic dissection of the photosynthetic parameters of maize (Zea mays L.) in drought–stressed and well–watered environments. Russ. J. Plant Physiol. 2021, 68, 1125–1134. [Google Scholar] [CrossRef]
- Goering, R.; Larsen, S.; Tan, J.; Whelan, J.; Makarevitch, I. QTL mapping of seedling tolerance to exposure to low temperature in the maize IBM RIL population. PLoS ONE 2021, 16, e0254437. [Google Scholar] [CrossRef]
- David, R.H.A.; Ramakrishnan, M.; Maharajan, T.; Barathikannan, K.; Babu, G.A.; Daniel, M.A.; Agastian, P.; Caesar, S.A.; Ignacimuthu, S. Mining QTL and genes for root traits and biochemical parameters under vegetative drought in South Indian genotypes of finger millet (Eleusine coracana (L.) Gaertn) by association mapping and in silico comparative genomics. Biocatal. Agric. Biotech. 2021, 32, 101935. [Google Scholar] [CrossRef]
- Wassom, J.J.; Mei, C.S.; Rocheford, T.R.; Widholm, J.M. Allelic variation in candidate genes associated with QTL for the maize anther culture response. Maydica 2000, 45, 267–276. [Google Scholar]
- EI–Mahdy, M.T.; Youssef, M.; Eissa, M.A. Impact of in vitro cold stress on two banana genotypes based on physio-biochemical evaluation. S. Afr. J. Bot. 2018, 119, 219–225. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Wang, E.M.; Zhao, T.F.; Wang, Q.Q.; Chen, L.J. Characteristics of chlorophyll fluorescence and antioxidant-oxidant balance in PEPC and PPDK transgenic rice under aluminum stress. Russ. J. Plant Physiol. 2018, 65, 49–56. [Google Scholar] [CrossRef]
- Choudhary, S.P.; Oral, H.V.; Bhardwaj, R.; Yu, J.Q.; Tran, L.S.P. Interaction of brassinosteroids and polyamines enhances copper stress tolerance in Raphanus sativus. J. Exp. Bot. 2012, 63, 5659–5675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, P.; Bhardwaj, R.; Kanwar, M.K. 24–epibrassinolide induced antioxidative defense system of Brassica juncea L. under Zn metal stress. Physiol. Mol. Biol. Plants 2010, 16, 285–293. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.Q.; Zhong, Y.; Zhou, W.Q. Molecular mechanisms of mesocotyl elongation induced by brassinosteroid in maize under deep-seeding stress by RNA-sequencing, microstructure observation, and physiological metabolism. Genomics 2021, 113, 3565–3581. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Niu, Y.N. The combination of conventional QTL analysis, bulked-segregant analysis, and RNA-sequencing provide new genetic insights into maize mesocotyl elongation under multiple deep-seeding environments. Int. J. Mol. Sci. 2022, 23, 4223. [Google Scholar] [CrossRef]

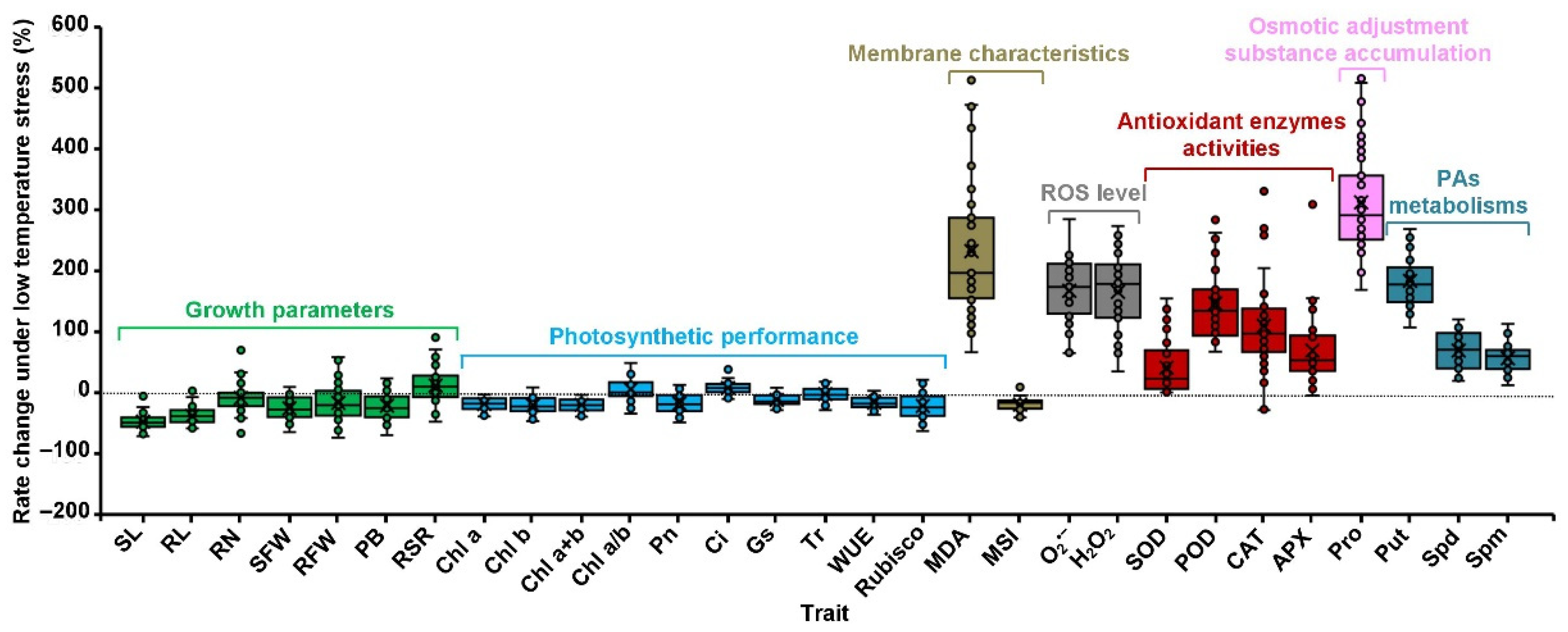
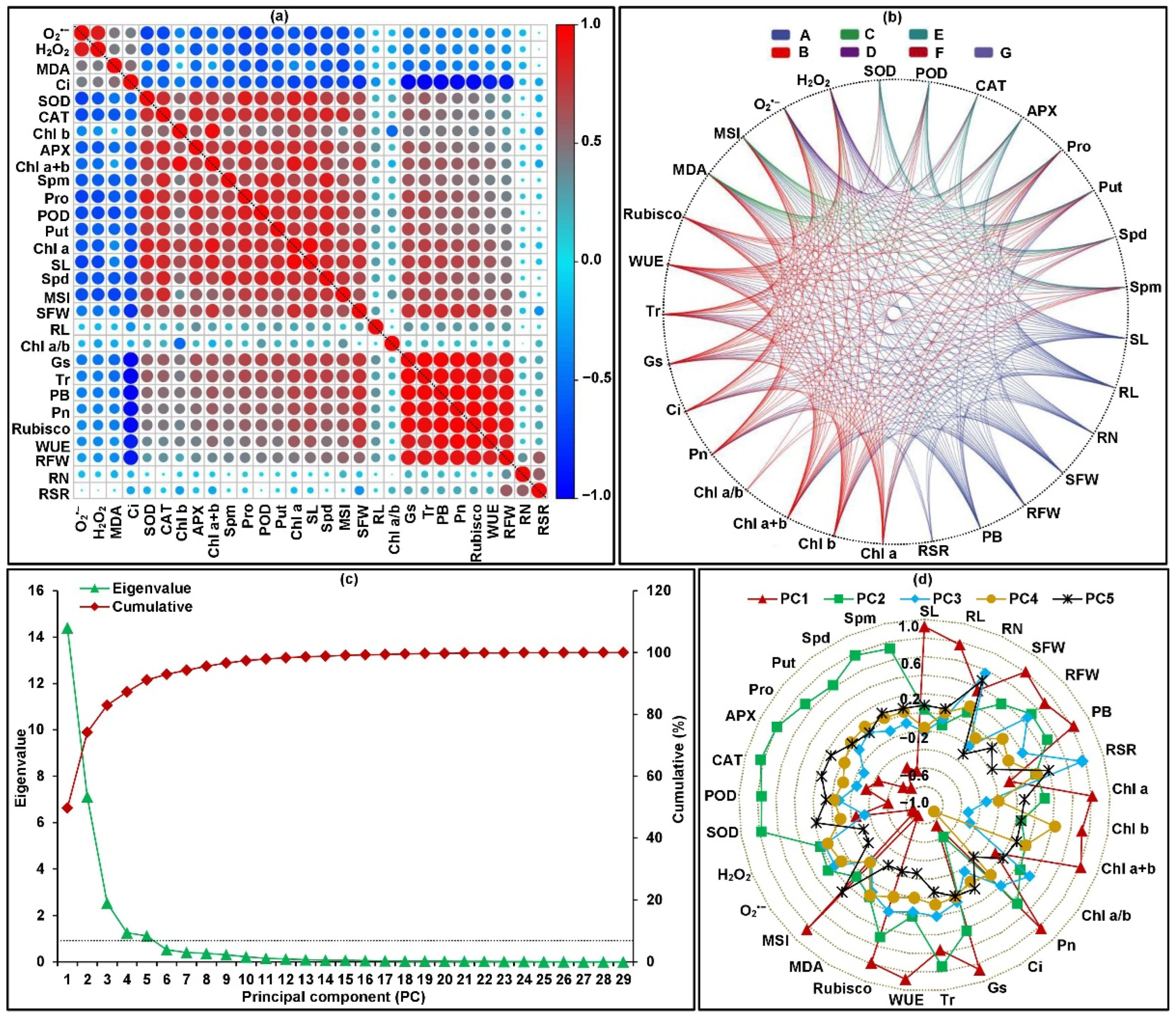
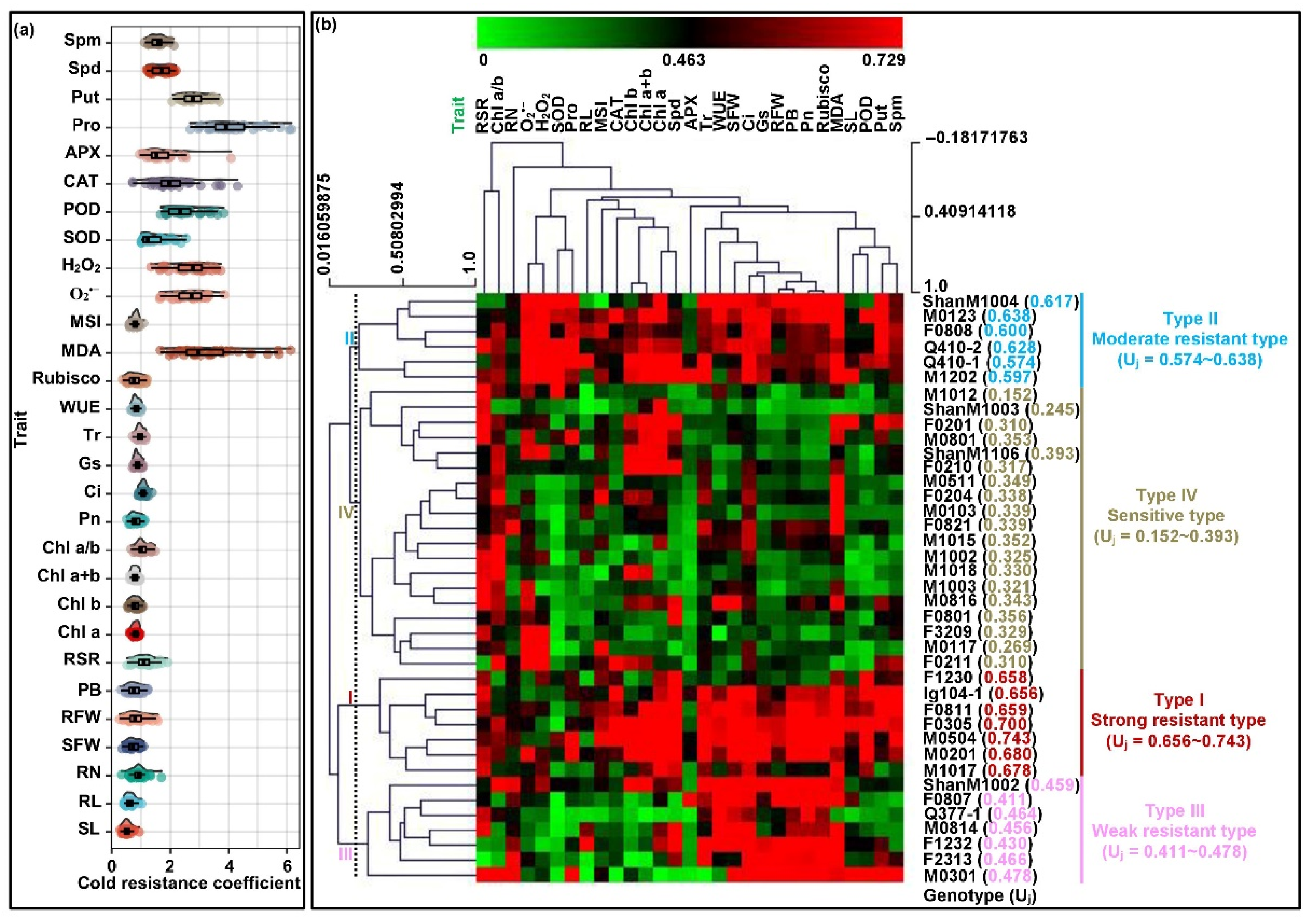

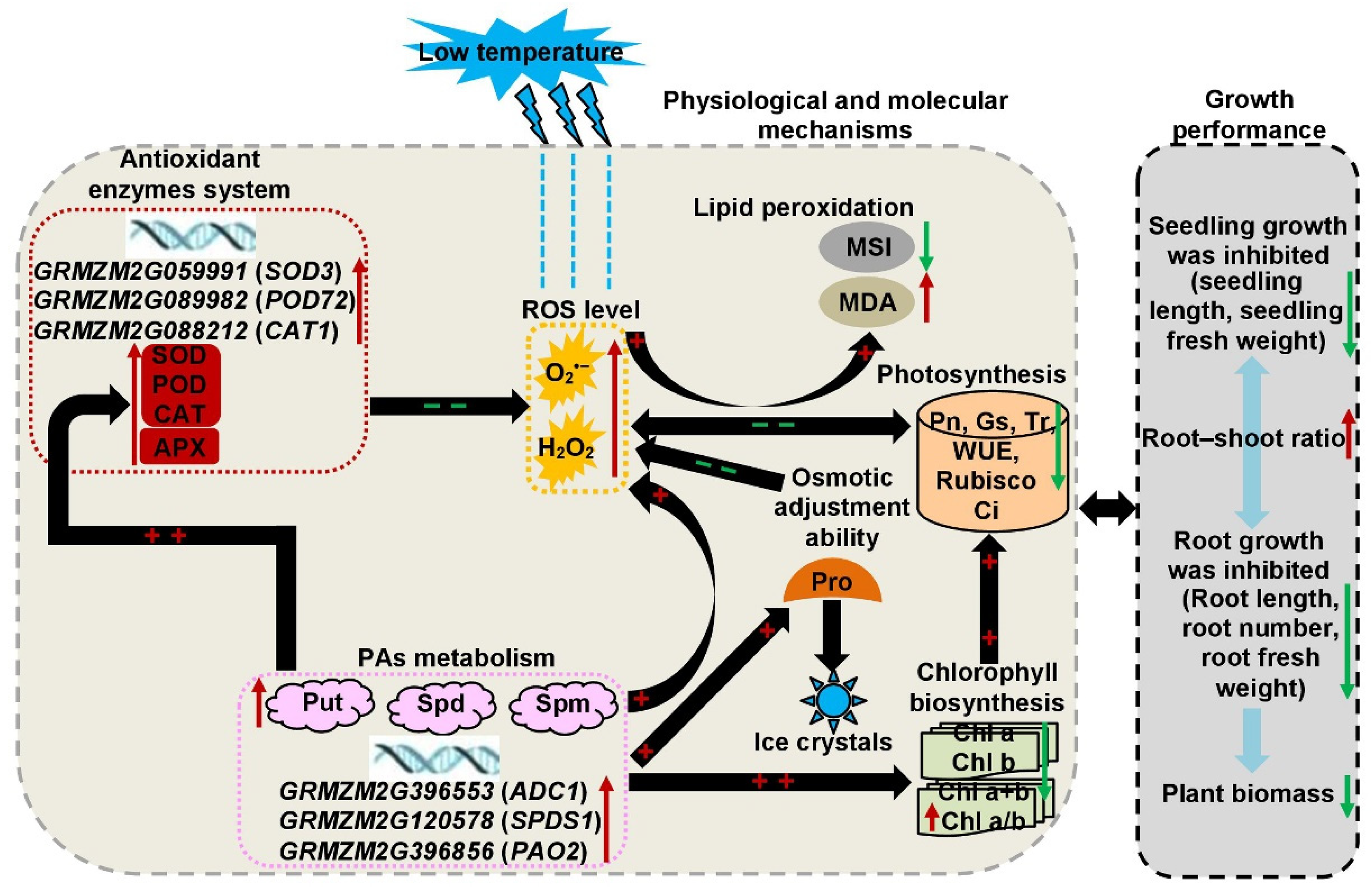
| Trait | Genotypes (G) | Temperature (T) | G × T Interaction | CVg (CK) (%) | CVg (LT) (%) |
|---|---|---|---|---|---|
| Growth parameter | |||||
| SL | F = 13.449 (p < 0.001) | F = 1335.996 (p < 0.001) | F = 4.637 (p < 0.001) | 20.58 | 27.56 |
| RL | F = 11.824 (p < 0.001) | F = 623.327 (p < 0.001) | F = 3.577 (p < 0.001) | 22.55 | 24.66 |
| RN | F = 4.237 (p < 0.001) | F = 17.938 (p < 0.001) | F = 2.409 (p < 0.001) | 16.96 | 23.99 |
| SFW | F = 14.358 (p < 0.001) | F = 241.485 (p < 0.001) | F = 3.906 (p < 0.001) | 27.29 | 28.38 |
| RFW | F = 9.652 (p < 0.001) | F = 67.389 (p < 0.001) | F = 3.621 (p < 0.001) | 21.08 | 34.54 |
| PB | F = 1297.621 (p < 0.001) | F = 211.363 (p < 0.001) | F = 13.034 (p < 0.01) | 26.54 | 27.92 |
| RSR | F = 7.794 (p < 0.001) | F = 15.104 (p < 0.001) | F = 2.318 (p < 0.001) | 24.15 | 31.21 |
| Photosynthetic performance | |||||
| Chl a | F = 623.203 (p < 0.001) | F = 182.376 (p < 0.001) | F = 12.377 (p < 0.01) | 10.88 | 16.75 |
| Chl b | F = 1025.780 (p < 0.001) | F = 481.451 (p < 0.001) | F = 4.053 (p < 0.05) | 15.46 | 17.90 |
| Chl a+b | F = 932.963 (p < 0.001) | F = 365.686 (p < 0.001) | F = 2.760 (p < 0.05) | 11.49 | 16.07 |
| Chl a/b | F = 146.129 (p < 0.001) | F = 172.080 (p < 0.001) | F = 30.104 (p < 0.001) | 13.45 | 12.02 |
| Pn | F = 28.235 (p < 0.001) | F = 9.676 (p < 0.001) | F = 2.172 (p < 0.001) | 21.44 | 18.63 |
| Ci | F = 163.418 (p < 0.001) | F = 13.466 (p < 0.001) | F = 9.302 (p < 0.001) | 8.67 | 9.21 |
| Gs | F = 210.699 (p < 0.001) | F = 78.650 (p < 0.001) | F = 4.512 (p < 0.05) | 6.52 | 11.82 |
| Tr | F = 99.425 (p < 0.001) | F = 38.087 (p < 0.001) | F = 9.110 (p < 0.001) | 9.19 | 10.87 |
| WUE | F = 98.802 (p < 0.001) | F = 38.862 (p < 0.001) | F = 9.176 (p < 0.001) | 13.16 | 8.43 |
| Rubisco | F = 78.860 (p < 0.001) | F = 22.607 (p < 0.001) | F = 7.635 (p < 0.001) | 32.15 | 29.16 |
| Membrane characteristics | |||||
| MDA | F = 311.124 (p < 0.001) | F = 1518.619 (p < 0.001) | F = 153.605 (p < 0.001) | 18.72 | 27.18 |
| MSI | F = 340.754 (p < 0.001) | F = 387.618 (p < 0.001) | F = 30.448 (p < 0.001) | 5.76 | 14.07 |
| ROS level | |||||
| O2•− | F = 1350.626 (p < 0.001) | F = 6675.706 (p < 0.001) | F = 513.397 (p < 0.001) | 11.82 | 16.49 |
| H2O2 | F = 1125.0331 (p < 0.001) | F = 5655.161 (p < 0.001) | F = 320.381 (p < 0.001) | 15.20 | 19.74 |
| Antioxidant enzymes activity | |||||
| SOD | F = 2457.673 (p < 0.001) | F = 3069.115 (p < 0.001) | F = 1400.441 (p < 0.001) | 7.43 | 34.85 |
| POD | F = 9.451 (p < 0.01) | F = 56.748 (p < 0.001) | F = 2.291 (p < 0.05) | 14.56 | 24.27 |
| CAT | F = 1023.800 (p < 0.001) | F = 360.449 (p < 0.001) | F = 10.329 (p < 0.01) | 37.59 | 40.13 |
| APX | F = 6469.477 (p < 0.001) | F = 7908.238 (p < 0.001) | F = 700.159 (p < 0.001) | 15.35 | 27.81 |
| Osmotic adjustment substance accumulation | |||||
| Pro | F = 138.342 (p < 0.001) | F = 10.049 (p < 0.01) | F = 5.876 (p < 0.01) | 9.69 | 22.48 |
| PAs metabolism | |||||
| Put | F = 555.242 (p < 0.001) | F = 32.162 (p < 0.01) | F = 11.102 (p < 0.001) | 8.87 | 12.72 |
| Spd | F = 78.228 (p < 0.001) | F = 11.436 (p < 0.01) | F = 7.118 (p < 0.001) | 8.32 | 20.02 |
| Spm | F = 35.699 (p < 0.001) | F = 31.052 (p < 0.001) | F = 15.870 (p < 0.001) | 4.59 | 15.62 |
| Gene ID (Encoded Protein) | Gene Position | Primer Sequence (5′ to 3′) | Gene Functional Annotation |
|---|---|---|---|
| GRMZM2G396553 (Arginine decarboxylase 1, ADC1) | Chromosome 9 (23396672_23399720 bp) | F: GCTACGGCTCAAGGTACCAG R: CCGAACTCCACAATGTCCTC | Arginine decarboxylase activity (GO:0008792); Arginine catabolic process (GO:0006527); Response to cold (GO:0009409) |
| GRMZM2G120578 (Spermidine synthase 1, SPDS1) | Chromosome 5 (72695353_72701535 bp) | F: TGTTCAATTCCATCCCCTAAA R: TCCACTGAACTGTGTCTGGAA | Catalytic activity (GO:0003824); Transferase activity (GO:0016740); Polyamine metabolic process (GO:0006595); Polyamine biosynthetic process (GO:0006596) |
| GRMZM2G396856 (Probable polyamine oxidase 2, PAO2) | Chromosome 10 (143652563_143657803 bp) | F: CACACACACCATCCGCTATT R: CATCAGCAGCAGCAAGACC | Oxidoreductase activity (GO:0016491); Polyamine oxidase activity (GO:0046592); Polyamine catabolic process (GO:0006598) |
| GRMZM2G059991 (Superoxide dismutase 3, SOD3) | Chromosome 6 (140001982_140006117 bp) | F: TCACCCAAGAGGGAGATG R: TTGCTCGCAGGATTGTAG | Removal of superoxide radicals (GO:0019430); Superoxide dismutase activity (GO:0004784); Oxidoreductase activity (GO:0016491) |
| GRMZM2G089982 (Peroxidase 72, POD72) | Chromosome 3 (40090724_40092823 bp) | F: GGATGTATCCTACGCCGCAA R: TTGTCAAACTTGGCAGGGGT | Peroxidase activity (GO:0004601); Oxidoreductase activity (GO:0016491); Response to oxidative stress (GO:0006979) |
| GRMZM2G088212 (Catalase 1, CAT1) | Chromosome 5 (65456774_65461269 bp) | F: CCGAATCCAAAGACCAAT R: ATGCCAACATCGTCAAAGAG | Catalase activity (GO:0004096); Peroxidase activity (GO:0004601); Response to oxidative stress (GO:0006979); Response to heat (GO:0009408) |
| GRMZM2G126010 (actin 1) | Chromosome 8 (102413768_102417536 bp) | F: CGATTGAGCATGGCATTGTCA R: CCCACTAGCGTACAACGAA | ATP binding (GO:0005524); Nucleotide binding (GO:0000166) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Zhao, C.; Niu, Y.; Chao, W.; He, W.; Wang, Y.; Mao, T.; Bai, X. Understanding and Comprehensive Evaluation of Cold Resistance in the Seedlings of Multiple Maize Genotypes. Plants 2022, 11, 1881. https://doi.org/10.3390/plants11141881
Zhao X, Zhao C, Niu Y, Chao W, He W, Wang Y, Mao T, Bai X. Understanding and Comprehensive Evaluation of Cold Resistance in the Seedlings of Multiple Maize Genotypes. Plants. 2022; 11(14):1881. https://doi.org/10.3390/plants11141881
Chicago/Turabian StyleZhao, Xiaoqiang, Cai Zhao, Yining Niu, Wun Chao, Wei He, Yifan Wang, Taotao Mao, and Xiaodong Bai. 2022. "Understanding and Comprehensive Evaluation of Cold Resistance in the Seedlings of Multiple Maize Genotypes" Plants 11, no. 14: 1881. https://doi.org/10.3390/plants11141881
APA StyleZhao, X., Zhao, C., Niu, Y., Chao, W., He, W., Wang, Y., Mao, T., & Bai, X. (2022). Understanding and Comprehensive Evaluation of Cold Resistance in the Seedlings of Multiple Maize Genotypes. Plants, 11(14), 1881. https://doi.org/10.3390/plants11141881






