Reproducibility and Sensitivity of High-Throughput Sequencing (HTS)-Based Detection of Citrus Tristeza Virus and Three Citrus Viroids
Abstract
:1. Introduction
2. Results
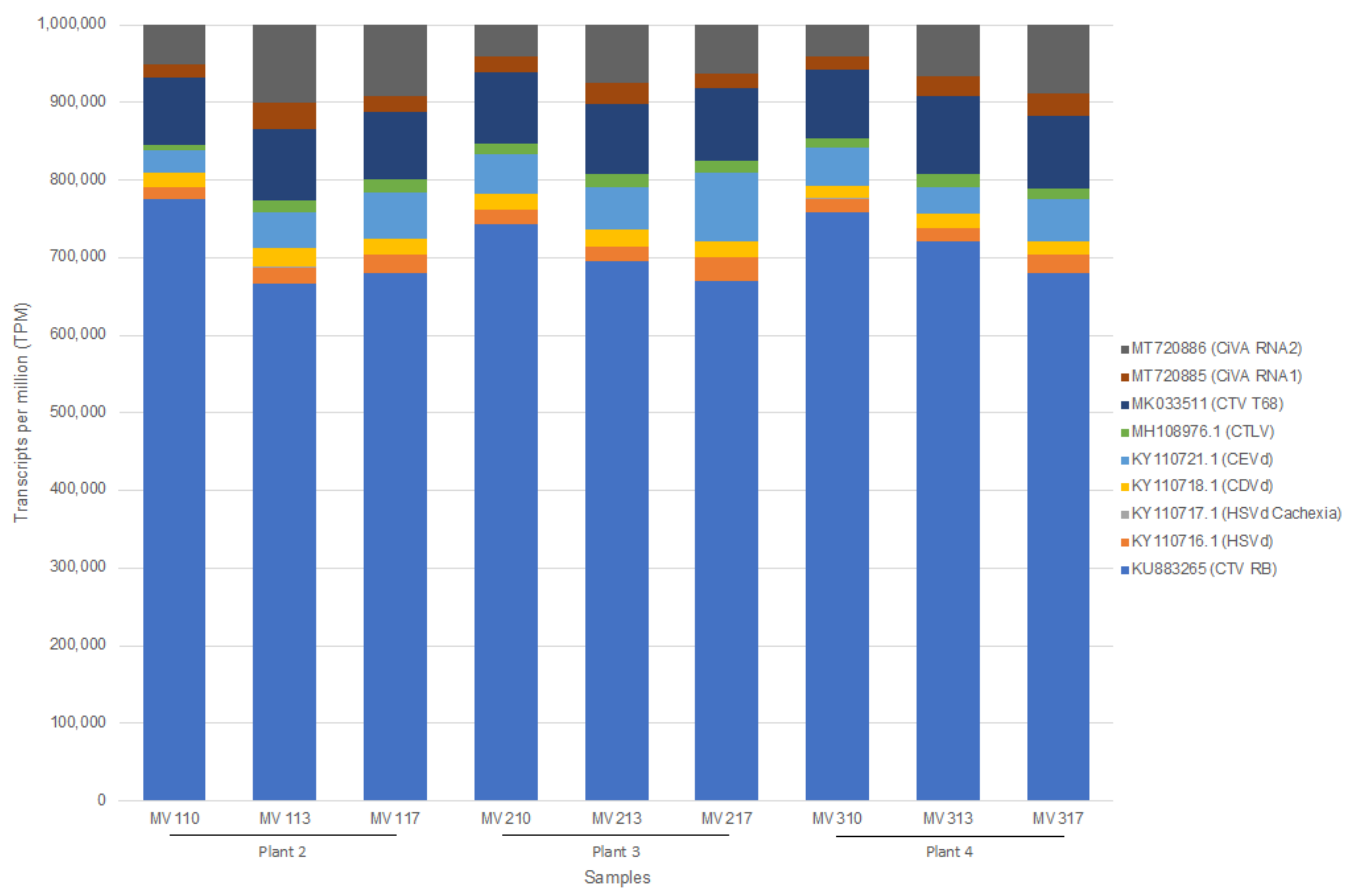
2.1. Reproducibility
2.2. Sensitivity
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Total RNA Extraction, RT-PCR and HTS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Massart, S.; Olmos, A.; Jijakli, H.; Candresse, T. Current Impact and Future Directions of High Throughput Sequencing in Plant Virus Diagnostics. Virus Res. 2014, 188, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Olmos, A.; Boonham, N.; Candresse, T.; Gentit, P.; Giovani, B.; Kutnjak, D.; Liefting, L.; Maree, H.J.; Minafra, A.; Moreira, A.; et al. High-Throughput Sequencing Technologies for Plant Pest Diagnosis: Challenges and Opportunities. EPPO Bull. 2018, 48, 219–224. [Google Scholar] [CrossRef] [Green Version]
- Maree, H.J.; Fox, A.; Al Rwahnih, M.; Boonham, N.; Candresse, T. Application of HTS for Routine Plant Virus Diagnostics: State of the Art and Challenges. Front. Plant Sci. 2018, 9, 1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villamor, D.E.V.; Ho, T.; Al Rwahnih, M.; Martin, R.R.; Tzanetakis, I.E. High Throughput Sequencing For Plant Virus Detection and Discovery. Phytopathology 2019, 109, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Maclot, F.; Candresse, T.; Filloux, D.; Malmstrom, C.M.; Roumagnac, P.; van der Vlugt, R.; Massart, S. Illuminating an Ecological Blackbox: Using High Throughput Sequencing to Characterize the Plant Virome Across Scales. Front. Microbiol. 2020, 11, 578064. [Google Scholar] [CrossRef] [PubMed]
- Soltani, N.; Stevens, K.A.; Klaassen, V.; Hwang, M.-S.; Golino, D.A.; Al Rwahnih, M. Quality Assessment and Validation of High-Throughput Sequencing for Grapevine Virus Diagnostics. Viruses 2021, 13, 1130. [Google Scholar] [CrossRef] [PubMed]
- Bester, R.; Cook, G.; Breytenbach, J.H.J.; Steyn, C.; De Bruyn, R.; Maree, H.J. Towards the Validation of High-Throughput Sequencing (HTS) for Routine Plant Virus Diagnostics: Measurement of Variation Linked to HTS Detection of Citrus Viruses and Viroids. Virol. J. 2021, 18, 61. [Google Scholar] [CrossRef] [PubMed]
- Rolland, M.; Villemot, J.; Marais, A.; Theil, S.; Faure, C.; Cadot, V.; Valade, R.; Vitry, C.; Rabenstein, F.; Candresse, T. Classical and next Generation Sequencing Approaches Unravel Bymovirus Diversity in Barley Crops in France. PLoS ONE 2017, 12, e0188495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagen, C.; Frizzi, A.; Gabriels, S.; Huang, M.; Salati, R.; Gabor, B.; Huang, S. Accurate and Sensitive Diagnosis of Geminiviruses through Enrichment, High-Throughput Sequencing and Automated Sequence Identification. Arch. Virol. 2012, 157, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Al Rwahnih, M.; Daubert, S.; Golino, D.; Islas, C.; Rowhani, A. Comparison of Next-Generation Sequencing Versus Biological Indexing for the Optimal Detection of Viral Pathogens in Grapevine. Phytopathology 2015, 105, 758–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rott, M.; Xiang, Y.; Boyes, I.; Belton, M.; Saeed, H.; Kesanakurti, P.; Hayes, S.; Lawrence, T.; Birch, C.; Bhagwat, B.; et al. Application of Next Generation Sequencing for Diagnostic Testing of Tree Fruit Viruses and Viroids. Plant Dis. 2017, 101, 1489–1499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bester, R.; Cook, G.; Maree, H.J. Citrus Tristeza Virus Genotype Detection Using High-Throughput Sequencing. Viruses 2021, 13, 168. [Google Scholar] [CrossRef] [PubMed]
- Santala, J.; Valkonen, J.P.T. Sensitivity of Small RNA-Based Detection of Plant Viruses. Front. Microbiol. 2018, 9, 939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecman, A.; Kutnjak, D.; Gutiérrez-Aguirre, I.; Adams, I.; Fox, A.; Boonham, N.; Ravnikar, M. Next Generation Sequencing for Detection and Discovery of Plant Viruses and Viroids: Comparison of Two Approaches. Front. Microbiol. 2017, 8, 1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visser, M.; Bester, R.; Burger, J.T.; Maree, H.J. Next-Generation Sequencing for Virus Detection: Covering All the Bases. Virol. J. 2016, 13, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-García, A.B.; Bester, R.; Olmos, A.; Maree, H.J. Bioinformatic Tools and Genome Analysis of Citrus Tristeza Virus. In Citrus Tristeza Virus: Methods and Protocols; Catara, A.F., Bar-Joseph, M., Licciardello, G., Eds.; Springer New York: New York, NY, USA, 2019; pp. 163–178. ISBN 978-1-4939-9558-5. [Google Scholar]
- Cook, G.; van Vuuren, S.P.; Breytenbach, J.H.J.; Burger, J.T.; Maree, H.J. Expanded Strain-Specific RT-PCR Assay for Differential Detection of Currently Known Citrus Tristeza Virus Strains: A Useful Screening Tool. J. Phytopathol. 2016, 164, 847–851. [Google Scholar] [CrossRef]
- Carra, A.; Gambino, G.; Schubert, A. A Cetyltrimethylammonium Bromide-Based Method to Extract Low-Molecular-Weight RNA from Polysaccharide-Rich Plant Tissues. Anal. Biochem. 2007, 360, 318–320. [Google Scholar] [CrossRef] [PubMed]
- Bester, R.; Pepler, P.T.; Burger, J.T.; Maree, H.J. Relative Quantitation Goes Viral: An RT-QPCR Assay for a Grapevine Virus. J. Virol. Methods 2014, 210, 67–75. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bester, R.; Steyn, C.; Breytenbach, J.H.J.; de Bruyn, R.; Cook, G.; Maree, H.J. Reproducibility and Sensitivity of High-Throughput Sequencing (HTS)-Based Detection of Citrus Tristeza Virus and Three Citrus Viroids. Plants 2022, 11, 1939. https://doi.org/10.3390/plants11151939
Bester R, Steyn C, Breytenbach JHJ, de Bruyn R, Cook G, Maree HJ. Reproducibility and Sensitivity of High-Throughput Sequencing (HTS)-Based Detection of Citrus Tristeza Virus and Three Citrus Viroids. Plants. 2022; 11(15):1939. https://doi.org/10.3390/plants11151939
Chicago/Turabian StyleBester, Rachelle, Chanel Steyn, Johannes H. J. Breytenbach, Rochelle de Bruyn, Glynnis Cook, and Hans J. Maree. 2022. "Reproducibility and Sensitivity of High-Throughput Sequencing (HTS)-Based Detection of Citrus Tristeza Virus and Three Citrus Viroids" Plants 11, no. 15: 1939. https://doi.org/10.3390/plants11151939





