A Proposed New Species Complex within the Cosmopolitan Ring Nematode Criconema annuliferum (de Man, 1921) Micoletzky, 1925
Abstract
:1. Introduction
2. Results
2.1. Species Delimitation Using Morphometry by Principal Component Analysis
2.2. Species Delimitation Based on Ribosomal and Mitochondrial DNA
2.3. Systematics
2.3.1. Criconema paraannuliferum sp. nov.
Description
Diagnosis and Relationships
Molecular Characterization
Type Habitat and Locality
Etymology
Type Material
2.3.2. Criconema plesioannuliferum sp. nov.
Description
Diagnosis and Relationships
Molecular Characterization
Type Habitat and Locality
Etymology
Type Material
2.4. Distribution of the Criconema annuliferum-Complex
2.5. Phylogenetic Analyses of the Criconema annuliferum-Complex
3. Discussion
4. Materials and Methods
4.1. Sampling Sites and Nematode Morphological Identification
4.2. DNA Extraction, PCR and Sequencing
4.3. Recognition of Putative Species within the Criconema annuliferum-Complex and Species Delimitation Approach
4.4. Phylogenetic Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taylor, A.L. The genera and species of Criconematinae, a subfamily of the Anguillulinidae (Nematoda). Trans. Am. Microsc. Soc. 1936, 55, 391–421. [Google Scholar] [CrossRef]
- Geraert, E. The Criconematidae of the World. Identification of the Family Criconematidae (Nematoda); Academia Press: Ghent, Belgium, 2010; 615p. [Google Scholar]
- Dropkin, V.H. The relations between nematodes and plants. Exp. Parasitol. 1955, 4, 282–322. [Google Scholar] [CrossRef]
- Ferris, H.; Zheng, L.; Walker, M.A. Resistance of grape rootstocks to plant-parasitic nematodes. J. Nematol. 2012, 4, 377–386. [Google Scholar]
- Westerdahl, B.B.; Duncan, R.A. Peach Nematodes. UC Pest Management Guidelines (Updated 9/15). UCIPM University of California Agriculture and Natural Resources, Statewide Integrated Pest Management Program. 2015. Available online: http://ipm.ucanr.edu/PMG/r602200111.html (accessed on 9 June 2022).
- Siddiqi, M.R. Tylenchida: Parasites of Plants and Insects, 2nd ed.; CAB International: Wallingford, UK, 2000; p. 848. [Google Scholar]
- Hofmänner, B.; Menzel, R. Neue arten freilebender Nematoden aus der Schweiz. Zool. Anz. 1914, 441, 80–91. [Google Scholar]
- De Grisse, A.T.; Loof, P.A.A. Revision of the genus Criconemoides (Nematoda). Meded. Fac. Landbouw. Opzoek. Gent 1965, 30, 577–603. [Google Scholar]
- Andrássy, I. Evolution as a Basis for the Systematization of Nematodes; Pitman Publishing: London, UK, 1976; 288p. [Google Scholar]
- Andrássy, I. Revision of the subfamily Criconematidae Taylor, 1936 (Nematoda). Opusc. Zool. 1979, 16, 11–57. [Google Scholar]
- Ebsary, B.A. Generic revision of Criconematidae (Nemata): Nothocriconema and related genera with proposal for Nothocriconemella n. gen. and Paracriconema n. gen. Can. J. Zool. 1981, 59, 1227–1236. [Google Scholar] [CrossRef]
- Raski, D.J.; Luc, M. A reappraisal of the genus Criconema Hofmänner & Menzel, 1914 (Nematoda: Criconematidae). Rev. Nématol. 1984, 7, 323–334. [Google Scholar]
- Subbotin, S.A.; Vovlas, N.; Crozzoli, R.; Sturhan, D.; Lamberti, F.; Moens, M.; Baldwin, J.G. Phylogeny of Criconematina Siddiqi, 1980 (Nematoda: Tylenchida) based on morphology and D2-D3 expansion segments of the 28S-rRNA gene sequences with application of a secondary structure model. Nematology 2005, 7, 927–944. [Google Scholar]
- Cordero, M.A.; Robbins, R.T.; Szalanski, A.L. Taxonomic and molecular identification of Bakernema, Criconema, Hemicriconemoides, Ogma and Xenocriconemella species (Nematoda: Criconematidae). J. Nematol. 2012, 44, 427–446. [Google Scholar]
- Powers, T.O.; Harris, T.S.; Higgins, R.S.; Mullin, P.G.; Powers, K.S. An 18S rDNA perspective on the classification of Criconematoidea. J. Nematol. 2017, 49, 236–244. [Google Scholar] [CrossRef] [Green Version]
- Etongwe, C.M.; Singh, P.R.; Bert, W.; Subbotin, S.A. Molecular characterisation of some plant-parasitic nematodes (Nematoda: Tylenchida) from Belgium. Russ. J. Nematol. 2020, 28, 1–28. [Google Scholar]
- Powers, T.O.; Harris, T.S.; Higgins, R.S.; Mullin, P.G.; Powers, K.S. Nematode biodiversity assessments need vouchered databases: A BOLD reference library for plant-parasitic nematodes in the superfamily Criconematoidea. Genome 2021, 64, 232–241. [Google Scholar] [CrossRef]
- Andrássy, I. Free-Living Nematodes of Hungary (Nematoda, Errantia), II; Pedozoologica, Hungarica; Hungarian Natural HistoryMuseum and Systematic Zoology Research Group of the Hungarian Academy of Sciences: Budapest, Hungary, 2007; Volume 4, 496p. [Google Scholar]
- Raski, D.J.; Golden, M.A. Studies on the genus Criconemoides Taylor, 1936 with descriptions of eleven new species and Bakernema variabile n. sp. (Criconematidae: Nematoda). Nematologica 1965, 11, 501–565. [Google Scholar]
- Escuer, M.; Lara, M.P.; Bello, A. Nematodos del suelo de la subfamilia Criconematinae (Nematoda: Criconematidae) en España peninsular. Orsis 1997, 12, 39–63. [Google Scholar]
- Katalan-Gateva, S.; Aleksiev, A.D.; Iliev, I.L.; Ilieva, Z.I. On the species composition and distribution of the family Criconematidae (Taylor, 1936) Thorne, 1949 (Nematoda) in Bulgaria. Acta Zool. Bulg. 1991, 41, 53–57. [Google Scholar]
- Lišková, M.; Vovlas, N.; Sasanelli, N. Criconematidae (Nematoda) in the Slovak Republic. Helminthologia 2004, 41, 161–170. [Google Scholar]
- Zapałowska, A.; Tomasz Skwiercz, A. Populations of parasitic nematodes colonizing Jerusalem artichoke (Helianthus tuberosus L.). Acta Soc. Bot. Pol. 2018, 87, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Bello, A.; Escuer, M.; Lara, M.P. La Familia Criconematidae en las Islas Canarias. Nematol. Mediterr. 1994, 22, 225–232. [Google Scholar]
- Eskandari, A.; Karegar, A.; Pourjam, E.; Alizadeh, A. Three new records of Criconematidae from Iran. Iranian J. Plant Pathol. 2006, 42, 134–157. [Google Scholar]
- Knight, K.W.L. Plant parasitic nematodes associated with six subtropical crops in New Zealand. N. Z. J. Crop Hortic. Sci. 2001, 29, 267–275. [Google Scholar] [CrossRef]
- Van den Berg, E.; Heyns, J. Notes on some plant-parasitic nematodes from South America. Nematologica 1997, 43, 131–144. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Millán, F.; Arias, M.; Bello, A.; López-Pedregal, J.M. Catálogo de los nematodos fitoparásitos y peri-radiculares encontrados en España. Bol. R. Soc. Esp. Hist. Natural 1965, 63, 47–104. [Google Scholar]
- Bello, A. Criconematidae. In Atlas of Plant Parasitic Nematodes of Spain; Alphey, T.J.W., Ed.; European Plan Parasitic Nematode Survey; Scottish Horticultural Research Institute: Dundee, UK, 1979; pp. 9–45. [Google Scholar]
- Palomo, A. Consideraciones biocenóticas sobre la nematofauna edáfica de las Sierras de Gata y Béjar. Bol. R. Soc. Esp. Hist. Nat. 1979, 77, 305–314. [Google Scholar]
- Bello, A.; Lara, M.P. Nematodos ectoparásitos de la Superfamilia Criconematoidea, Taylor, 1936 (Geraert, 1966) encontrados en España Continental. Bol. San. Veg. Plagas 1986, 12, 51–93. [Google Scholar]
- Gomez-Barcina, A.; Castillo, P.; González-País, M.A. Nematodos fitoparásitos de la subfamilia Criconematinae Taylor, 1936 en la Sierra de Cazorla. Rev. Ibér. Parasitol. 1989, 49, 241–255. [Google Scholar]
- Peña-Santiago, R. Plant-parasitic nematodes associated with olive (Olea europea L.) in the province of Jaén, Spain. Rev. Nématol. 1990, 13, 113–115. [Google Scholar]
- Escuer, M.; Palomo, A. Nematodos asociados a melocotonero, peral y manzano en el bajo Cinca (Aragón). Orsis 1991, 6, 75–81. [Google Scholar]
- Escuer, M. Els Nematodes. In El Patrimoni Biològic del Montseny. Catàleg de Flora i Fauna; Servei de Parcs Naturals; Diputació de Barcelona: Barcelona, Spain, 1995; Volume 2, pp. 17–22. [Google Scholar]
- Escuer, M.; Cano, A.; Bello, A. Nematodos fitoparásitos de la Región de Murcia y alternativas de control. In Serie Jornadas y Congresos, 16. Desinfección de Suelos en Invernaderos de Pimientos; Consejería de Agricultura, Agua y Medioambiente: Región de Murcia, Spain, 2004; pp. 27–57. [Google Scholar]
- Archidona-Yuste, A.; Wiegand, T.; Castillo, P.; Navas-Cortés, J.A. Spatial structure and soil properties shape local community structure of plant-parasitic nematodes in cultivated olive trees in southern Spain. Agric. Ecosyst. Environ. 2020, 287, 106688. [Google Scholar] [CrossRef]
- Archidona-Yuste, A.; Wiegand, T.; Eisenhauer, N.; Cantalapiedra-Navarrete, C.; Palomares-Rius, J.E.; Castillo, P. Agriculture causes homogenization of plant-feeding nematode communities at the regional scale. J. Appl. Ecol. 2021, 58, 2881–2891. [Google Scholar] [CrossRef]
- Micoletzky, H. Die freilebende Süsswasser- und Moornematoden Dänemarks. Nebst Anhang: Über Amöbosporidien und andere Parasiten bei freilebenden Nematoden. K. Danske Vidensk. Selsk. Skr. 1925, 10, 57–310. [Google Scholar]
- Peneva, V.; Neilson, R.; Boag, B.; Brown, D.J.F. Criconematidae (Nematoda) from oak forests in two nature reserves in Russia. Syst. Parasitol. 2000, 46, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Hunt, D.J.; Palomares-Rius, J.E. General morphology and morphometries of plant-parasitic nematodes. In Practical Plant Nematology; Biblioteca Basica de Agricultura: Texcoco, Mexico, 2012; pp. 25–64. [Google Scholar]
- Masters, B.C.; Fan, V.; Ross, H.A. Species delimitation a Geneious plugin for the exploration of species boundaries. Mol. Ecol. Resour. 2011, 11, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, C.A.; Hendrixson, B.E.; Brewer, M.S.; Bond, J.E. An evaluation of sampling effects on multiple DNA barcoding methods leads to an integrative approach for delimiting species: A case study of the North American tarantula genus Aphonopelma (Araneae, Mygalomorphae, Theraphosidae). Mol. Phylogen. Evol. 2014, 71, 79–93. [Google Scholar] [CrossRef]
- Ross, H.A.; Murugan, S.; Li, W.L.S. Testing the reliability of genetic methods of species identification via simulation. Syst. Biol. 2008, 57, 216–230. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, N.A. Statistical tests for taxonomic distinctiveness from observations of monophyly. Evolution 2007, 61, 317–323. [Google Scholar] [CrossRef]
- Cobb, N.A. Iota crotaloides n. sp. and the amphids of the triplonchs. J. Parasitol. 1924, 11, 102. [Google Scholar]
- Schuurmans Stekhoven, J.H.; Teunissen, R.J.H. Nématodes libres terrestres. Explor. Parc Nat. Albert Mission Witte 1938, 22, 1–229. [Google Scholar]
- Azimi, S.; Pedram, M. Description of Criconema iranicum n. sp. (Nematoda: Criconematidae) from Iran. J. Crop Prot. 2020, 9, 497–505. [Google Scholar]
- Afshar, F.J.; Pourjam, E.; Mokhtassi-Bidgoli, A.; Pedram, M. Three new records of criconematids (Criconematidae: Criconematinae) from Iran. J. Crop Prot. 2020, 9, 355–366. [Google Scholar]
- Archidona-Yuste, A.; Cantalapiedra-Navarrete, C.; Castillo, P.; Palomares-Rius, J.E. Molecular phylogenetic analysis and comparative morphology reveals the diversity and distribution of needle nematodes of the genus Longidorus (Dorylaimida: Longidoridae) from Spain. Contrib. Zool. 2019, 88, 1–41. [Google Scholar] [CrossRef] [Green Version]
- Cai, R.; Archidona-Yuste, A.; Cantalapiedra-Navarrete, C.; Palomares-Rius, J.E.; Castillo, P. New evidence of cryptic speciation in the family Longidoridae (Nematoda: Dorylaimida). J. Zool. Syst. Evol. Res. 2020, 58, 869–899. [Google Scholar] [CrossRef]
- Dey, A.; Chan, C.K.; Thomas, C.G.; Cutter, A.D. Molecular hyperdiversity defines populations of the nematode Caenorhabditis brenneri. Proc. Natl. Acad. Sci. USA 2013, 110, 11056–11060. [Google Scholar] [CrossRef] [Green Version]
- Olson, M.; Harris, T.; Higgins, R.; Mullin, P.; Powers, K.; Olson, S.; Powers, T.O. Species delimitation and description of Mesocriconema nebraskense n. sp. (Nematoda: Criconematidae), a morphologically cryptic, parthenogenetic species from north American grasslands. J. Nematol. 2017, 49, 42–66. [Google Scholar] [CrossRef] [Green Version]
- Maria, M.; Miao, W.; Cai, R.; Tian, Z.; Castillo, P.; Zheng, J. Species diversity of ring nematodes of the genus Criconemoides (Nematoda: Criconematidae) based on three new species from China, using integrative taxonomy. Eur. J. Plant. Pathol. 2020, 157, 119–139. [Google Scholar] [CrossRef]
- Mueller, L. Conceptual Breakthroughs in Evolutionary Ecology; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Cho, M.R.; Robbins, R.T. Morphological variation among 23 Xiphinema americanum populations. J. Nematol. 1991, 23, 134–144. [Google Scholar]
- Archidona-Yuste, A.; Navas-Cortés, J.A.; Cantalapiedra-Navarrete, C.; Palomares-Rius, J.E.; Castillo, P. Cryptic diversity and species delimitation in the Xiphinema americanum-group complex (Nematoda: Longidoridae) as inferred from morphometrics and molecular markers. Zool. J. Linn. Soc. 2016, 176, 231–265. [Google Scholar] [CrossRef] [Green Version]
- Cai, R.; Archidona-Yuste, A.; Cantalapiedra-Navarrete, C.; Palomares-Rius, J.E.; Castillo, P. Integrative descriptions and molecular phylogeny of two new needle nematodes of the genus Longidorus (Nematoda: Longidoridae) from Spain. Eur. J. Plant Pathol. 2020, 156, 67–86. [Google Scholar] [CrossRef]
- Gittenberger, E. What about non-adaptive radiation? Biol. J. Linn. Soc. 1991, 3, 263–272. [Google Scholar] [CrossRef]
- Powers, T.O.; Mullin, P.G.; Higgins, R.S.; Harris, T.S.; Powers, K.S. Description of Mesocriconema ericaceum n. sp. (Nematoda: Criconematidae) and notes on other nematode species discovered in an ericaceous heath bald community in Great Smoky Mountains National Park, USA. Nematology 2016, 18, 879–903. [Google Scholar] [CrossRef] [Green Version]
- Munawar, M.; Powers, T.O.; Tian, Z.; Harris, T.; Higgins, R.; Zheng, J. Description and distribution of three criconematid nematodes from Hangzhou, Zhejiang Province, China. J. Nematol. 2017, 50, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Clavero-Camacho, I.; Cantalapiedra-Navarrete, C.; Archidona-Yuste, A.; Castillo, P.; Palomares-Rius, J.E. Remarkable cryptic diversity of Paratylenchus spp. (Nematoda: Tylenchulidae) in Spain. Animals 2021, 11, 1161. [Google Scholar] [CrossRef]
- Clavero-Camacho, I.; Palomares-Rius, J.E.; Cantalapiedra-Navarrete, C.; León-Ropero, G.; Martín-Barbarroja, J.; Archidona-Yuste, A.; Castillo, P. Integrative taxonomy reveals hidden cryptic diversity within pin nematodes of the genus Paratylenchus (Nematoda: Tylenchulidae). Plants 2021, 10, 1454. [Google Scholar] [CrossRef]
- Coolen, W.A. Methods for extraction of Meloidogyne spp. and other nematodes from roots and soil. In Root-Knot Nematodes (Meloidogyne Species). Systematics, Biology and Control; Lamberti, F., Taylor, C.E., Eds.; Academic Press: New York, NY, USA, 1979; pp. 317–329. [Google Scholar]
- Seinhorst, J.W. Killing nematodes for taxonomic study with hot F.A. 4:1. Nematologica 1966, 12, 178. [Google Scholar] [CrossRef]
- De Grisse, A.T. Redescription ou modifications de quelques techniques utilisées dans l’étude de nématodes phytoparasitaires. Meded. Rijksfac. Landbouwwet. Gent 1969, 34, 315–359. [Google Scholar]
- Abolafia, J. A low-cost technique to manufacture a container to process meiofauna for scanning electron microscopy. Microsc. Res. Tech. 2015, 78, 771–776. [Google Scholar] [CrossRef]
- Palomares-Rius, J.E.; Clavero-Camacho, I.; Archidona-Yuste, A.; Cantalapiedra-Navarrete, C.; León-Ropero, G.; Braun Miyara, S.; Karssen, G.; Castillo, P. Global distribution of the reniform nematode genus Rotylenchulus with the synonymy of Rotylenchulus macrosoma with Rotylenchulus borealis. Plants 2021, 10, 7. [Google Scholar] [CrossRef]
- De Ley, P.; Felix, M.A.; Frisse, L.; Nadler, S.; Sternberg, P.; Thomas, W.K. Molecular and morphological characterisation of two reproductively isolated species with mirror-image anatomy (Nematoda: Cephalobidae). Nematology 1999, 1, 591–612. [Google Scholar] [CrossRef]
- Subbotin, S.A.; Vierstraete, A.; De Ley, P.; Rowe, J.; Waeyenberge, L.; Moens, M.; Vanfleteren, J.R. Phylogenetic relationships within the cyst-forming nematodes (Nematoda, Heteroderidae) based on analysis of sequences from the ITS regions of ribosomal DNA. Mol. Phylogenet. Evol. 2001, 21, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Holterman, M.; Van der Wurff, A.; Van den Elsen, S.; Van Megen, H.; Bongers, T.; Holovachov, O.; Bakker, J.; Helder, J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Biol. Evol. 2006, 23, 1792–1800. [Google Scholar] [CrossRef]
- Powers, T.O.; Bernard, E.C.; Harris, T.; Higgins, R.; Olson, M.; Lodema, M.; Mullin, P.; Sutton, L.; Powers, K.S. COI haplotype groups in Mesocriconema (Nematoda: Criconematidae) and their morphospecies associations. Zootaxa 2014, 3827, 101–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Legendre, P.; Legendre, L. Numerical Ecology; Elsevier: Amsterdam, The Netherlands, 2012; 990p. [Google Scholar]
- Zuur, A.F.; Ieno, E.N.; Elphick, C.S. A protocol for data exploration to avoid common statistical problems. Methods Ecol. Evol. 2010, 1, 3–14. [Google Scholar] [CrossRef]
- Montgomery, D.C.; Peck, E.A.; Vining, G.G. Introduction to Linear Regression Analysis; Wiley: New York, NJ, USA, 2012. [Google Scholar]
- Revelle, W. Psych: Procedures for Personality and Psychological Research; Northwestern University, Evanston, Illinois. R Package Version 2.2.5. 2022. Available online: https://CRAN.R-project.org/package=psychTools (accessed on 9 June 2022).
- Quadros, A. emstreeR: Tools for Fast Computing and Plotting Euclidean Minimum Spanning Trees. R Package Version 3.0.0. 2022. Available online: https://CRAN.R-project.org/package=emstreeR (accessed on 9 June 2022).
- R_Core_Team. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria. 2021. Available online: https://www.R-project.org/ (accessed on 9 June 2022).
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. FigTree v.1.4.3, A Graphical Viewer of Phylogenetic Trees. 2016. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 28 April 2022).
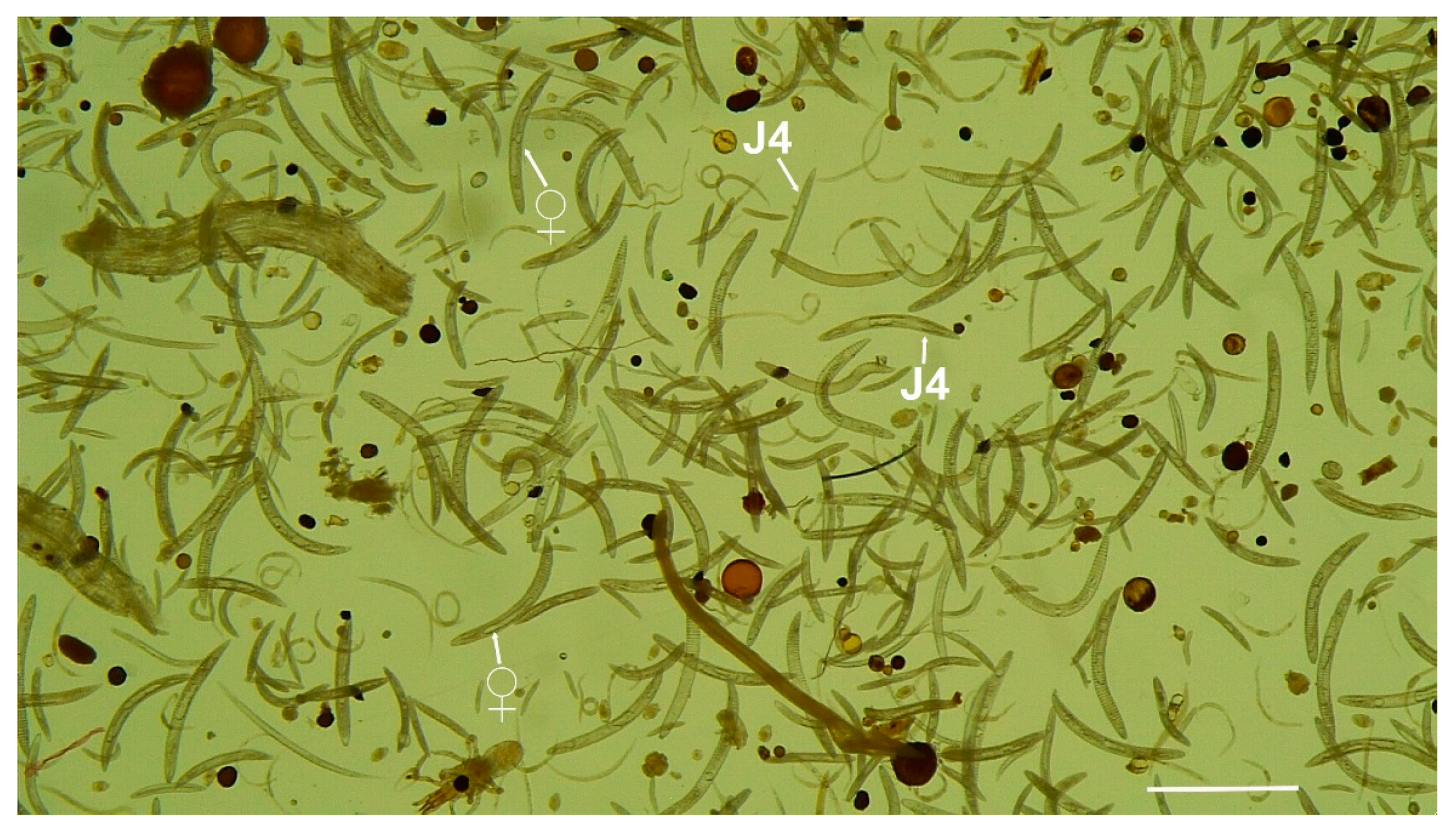








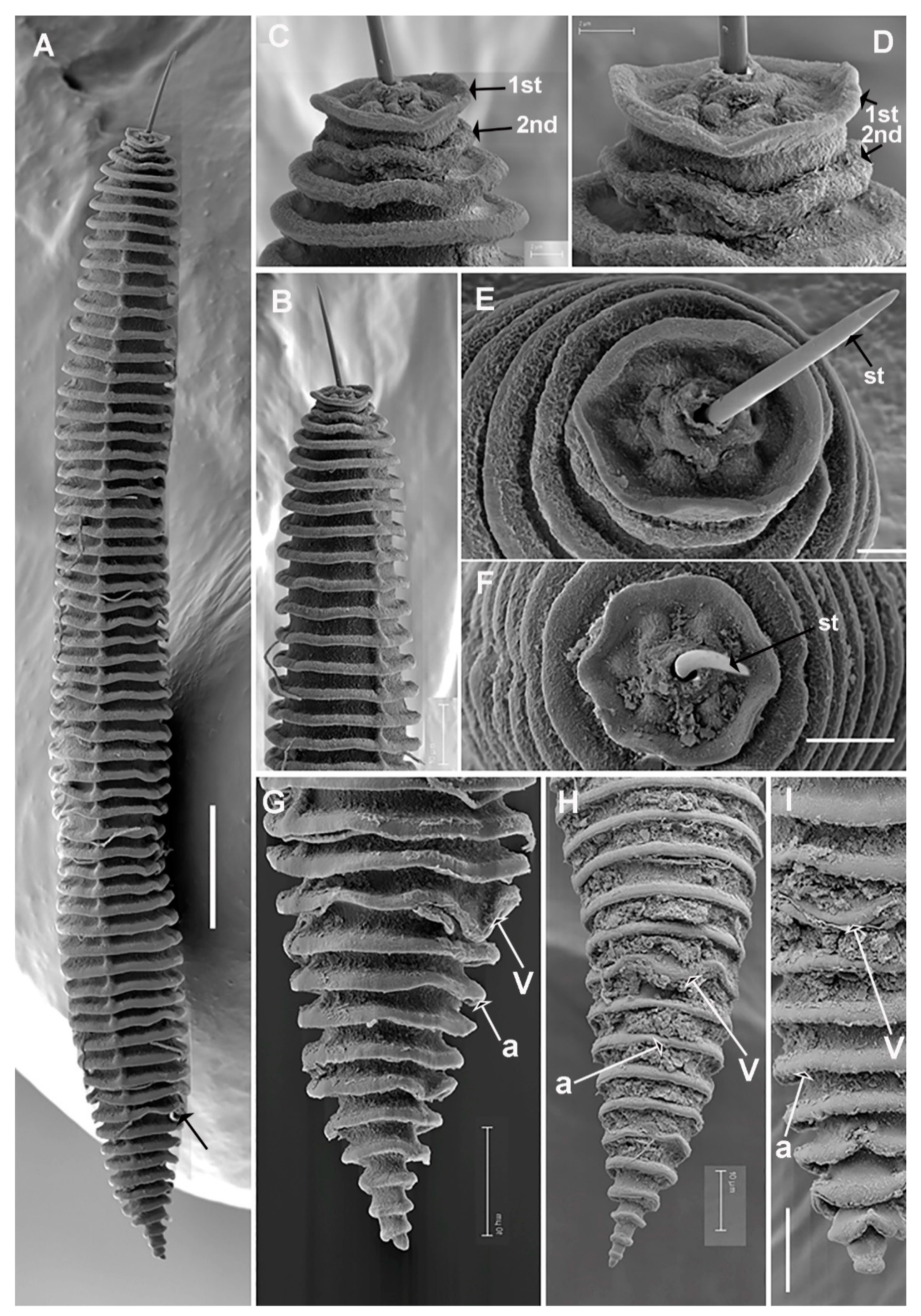






| Species | Sample Code | Locality, Province | Host-Plant | D2–D3 | ITS | 18S | COI |
|---|---|---|---|---|---|---|---|
| C. paraannuliferum sp. nov. | PR-129 | Calasparra, Murcia | peach | ON705053–ON705054 | ON705081–ON705084 | ON705034 | ON648825–ON648828 |
| C. paraannuliferum sp. nov. | PR-125 | Barranda, Murcia | apricot | ON705055 | - | ON705035 | - |
| C. paraannuliferum sp. nov. | PR-141 | Cieza, Murcia | almond | ON705056–ON705057 | - | - | - |
| C. paraannuliferum sp. nov. | PR-203 | Ricla, Zaragoza | peach | ON705058–ON705059 | ON705085–ON705086 | ON705036 | ON648829–ON648830 |
| C. paraannuliferum sp. nov. | PR-217 | Quinto de Ebro, Zaragoza | plum | ON705060–ON705061 | ON705088–ON705089 | ON705037 | ON648832–ON648835 |
| C. paraannuliferum sp. nov. | PR-201 | La Almunia, Zaragoza | almond | ON705062–ON705063 | - | ON705038 | - |
| C. paraannuliferum sp. nov. | PR-208 | Sástago, Zaragoza | apricot | ON705064 | ON705087 | ON705039 | ON648831 |
| C. paraannuliferum sp. nov. | NEV-22 | Castillo de Locubín, Jaén | cultivated olive | ON705065–ON705066 | ON705094–ON705096 | ON705040–ON705044 | ON648840–ON648845 |
| C. paraannuliferum sp. nov. | AR-086 | Prado del Rey, Cádiz | wild olive | ON705067–ON705070 | ON705090–ON705093 | ON705045–ON705047 | ON648836–ON648839 |
| C. paraannuliferum sp. nov. | VAL-22 | Valdepeñas, Jaén | common yew | ON705071–ON705072 | ON705097–ON705102 | - | ON648846–ON648860 |
| C. plesioannuliferum sp. nov. | VAL-22 | Valdepeñas, Jaén | common yew | ON705073–ON705080 | ON705103–ON705116 | ON705048–ON705051 | ON648861–ON648884 |
| Character/Ratio a,b | MLC1 | MLC2 | MLC3 |
|---|---|---|---|
| L | −0.29 | 0.89 | −0.11 |
| Stylet length | 0.00 | 0.66 | −0.11 |
| R | 0.37 | 0.48 | −0.25 |
| RV | 0.86 | −0.19 | 0.02 |
| Ran | 0.90 | −0.23 | 0.26 |
| a | −0.09 | 0.54 | 0.35 |
| b | −0.30 | 0.61 | −0.17 |
| c’ | 0.73 | 0.06 | 0.67 |
| V | −0.12 | 0.15 | −0.53 |
| SS loadings | 2.42 | 2.24 | 1.04 |
| % of total variance | 0.27 | 0.25 | 0.12 |
| Cumulative % of total variance | 0.27 | 0.52 | 0.63 |
| Species | Gene | Intra/Inter a | P ID (Liberal) b | Clade Support c | Rosenberg’s PAB d |
|---|---|---|---|---|---|
| Criconema annuliferum | D2–D3 | 0.12 | 0.97 (0.87,1.0) e | 1.00 | 1.06 × 10−3 |
| ITS | - | - | - | - | |
| COI | 0.02 | 1.00 (0.95,1.0) | 1.00 | 5.3 × 10−16 | |
| Criconema paraannuliferum sp. nov. | D2–D3 | 0.13 | 0.98 (0.95,1.0) | 1.00 | 7.1 × 10−9 |
| ITS | 0.02 | 1.00 (0.95,1.0) | 1.00 | 2.4 × 10−6 | |
| COI | 0.04 | 0.98 (0.93,1.0) | 1.00 | 6.5 × 10−0 | |
| Criconema plesioannuliferum sp. nov. | D2–D3 | 0.09 | 0.98 (0.93,1.0) | 0.99 | 2.2 × 10−6 |
| ITS | 0.02 | 1.00 (0.95,1.0) | 1.00 | 2.4 × 10−6 | |
| COI | 0.01 | 1.00 (0.98,1.0) | 1.00 | 2.6 × 10−5 |
| Locality | Calasparra, Murcia, Peach (PR-129) | Ricla, Zaragoza, Peach (PR-203) | Quinto de Ebro, Zaragoza, Peach (PR-217) | Valdepeñas, Jaén, Common Yew | Castillo de Locubín, Jaén, Cultivated Olive | Prado del Rey, Cádiz, Wild Olive | |||
|---|---|---|---|---|---|---|---|---|---|
| Character/Ratio a | Holotype | Female Paratypes | Fourth-Stage Juveniles | Females | Males | Females | |||
| n | 1 | 20 | 4 | 5 | 5 | 11 | 8 | 5 439.4 ± 31.5 (409–490) | 8 |
| L | 496 | 482.1 ± 34.9 (396–537) | 333.5 ± 48.5 (305–406) | 561.2 ± 61.8 (503–657) | 575.6 ± 33.3 (542–624) | 508.1 ± 45.4 (446–619) | 585.6 ± 18.9 (563–601) | 618.2 ± 21.7 (592–651) | |
| R | 57 | 57.8 ± 0.8 (57–59) | 61.0 ± 0.8 (60–62) | 59.2 ± 1.3 (58–61) | 60.4 ± 1.9 (59–63) | 60.7 ± 2.6 (57–66) | 63.4 ± 1.1 (62–65) | - | 63.4 ± 1.1 (62–65) |
| Rst | 12 | 13.5 ± 1.1 (12–15) | 16.0 ± 1.4 (15–18) | 13.2 ± 0.8 (12–14) | 13.6 ± 0.9 (13–15) | 14.1 ± 0.9 (12–15) | 13.4 ± 1.1 (12–15) | - | 12.8 ± 0.4 (12–13) |
| Roes | 21 | 18.6 ± 1.8 (15–23) | 21.0 ± 2.4 (19–24) | 18.8 ± 0.8 (18–20) | 18.4 ± 1.1 (17–20) | 18.2 ± 1.2 (16–20) | 17.6 ± 1.1 (16–19) | - | 17.8 ± 1.6 (16–20) |
| Rex | 20 | 17.8 ± 1.9 (14–22) | 20.0 ± 2.4 (18–23) | 17.8 ± 0.8 (17–19) | 17.6 ± 1.1 (16–19) | 19.0 ± 1.4 (17–21) | 18.8 ± 1.3 (17–20) | - | 18.8 ± 1.6 (17–21) |
| RV | 7 | 8.5 ± 0.6 (7–10) | - | 8.8 ± 0.4 (8–9) | 8.8 ± 0.8 (8–10) | 9.1 ± 0.5 (8–10) | 8.4 ± 0.5 (8–9) | - | 8.2 ± 0.8 (7–9) |
| Rvan | 3 | 4.3 ± 0.7 (3–5) | - | 4.4 ± 0.5 (4–5) | 4.6 ± 0.5 (4–5) | 4.4 ± 0.5 (4–5) | 4.2 ± 0.8 (3–5) | - | 4.4 ± 0.5 (4–5) |
| Ran | 4 | 4.2 ± 0.6 (3–6) | 4.3 ± 0.5 (4–5) | 4.2 ± 0.4 (4–5) | 4.2 ± 0.4 (4–5) | 4.7 ± 0.5 (4–5) | 4.2 ± 0.4 (4–5) | - | 4.0 ± 0.7 (3–5) |
| O | 10.6 | 9.7 ± 1.2 (8.2–13.3) | 10.8 ± 2.4 (8.0–13.2) | 8.9 ± 0.6 (8.3–9.8) | 8.9 ± 0.4 (8.3–9.5) | 11.2 ± 3.0 (8.6–14.4) | 8.4 ± 0.9 (7.5–9.1) | - | 8.5 ± 1.0 (7.8–10.3) |
| a | 11.8 | 10.1 ± 0.9 (8.6–11.8) | 8.3 ± 0.6 (7.6–9.0) | 10.3 ± 0.6 (9.7–11.1) | 10.8 ± 0.5 (10.4–11.6) | 9.4 ± 0.6 (8.7–10.4) | 11.5 ± 1.1 (10.2–12.9) | 16.3 ± 1.8 (13.9–18.8) | 10.7 ± 1.5 (9.2–13.0) |
| b | 3.4 | 3.5 ± 0.3 (3.0–3.9) | 3.0 ± 0.5 (2.6–3.5) | 3.9 ± 0.3 (3.5–4.4) | 4.1 ± 0.4 (3.8–4.8) | 3.6 ± 0.2 (3.2–3.9) | 3.9 ± 0.1 (3.7–4.1) | 4.5 ± 0.4 (4.0–4.9) | 4.0 ± 0.2 (3.6–4.2) |
| c | 24.8 | 23.7 ± 1.5 (20.3–26.3) | 16.6 ± 1.4 (15.3–18.5) | 27.2 ± 2.5 (24.0–29.9) | 28.3 ± 2.5 (24.6–31.2) | 24.4 ± 2.0 (22.2–28.1) | 24.3 ± 1.2 (23.1–26.1) | 11.1 ± 0.8 (10.3–12.3) | 20.7 ± 3.2 (18.4–26.0) |
| c’ | 1.1 | 1.1 ± 0.1 (1.0–1.2) | 1.1 ± 0.1 (1.1–1.2) | 1.1 ± 0.1 (1.0–1.2) | 1.1 ± 0.1 (1.1–1.2) | 1.1 ± 0.1 (1.0–1.2) | 1.1 ± 0.1 (1.0–1.2) | 2.0 ± 0.1 (1.8–2.1) | 1.3 ± 0.1 (1.2–1.5) |
| V or T | 88.9 | 86.9 ± 1.6 (82.8–89.1) | - | 87.6 ± 2.0 (84.1–89.1) | 87.8 ± 0.6 (87.3–88.7) | 87.4 ± 1.5 (84.6–89.8) | 87.8 ± 0.5 (87.3–88.6) | - | 85.5 ± 1.4 (83.9–86.9) |
| VL/VB | 1.7 | 1.8 ± 0.1 (1.6–2.0) | - | 1.7 ± 0.1 (1.6–1.9) | 1.8 ± 0.1 (1.6–1.9) | 1.6 ± 0.1 (1.4–1.8) | 1.8 ± 0.1 (1.7–1.9) | - | 1.7 ± 0.1 (1.5–1.9) |
| First annulus | 15.0 | 18.0 ± 1.2 (15.0–20.5) | 12.6 ± 1.1 (11.5–14.0) | 18.0 ± 1.0 (17–19) | 19.6 ± 0.9 (19.0–21.0) | 18.7 ± 1.3 (16.5–21.0) | 19.1 ± 1.7 (17.0–21.0) | - | 19.7 ± 1.0 (18.0–20.5) |
| Second annulus | 13.0 | 15.9 ± 1.0 (13.0–18.0) | 12.9 ± 0.9 (12.0–14.0) | 16.0 ± 0.7 (15–17) | 18.1 ± 0.7 (17.0–19.0) | 16.6 ± 1.3 (14.5–19.0) | 16.8 ± 2.0 (15.0–19.0) | - | 16.6 ± 1.1 (15.0–18.0) |
| Stylet | 90.0 | 92.5 ± 3.2 (85.0–99.0) | 72.1 ± 4.9 (65.0–75.5) | 93.6 ± 4.2 (88.0–99.0) | 100.2 ± 6.8 (95.0–112.0) | 100.8 ± 5.7 (91.0–113.0) | 103.7 ± 3.0 (98.5–106.0) | - | 106.4 ± 8.1 (92.0–112.0) |
| conus | 77.0 | 77.8 ± 3.1 (72.0–84.0) | 60.1 ± 5.3 (53.0–65.5) | 80.0 ± 2.7 (77.0–84.00) | 83.4 ± 6.6 (79.0–95.0) | 86.5 ± 3.8 (80.0–94.0) | 91.0 ± 3.2 (86.0–94.0) | - | 92.6 ± 6.1 (82.0–97.0) |
| Pharynx | 148.0 | 138.0 ± 12.2 (115–168) | 111.3 ± 13.6 (91–119) | 145.4 ± 3.6 (140–150) | 141.8 ± 9.8 (130–157) | 142.5 ± 11.8 (132–172) | 151.4 ± 5.0 (146–159) | 97.8 ± 7.3 (85–102) | 155.8 ± 7.4 (148–168) |
| Max. body width | 42.0 | 47.9 ± 3.0 (42.0–54.0) | 40.0 ± 3.6 (37.0–45.0) | 54.4 ± 4.5 (48.0–59.0) | 53.2 ± 2.8 (50.0–57.0) | 54.0 ± 4.8 (45.0–51.0) | 51.2 ± 6.1 (44.0–59.0) | 27.1 ± 2.3 (24.0–30.0) | 58.2 ± 6.3 (50.0–66.0) |
| Anal body diam. | 18.5 | 18.7 ± 0.5 (17.0–19.0) | 17.5 ± 1.3 (16.0–19.0) | 18.8 ± 0.4 (18.0–19.0) | 18.7 ± 0.4 (18.0–19.0) | 19.0 ± 0.7 (18.0–20.0) | 22.1 ± 1.2 (23.0–25.0) | 19.8 ± 1.3 (18.0–21.0) | 22.5 ± 2.3 (19.0–25.0) |
| Vulva to anus distance | 31.0 | 29.8 ± 1.5 (27.0–32.0) | - | 29.6 ± 1.7 (28.0–32.0) | 31.0 ± 1.0 (30.0–32.0) | 30.3 ± 2.1 (28.0–32.0) | 48.0 ± 5.8 (41.0–54.0) | - | 35.8 ± 6.0 (28.0–43.0) |
| Tail | 20.0 | 20.4 ± 0.9 (18.5–22.0) | 20.0 ± 1.6 (18.0–22.0) | 20.6 ± 1.1 (19.5–22.0) | 20.4 ± 0.9 (20.0–22.0) | 20.8 ± 1.3 (19.0–23.0) | 24.1 ± 0.7 (23.0–25.0) | 39.8 ± 2.0 (38.0–43.0) | 30.4 ± 4.0 (24.0–34.0) |
| Spicules | - | - | - | - | - | - | - | 33.1 ± 2.8 (30.5–37.0) | - |
| Gubernaculum | - | - | - | - | - | - | - | 12.1 ± 1.8 (9.0–13.5) | - |
| Character/Ratio a,b | Holotype | Paratypes | ||
|---|---|---|---|---|
| Females | Males | Fourth-Stage Juveniles | ||
| n | 1 | 20 | 3 | 3 |
| L | 489 | 460.1 ± 65.3 (372–658) | 340.7 ± 9.0 (332–350) | 302.0 ± 13.7 (287–314) |
| R | 61 | 61.0 ± 2.6 (57–67) | - | 62.3 ± 1.5 (61–64) |
| Rst | 15 | 14.6 ± 1.4 (12–18) | - | 16.7 ± 0.6 (16–17) |
| Roes | 19 | 20.4 ± 1.0 (19–23) | - | 22.3 ± 0.6 (22–23) |
| Rex | 20 | 20.1 ± 1.5 (17–22) | - | 20.3 ± 0.6 (20–21) |
| RV | 11 | 10.4 ± 0.9 (9–12) | - | - |
| Rvan | 1 | 1.5 ± 0.5 (1–2) | - | - |
| Ran | 10 | 8.9 ± 0.8 (8–10) | - | 7.0 ± 1.0 (6–8) |
| O | 9.2 | 10.4 ± 2.1 (8.9–17.0) | - | 9.8 ± 0.9 (9.3–10.8) |
| a | 10.4 | 10.1 ± 1.0 (8.1–12.4) | 15.9 ± 0.7 (15.1–16.6) | 7.4 ± 0.3 (7.2–7.7) |
| b | 3.6 | 3.3 ± 0.3 (2.7–4.0) | 3.6 ± 0.2 (3.4–3.8) | 2.9 ± 0.1 (2.8–2.9) |
| c | 10.4 | 10.6 ± 1.2 (8.8–13.7) | 10.7 ± 1.0 (9.7–11.7) | 10.8 ± 0.9 (9.8–11.6) |
| c’ | 1.7 | 1.6 ± 0.1 (1.3–1.8) | 2.0 ± 0.2 (1.8–2.3) | 1.2 ± 0.1 (1.1-1.3) |
| V or T | 88.5 | 86.0 ± 1.5 (81.7–88.5) | 40.1 ± 4.1 (35.6–43.4) | - |
| VL/VB | 1.8 | 1.7 ± 0.1 (1.6–2.0) | - | - |
| First annulus | 15.0 | 16.2 ± 1.3 (14.0–19.0) | - | 10.7 ± 0.3 (10.5–11.0) |
| Second annulus | 13.0 | 14.5 ± 1.1 (13.0–16.5) | - | 9.5 ± 0.5 (9.0–10.0) |
| Stylet | 92.0 | 95.7 ± 5.7 (86.0–108.0) | - | 70.2 ± 5.3 (65.0–75.5) |
| conus | 81.0 | 83.6 ± 4.6 (76.0–92.0) | - | 61.5 ± 3.8 (58.0–65.5) |
| Pharynx | 134.0 | 139.8 ± 26.7 (103–215) | 94.0 ± 3.6 (90–97) | 105.3 ± 4.0 (101–109) |
| Max. body width | 47.0 | 45.9 ± 6.8 (36.0–60.5) | 21.5 ± 0.9 (20.5–22.0) | 41.0 ± 1.0 (40.0–42.0) |
| Anal body diam. | 27.0 | 27.0 ± 4.8 (22.0–39.0) | 15.7 ± 1.5 (14.0–17.0) | 23.7 ± 0.6 (23.0–24.0) |
| Vulva to anus distance | 15.0 | 16.3 ± 2.6 (11.0–21.0) | - | - |
| Tail | 47.0 | 43.9 ± 8.8 (33.5–66.0) | 32.0 ± 3.6 (29.0–36.0) | 28.0 ± 2.6 (26.0–31.0) |
| Spicules | - | - | 31.0 ± 1.7 (29.0–32.0) | - |
| Gubernaculum | - | - | 7.0 ± 0.5 (6.5–7.5) | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clavero-Camacho, I.; Palomares-Rius, J.E.; Cantalapiedra-Navarrete, C.; Castillo, P.; Liébanas, G.; Archidona-Yuste, A. A Proposed New Species Complex within the Cosmopolitan Ring Nematode Criconema annuliferum (de Man, 1921) Micoletzky, 1925. Plants 2022, 11, 1977. https://doi.org/10.3390/plants11151977
Clavero-Camacho I, Palomares-Rius JE, Cantalapiedra-Navarrete C, Castillo P, Liébanas G, Archidona-Yuste A. A Proposed New Species Complex within the Cosmopolitan Ring Nematode Criconema annuliferum (de Man, 1921) Micoletzky, 1925. Plants. 2022; 11(15):1977. https://doi.org/10.3390/plants11151977
Chicago/Turabian StyleClavero-Camacho, Ilenia, Juan Emilio Palomares-Rius, Carolina Cantalapiedra-Navarrete, Pablo Castillo, Gracia Liébanas, and Antonio Archidona-Yuste. 2022. "A Proposed New Species Complex within the Cosmopolitan Ring Nematode Criconema annuliferum (de Man, 1921) Micoletzky, 1925" Plants 11, no. 15: 1977. https://doi.org/10.3390/plants11151977
APA StyleClavero-Camacho, I., Palomares-Rius, J. E., Cantalapiedra-Navarrete, C., Castillo, P., Liébanas, G., & Archidona-Yuste, A. (2022). A Proposed New Species Complex within the Cosmopolitan Ring Nematode Criconema annuliferum (de Man, 1921) Micoletzky, 1925. Plants, 11(15), 1977. https://doi.org/10.3390/plants11151977







