Engineering Ribosomes to Alleviate Abiotic Stress in Plants: A Perspective
Abstract
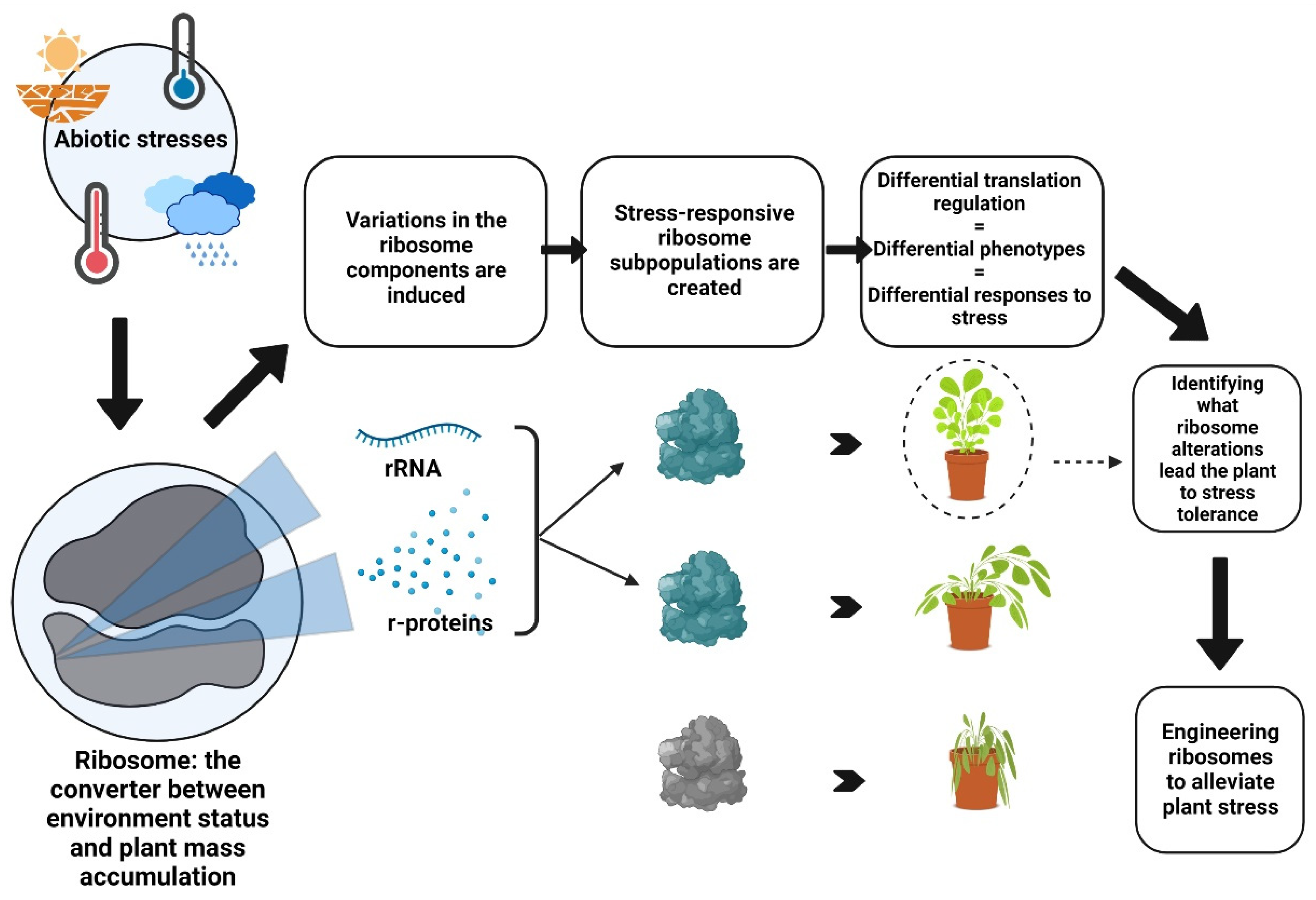
:1. Introduction
2. Ribosome Heterogeneity: A Platform for Custom Stress-Responsive Ribosomes
3. The Plant Ribosome and Its Vocation to Stress Response Regulation
3.1. Ribosomal Proteins Heterogeneity: Variety to Face Adversity
3.2. Plant Ribosome Stress Response: Finer, as It Should Be
3.3. Plant Nucleolar Vacuole: A Cavity Stuffed with Ribosome Heterogeneity
3.4. TOR and SnRKs: The Connectors of Stress Signaling and Ribosome Biogenesis Regulation
4. Ribosome Heterogeneity and Abiotic Stress Regulation in Plants: What Is Already Known?
5. Ribosome Heterogeneity and Abiotic Stress Regulation in Plants: What Is Next?
- Manipulating the ribosomes is modifying a system:The role of a heterogeneous ribosome subgroup in the plant response to abiotic stress can only be fairly characterized under the prism of a whole regulatory system.The ribosome heterogeneity applies a translational selection on mRNA pools which are themselves the product of transcriptional regulation. Therefore, to identify which ribosome feature is worth being engineered, it is necessary not only to understand its impact on a certain translation outcome but also how it relates to the other regulation layers.
- The complexity of abiotic stressors must be addressed:Although they meet the goals of basic science, most of the studies limit the spectrum of plant response to abiotic stresses by analyzing each type of them individually. A single kind of stress at a time is the opposite scenario of what happens in nature. The environmental conditions are essentially unstable. The plants usually have to tackle different abiotic stressors simultaneously. Thus, their reaction in this case is surely not the same as when only one stress is applied, as demonstrated by Rasmussen et al. (2013) [189]. So, for plant breeding purposes, the investigations of ribosome heterogeneity induced by a combination of environmental stress must be intensified. This way, the ribosome engineering can be centered on data that reflect more consistently the conditions found in the field.
- Synthetic biology techniques should be optimized for plant research:While techniques such as CRISPR/Cas have been broadly used to precisely edit the genome of crop species and develop more sustainable cultivars [190], other ones remain to be mastered by plant research. Methods of in vitro ribosome synthesis such as the “Integrated synthesis, assembly, and translation” (iSAT) [191,192] and “In vitro ribosome synthesis and evolution” (RISE) [193] enable the production and investigation of ribosome variants. Once optimized and fully explored by plant biologists, significant advances in the knowledge of plant ribosomes can be achieved through them.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yadav, S.; Modi, P.; Dave, A.; Vijapura, A.; Patel, D.; Patel, M. Effect of Abiotic Stress on Crops. In Sustainable Crop Production; Hasanuzzaman, M., Fujita, M., Teixeira Filho, M.C.M., Nogueira, T.A.R., Galindo, F.S., Eds.; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Boyer, J.S. Plant Productivity and Environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Uemura, M.; Bailey-Serres, J.; Bray, E.A.; Weretilnyk, E. Responses to abiotic stress. In Biochemistry and Molecular Biology of Plants; Buchanan, B.B., Gruissem, W., Jones, R.L., Eds.; Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 1051–1100. [Google Scholar]
- Vij, S.; Tyagi, A.K. Emerging trends in the functional genomics of the abiotic stress response in crop plants. Plant Biotechnol. J. 2007, 5, 361–380. [Google Scholar] [CrossRef] [PubMed]
- Zurbriggen, M.D.; Hajirezaei, M.-R.; Carrillo, N. Engineering the future. Development of transgenic plants with enhanced tolerance to adverse environments. Biotechnol. Genet. Eng. Rev. 2010, 27, 33–56. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, J.C. Plant synthetic biology could drive a revolution in biofuels and medicine. Exp. Biol. Med. 2019, 244, 323–331. [Google Scholar] [CrossRef] [PubMed]
- D’Aquino, A.E.; Kim, D.S.; Jewett, M.C. Engineered Ribosomes for Basic Science and Synthetic Biology. Annu. Rev. Chem. Biomol. Eng. 2018, 9, 311–340. [Google Scholar] [CrossRef]
- Prabhakar, A.; Choi, J.; Wang, J.; Petrov, A.; Puglisi, J.D. Dynamic basis of fidelity and speed in translation: Coordinated multistep mechanisms of elongation and termination: Dynamic Basis of Fidelity and Speed in Translation. Protein Sci. 2017, 26, 1352–1362. [Google Scholar] [CrossRef]
- Ohta, A.; Murakami, H.; Higashimura, E.; Suga, H. Synthesis of Polyester by Means of Genetic Code Reprogramming. Chem. Biol. 2007, 14, 1315–1322. [Google Scholar] [CrossRef]
- Liu, C.C.; Schultz, P.G. Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef]
- Neumann, H.; Wang, K.; Davis, L.; Garcia-Alai, M.; Chin, J.W. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 2010, 464, 441–444. [Google Scholar] [CrossRef]
- Zhang, Y.; Ptacin, J.L.; Fischer, E.C.; Aerni, H.R.; Caffaro, C.E.; Jose, K.S.; Feldman, A.W.; Turner, C.R.; Romesberg, F.E. A semi-synthetic organism that stores and retrieves increased genetic information. Nature 2017, 551, 644–647. [Google Scholar] [CrossRef]
- O’Donoghue, P.; Ling, J.; Wang, Y.-S.; Söll, D. Upgrading protein synthesis for synthetic biology. Nat. Chem. Biol. 2013, 9, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-Y.; Park, S.-W.; Chung, Y.-S.; Chung, C.-H.; Kim, J.-I.; Lee, J.-H. Molecular cloning of low-temperature-inducible ribosomal proteins from soybean. J. Exp. Bot. 2004, 55, 1153–1155. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lan, P.; Gao, H.; Zheng, L.; Li, W.; Schmidt, W. Expression changes of ribosomal proteins in phosphate- and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genom. 2013, 14, 783. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Das, S.; Moin, M.; Dutta, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Genome-Wide Identification and Comprehensive Expression Profiling of Ribosomal Protein Small Subunit (RPS) Genes and their Comparative Analysis with the Large Subunit (RPL) Genes in Rice. Front. Plant Sci. 2017, 8, 1553. [Google Scholar] [CrossRef]
- Cherepneva, G.N.; Schmidt, K.-H.; Kulaeva, O.N.; Oelmüller, R.; Kusnetsov, V. Expression of the ribosomal proteins S14, S16, L13a and L30 is regulated by cytokinin and abscisic acid: Implication of the involvement of phytohormones in translational processes. Plant Sci. 2003, 165, 925–932. [Google Scholar] [CrossRef]
- Kawasaki, S.; Borchert, C.; Deyholos, M.; Wang, H.; Brazille, S.; Kawai, K.; Galbraith, D.; Bohnert, H.J. Gene Expression Profiles during the Initial Phase of Salt Stress in Rice. Plant Cell 2001, 13, 889–905. [Google Scholar] [CrossRef]
- Wilson, D.; Cate, J.H.D. The Structure and Function of the Eukaryotic Ribosome. Cold Spring Harb. Perspect. Biol. 2012, 4, a011536. [Google Scholar] [CrossRef]
- Genuth, N.R.; Barna, M. Heterogeneity and specialized functions of translation machinery: From genes to organisms. Nat. Rev. Genet. 2018, 19, 431–452. [Google Scholar] [CrossRef]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef]
- Calamita, P.; Gatti, G.; Miluzio, A.; Scagliola, A.; Biffo, S. Translating the Game: Ribosomes as Active Players. Front. Genet. 2018, 9, 533. [Google Scholar] [CrossRef]
- Emmott, E.; Jovanovic, M.; Slavov, N. Ribosome Stoichiometry: From Form to Function. Trends Biochem. Sci. 2019, 44, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Fujii, K.; Kovary, K.M.; Genuth, N.R.; Röst, H.L.; Teruel, M.N.; Barna, M. Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol. Cell 2017, 67, 71–83.e7. [Google Scholar] [CrossRef] [PubMed]
- Gunderson, J.H.; Sogin, M.L.; Wollett, G.; Hollingdale, M.; de la Cruz, V.F.; Waters, A.P.; McCutchan, T.F. Structurally Distinct, Stage-Specific Ribosomes Occur in Plasmodium. Science 1987, 238, 933–937. [Google Scholar] [CrossRef]
- Ramagopal, S. Induction of cell-specific ribosomal proteins in aggregation-competent nonmorphogenetic Dictyostelium discoideum. Biochem. Cell Biol. 1990, 68, 1281–1287. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.; Sussex, I.M. Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J. 1995, 8, 65–76. [Google Scholar] [CrossRef]
- Milne, A.N.; Mak, W.W.; Wong, J.T.-F. Variation of ribosomal proteins with bacterial growth rate. J. Bacteriol. 1975, 122, 89–92. [Google Scholar] [CrossRef]
- Bortoluzzi, S.; D’Alessi, F.; Romualdi, C.; Danieli, G.A. Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics 2001, 17, 1152–1157. [Google Scholar] [CrossRef]
- Pb, K.; Moin, M.; Bakshi, A.; Saha, A. Ribosomal Proteins and their Extra Ribosomal Functions in Abiotic Stress Tolerance of Plants. Mod. Concepts Dev. Agron. 2019, 4, 1024–1038. [Google Scholar] [CrossRef]
- Salih, K.J.; Duncan, O.; Li, L.; O’Leary, B.; Fenske, R.; Trösch, J.; Millar, A.H. Impact of oxidative stress on the function, abundance, and turnover of the Arabidopsis 80S cytosolic ribosome. Plant J. 2020, 103, 128–139. [Google Scholar] [CrossRef]
- Terhorst, A.; Sandikci, A.; Neurohr, G.E.; Whittaker, C.A.; Szórádi, T.; Holt, L.J.; Amon, A. The Environmental Stress Response Regulates Ribosome Content in Cell Cycle-Arrested S. Cerevisiae. bioRxiv 2021. [Google Scholar] [CrossRef]
- El-Sharoud, W.M. Ribosome Inactivation for Preservation: Concepts and Reservations. Sci. Prog. 2004, 87, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Wigge, P.A.; Guillaume-Schoepfer, D.; Jaeger, K.E.; Geng, F.; Doccula, F.G.; Costa, A.; Webb, A.A. Ribosomes Act as Cryosensors in Plants. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hang, R.; Wang, Z.; Deng, X.; Liu, C.; Yan, B.; Yang, C.; Song, X.; Mo, B.; Cao, X. Ribosomal RNA Biogenesis and Its Response to Chilling Stress in Oryza sativa. Plant Physiol. 2018, 177, 381–397. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yan, P.; Du, Q.; Wang, Y.; Guo, Y.; Fu, Z.; Wang, H.; Tang, J. Pre-rRNA processing and its response to temperature stress in maize. J. Exp. Bot. 2019, 71, 1363–1374. [Google Scholar] [CrossRef]
- Camacho, R.A.U.; Lokdarshi, A.; von Arnim, A.G. Translational gene regulation in plants: A green new deal. Wiley Interdiscip. Rev. RNA 2020, 11, e1597. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Huey, R.; Carlson, M.; Crozier, L.G.; Frazier, M.R.; Hamilton, H.; Harley, C.; Hoang, A.; Kingsolver, J.G. Plants Versus Animals: Do They Deal with Stress in Different Ways? Integr. Comp. Biol. 2002, 42, 415–423. [Google Scholar] [CrossRef]
- Graber, T.E.; Holcik, M. Cap-independent regulation of gene expression in apoptosis. Mol. BioSyst. 2007, 3, 825–834. [Google Scholar] [CrossRef]
- Muñoz, A.; Castellano, M.M. Regulation of Translation Initiation under Abiotic Stress Conditions in Plants: Is It a Conserved or Not so Conserved Process among Eukaryotes? Comp. Funct. Genom. 2012, 2012, 406357. [Google Scholar] [CrossRef]
- Floris, M.; Mahgoub, H.; Lanet, E.; Robaglia, C.; Menand, B. Post-transcriptional Regulation of Gene Expression in Plants during Abiotic Stress. Int. J. Mol. Sci. 2009, 10, 3168–3185. [Google Scholar] [CrossRef]
- Juntawong, P.; Bailey-Serres, J. Dynamic Light Regulation of Translation Status in Arabidopsis thaliana. Front. Plant Sci. 2012, 3, 66. [Google Scholar] [CrossRef] [PubMed]
- Piques, M.; Schulze, W.X.; Höhne, M.; Usadel, B.; Gibon, Y.; Rohwer, J.; Stitt, M. Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol. Syst. Biol. 2009, 5, 314. [Google Scholar] [CrossRef] [PubMed]
- I Zandalinas, S.; Balfagón, D.; Gómez-Cadenas, A.; Mittler, R. Plant responses to climate change: Metabolic changes under combined abiotic stresses. J. Exp. Bot. 2022, 73, 3339–3354. [Google Scholar] [CrossRef] [PubMed]
- Sormani, R.; Masclaux-Daubresse, C.; Daniele-Vedele, F.; Chardon, F. Transcriptional Regulation of Ribosome Components Are Determined by Stress According to Cellular Compartments in Arabidopsis thaliana. PLoS ONE 2011, 6, e28070. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Y.; Zhu, J.-K. Thriving under Stress: How Plants Balance Growth and the Stress Response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Seidel, F.; Golovchuk, O.B.; Hsieh, Y.-C.; Kopka, J. Systematic Review of Plant Ribosome Heterogeneity and Specialization. Front. Plant Sci. 2020, 11, 948. [Google Scholar] [CrossRef]
- Horiguchi, G.; Van Lijsebettens, M.; Candela, H.; Micol, J.L.; Tsukaya, H. Ribosomes and translation in plant developmental control. Plant Sci. 2012, 191–192, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Gerst, J.E. Pimp My Ribosome: Ribosomal Protein Paralogs Specify Translational Control. Trends Genet. 2018, 34, 832–845. [Google Scholar] [CrossRef]
- Ferretti, M.; Karbstein, K. Does functional specialization of ribosomes really exist? RNA 2019, 25, 521–538. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, R. mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 2005, 33, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986, 44, 283–292. [Google Scholar] [CrossRef]
- Hernández, G.; Osnaya, V.G.; Pérez-Martínez, X. Conservation and Variability of the AUG Initiation Codon Context in Eukaryotes. Trends Biochem. Sci. 2019, 44, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, J.M.; Hoermann, B.; Schlimbach, T.; Teleman, A.A. Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence. Sci. Rep. 2018, 8, 4018. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Rangan, L.; Ramesh, T.V.; Gupta, M. Comparative analysis of contextual bias around the translation initiation sites in plant genomes. J. Theor. Biol. 2016, 404, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Lu, Y.; Zinta, G.; Lang, Z.; Zhu, J.-K. UTR-Dependent Control of Gene Expression in Plants. Trends Plant Sci. 2018, 23, 248–259. [Google Scholar] [CrossRef]
- Lei, L.; Shi, J.; Chen, J.; Zhang, M.; Sun, S.; Xie, S.; Li, X.; Zeng, B.; Peng, L.; Hauck, A.; et al. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 2015, 84, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-J.; Wu, S.-H.; Wu, J.-F.; Lin, W.-D.; Wu, Y.-C.; Tsai, T.-Y.; Tsai, H.-L.; Wu, S.-H. Translational Landscape of Photomorphogenic Arabidopsis. Plant Cell 2013, 25, 3699–3710. [Google Scholar] [CrossRef] [PubMed]
- Juntawong, P.; Girke, T.; Bazin, J.; Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, E203–E212. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, A.; Yue, Y.; Zhao, Y. uORFs: Important Cis-Regulatory Elements in Plants. Int. J. Mol. Sci. 2020, 21, 6238. [Google Scholar] [CrossRef]
- von Arnim, A.G.; Jia, Q.; Vaughn, J.N. Regulation of plant translation by upstream open reading frames. Plant Sci. 2014, 214, 1–12. [Google Scholar] [CrossRef]
- Um, T.; Park, T.; Shim, J.; Kim, Y.; Lee, G.-S.; Choi, I.-Y.; Kim, J.-K.; Seo, J.; Park, S. Application of Upstream Open Reading Frames (uORFs) Editing for the Development of Stress-Tolerant Crops. Int. J. Mol. Sci. 2021, 22, 3743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Si, X.; Ji, X.; Fan, R.; Liu, J.; Chen, K.; Wang, D.; Gao, C. Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 2018, 36, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, Y. uORF Shuffling Fine-Tunes Gene Expression at a Deep Level of the Process. Plants 2020, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- Young, S.K.; Wek, R.C. Upstream Open Reading Frames Differentially Regulate Gene-specific Translation in the Integrated Stress Response. J. Biol. Chem. 2016, 291, 16927–16935. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.W.; Green, R. Ribosomopathies: There’s strength in numbers. Science 2017, 358, eaan2755. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.E. A role for the ribosome in development. Trends Plant Sci. 2009, 14, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Weis, B.L.; Kovacevic, J.; Missbach, S.; Schleiff, E. Plant-Specific Features of Ribosome Biogenesis. Trends Plant Sci. 2015, 20, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Barakat, A.; Szick-Miranda, K.; Chang, I.-F.; Guyot, R.; Blanc, G.; Cooke, R.; Delseny, M.; Bailey-Serres, J. The Organization of Cytoplasmic Ribosomal Protein Genes in the Arabidopsis Genome. Plant Physiol. 2001, 127, 398–415. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.J.; Heazlewood, J.L.; Ito, J.; Millar, A.H. Analysis of the Arabidopsis Cytosolic Ribosome Proteome Provides Detailed Insights into Its Components and Their Post-translational Modification. Mol. Cell. Proteom. 2008, 7, 347–369. [Google Scholar] [CrossRef]
- Wool, I.G.; Chan, Y.-L.; Glück, A. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 1995, 73, 933–947. [Google Scholar] [CrossRef]
- Harrison, P.M.; Hegyi, H.; Balasubramanian, S.; Luscombe, N.M.; Bertone, P.; Echols, N.; Johnson, T.; Gerstein, M. Molecular Fossils in the Human Genome: Identification and Analysis of the Pseudogenes in Chromosomes 21 and 22. Genome Res. 2002, 12, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Harrison, P.; Gerstein, M. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res. 2002, 12, 1466–1482. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Matsui, E.; Yamamoto, K.; Nagamura, Y.; Kurata, N.; Takuji, S.; Minobe, Y. Genomic organization of 57 ribosomal protein genes in rice (Oryza sativa L.) through RFLP mapping. Genome 1995, 38, 1189–1200. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Cheng, F. Plant Polyploidy: Origin, Evolution, and Its Influence on Crop Domestication. Hortic. Plant J. 2019, 5, 231–239. [Google Scholar] [CrossRef]
- Ohno, S. Evolution by Gene Duplication; Springer: Berlin/Heidelberg, Germany, 1970. [Google Scholar]
- Cheng, F.; Wu, J.; Cai, X.; Liang, J.; Freeling, M.; Wang, X. Gene retention, fractionation and sub genome differences in polyploid plants. Nat. Plants 2018, 4, 258–268. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K. Functional Divergence of Duplicated Genes Formed by Polyploidy during Arabidopsis Evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef]
- Thomas, B.C.; Pedersen, B.; Freeling, M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006, 16, 934–946. [Google Scholar] [CrossRef]
- Brodersen, D.E.; Nissen, P. The social life of ribosomal proteins: The Social Life of Ribosomal Proteins. FEBS J. 2005, 272, 2098–2108. [Google Scholar] [CrossRef]
- McIntosh, K.B.; Bonham-Smith, P.C. Ribosomal protein gene regulation: What about plants? Can. J. Bot. 2006, 84, 342–362. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome. Front. Plant Sci. 2018, 9, 122. [Google Scholar] [CrossRef]
- Salih, K.J.; Duncan, O.; Li, L.; Trösch, J.; Millar, A.H. The composition and turnover of the Arabidopsis thaliana 80S cytosolic ribosome. Biochem. J. 2020, 477, 3019–3032. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Yi, H.; Chen, C.; Yan, S.; Yao, H.; He, G.; Li, G.; Jiang, Y.; Deng, T.; Deng, X. Nucleolar stress: Is there a reverse version? J. Cancer 2018, 9, 3723–3727. [Google Scholar] [CrossRef] [PubMed]
- Moss, T.; Stefanovsky, V.Y. At the Center of Eukaryotic Life. Cell 2002, 109, 545–548. [Google Scholar] [CrossRef]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.-M.; Lamond, A.I. The Nucleolus under Stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef]
- Sáez-Vásquez, J.; Delseny, M. Ribosome Biogenesis in Plants: From Functional 45S Ribosomal DNA Organization to Ribosome Assembly Factors. Plant Cell 2019, 31, 1945–1967. [Google Scholar] [CrossRef]
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2018, 2, 125–140. [Google Scholar] [CrossRef]
- Kalinina, N.O.; Makarova, S.; Makhotenko, A.; Love, A.J.; Taliansky, M. The Multiple Functions of the Nucleolus in Plant Development, Disease and Stress Responses. Front. Plant Sci. 2018, 9, 132. [Google Scholar] [CrossRef]
- Lane, D.P. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Golomb, L.; Volarevic, S.; Oren, M. p53 and ribosome biogenesis stress: The essentials. FEBS Lett. 2014, 588, 2571–2579. [Google Scholar] [CrossRef]
- Garapati, P.; Xue, G.-P.; Munné-Bosch, S.; Balazadeh, S. Transcription Factor ATAF1 in Arabidopsis Promotes Senescence by Direct Regulation of Key Chloroplast Maintenance and Senescence Transcriptional Cascades. Plant Physiol. 2015, 168, 1122–1139. [Google Scholar] [CrossRef]
- Lee, S.; Seo, P.J.; Lee, H.-J.; Park, C.-M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012, 70, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Wu, A.; Mueller-Roeber, B. Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal. Behav. 2010, 5, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Yoshiyama, K.O. SOG1: A master regulator of the DNA damage response in plants. Genes Genet. Syst. 2015, 90, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Yoshiyama, K.O.; Kobayashi, J.; Ogita, N.; Ueda, M.; Kimura, S.; Maki, H.; Umeda, M. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 2013, 14, 817–822. [Google Scholar] [CrossRef]
- Ohbayashi, I.; Sugiyama, M. Plant Nucleolar Stress Response, a New Face in the NAC-Dependent Cellular Stress Responses. Front. Plant Sci. 2018, 8, 2247. [Google Scholar] [CrossRef]
- Shaw, P.; Brown, J. Nucleoli: Composition, Function, and Dynamics. Plant Physiol. 2012, 158, 44–51. [Google Scholar] [CrossRef]
- Beven, A.F.; Lee, R.; Razaz, M.; Leader, D.J.; Brown, J.W.; Shaw, P.J. The organization of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs. J. Cell Sci. 1996, 109, 1241–1251. [Google Scholar] [CrossRef]
- Beven, A.F.; Simpson, G.G.; Brown, J.W.S.; Shaw, P.J. The organization of spliceosomal components in the nuclei of higher plants. J. Cell Sci. 1995, 108, 509–551. [Google Scholar] [CrossRef] [PubMed]
- Lorković, Z.J.; Barta, A. Role of Cajal Bodies and Nucleolus in the Maturation of the U1 snRNP in Arabidopsis. PLoS ONE 2008, 3, e3989. [Google Scholar] [CrossRef]
- Stępiński, D. Immunofluorescent localization of ubiquitin and proteasomes in nucleolar vacuoles of soybean root meristematic cells. Eur. J. Histochem. 2012, 56, e13. [Google Scholar] [CrossRef] [PubMed]
- Stępiński, D. Functional ultrastructure of the plant nucleolus. Protoplasma 2014, 251, 1285–1306. [Google Scholar] [CrossRef]
- Terns, M.P.; Terns, R.M. Small nucleolar RNAs: Versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002, 10, 17–39. [Google Scholar] [PubMed]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.-F.; Chakraborty, A.; Gleizes, P.-E. An overview of pre-ribosomal RNA processing in eukaryotes: Pre-Ribosomal RNA Processing in Eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.-D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; León-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide Mapping Reveals Widespread Dynamic-Regulated Pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Xiao, M.; Yang, C.; Yu, Y.-T. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 2011, 30, 79–89. [Google Scholar] [CrossRef]
- Yu, Y.-T.; Meier, U.T. RNA-guided isomerization of uridine to pseudouridine—pseudouridylation. RNA Biol. 2014, 11, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR Signaling in Growth and Metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wu, Y.; Sheen, J. TOR signaling in plants: Conservation and innovation. Development 2018, 145, dev160887. [Google Scholar] [CrossRef] [PubMed]
- Caldana, C.; Li, Y.; Leisse, A.; Zhang, Y.; Bartholomaeus, L.; Fernie, A.R.; Willmitzer, L.; Giavalisco, P. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 2013, 73, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Kunz, J.; Henriquez, R.; Schneider, U.; Deuter-Reinhard, M.; Movva, N.R.; Hall, M.N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 1993, 73, 585–596. [Google Scholar] [CrossRef]
- Sabatini, D.M.; Erdjument-Bromage, H.; Lui, M.; Tempst, P.; Snyder, S.H. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994, 78, 35–43. [Google Scholar] [CrossRef]
- Chiu, M.I.; Katz, H.; Berlin, V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. USA 1994, 91, 12574–12578. [Google Scholar] [CrossRef]
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for Cell Cycle Arrest by the Immunosuppressant Rapamycin in Yeast. Science 1991, 253, 905–909. [Google Scholar] [CrossRef]
- Dong, P.; Xiong, F.; Que, Y.; Wang, K.; Yu, L.; Li, Z.; Ren, M. Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front. Plant Sci. 2015, 6, 677. [Google Scholar] [CrossRef] [PubMed]
- Dobrenel, T.; Marchive, C.; Sormani, R.; Moreau, M.; Mozzo, M.; Montané, M.; Menand, B.; Robaglia, C.; Meyer, C. Regulation of plant growth and metabolism by the TOR kinase. Biochem. Soc. Trans. 2011, 39, 477–481. [Google Scholar] [CrossRef]
- Fu, L.; Wang, P.; Xiong, Y. Target of Rapamycin Signaling in Plant Stress Responses. Plant Physiol. 2020, 182, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Qiu, S.; Venglat, P.; Xiang, D.; Feng, L.; Selvaraj, G.; Datla, R. Target of Rapamycin Regulates Development and Ribosomal RNA Expression through Kinase Domain in Arabidopsis. Plant Physiol. 2011, 155, 1367–1382. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-K.; Kim, S.; Shin, Y.-J.; Hur, Y.-S.; Kim, W.-Y.; Lee, M.-S.; Cheon, C.-I.; Verma, D.P.S. Ribosomal Protein S6, a Target of Rapamycin, Is Involved in the Regulation of rRNA Genes by Possible Epigenetic Changes in Arabidopsis. J. Biol. Chem. 2014, 289, 3901–3912. [Google Scholar] [CrossRef]
- Menand, B.; Desnos, T.; Nussaume, L.; Berger, F.; Bouchez, D.; Meyer, C.; Robaglia, C. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 2002, 99, 6422–6427. [Google Scholar] [CrossRef]
- Rodriguez, M.S.; Parola, R.; Andreola, S.; Pereyra, C.; Martínez-Noël, G. TOR and SnRK1 signaling pathways in plant response to abiotic stresses: Do they always act according to the “yin-yang” model? Plant Sci. 2019, 288, 110220. [Google Scholar] [CrossRef]
- Ren, M.; Venglat, P.; Qiu, S.; Feng, L.; Cao, Y.; Wang, E.; Xiang, D.; Wang, J.; Alexander, D.; Chalivendra, S.; et al. Target of Rapamycin Signaling Regulates Metabolism, Growth, and Life Span in Arabidopsis. Plant Cell 2013, 24, 4850–4874. [Google Scholar] [CrossRef]
- Urban, J.; Soulard, A.; Huber, A.; Lippman, S.; Mukhopadhyay, D.; Deloche, O.; Wanke, V.; Anrather, D.; Ammerer, G.; Riezman, H.; et al. Sch9 Is a Major Target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 2007, 26, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Hay, N.; Sonenberg, N. Upstream and Downstream of mTOR. Genes Dev. 2004, 18, 1926–1945. [Google Scholar] [CrossRef]
- Xiong, Y.; McCormack, M.P.; Li, L.; Hall, Q.; Xiang, C.; Sheen, J. Glucose–TOR signalling reprograms the transcriptome and activates meristems. Nature 2013, 496, 181–186. [Google Scholar] [CrossRef]
- Dobrenel, T.; Mancera-Martínez, E.; Forzani, C.; Azzopardi, M.; Davanture, M.; Moreau, M.; Schepetilnikov, M.; Chicher, J.; Langella, O.; Zivy, M.; et al. The Arabidopsis TOR Kinase Specifically Regulates the Expression of Nuclear Genes Coding for Plastidic Ribosomal Proteins and the Phosphorylation of the Cytosolic Ribosomal Protein S6. Front. Plant Sci. 2016, 7, 1611. [Google Scholar] [CrossRef]
- Bakshi, A.; Moin, M.; Madhav, M.S.; Gayatri, M.B.; Reddy, A.B.M.; Datla, R.; Kirti, P.B. Target of Rapamycin: Function in Abiotic Stress Tolerance in Arabidopsis and Its Involvement in a Possible Cross-Talk with Ribosomal Proteins. bioRxiv 2020. [Google Scholar] [CrossRef]
- Busche, M.; Scarpin, M.R.; Hnasko, R.; Brunkard, J.O. TOR coordinates nucleotide availability with ribosome biogenesis in plants. Plant Cell 2021, 33, 1615–1632. [Google Scholar] [CrossRef]
- Halford, N.G.; Hardie, G. SNF1-related protein kinases: Global regulators of carbon metabolism in plants? Plant Mol. Biol. 1998, 37, 735–748. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.G.; Kudla, J.; Luan, S.; Nimmo, H.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK Superfamily of Protein Kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef]
- Halford, N.G.; Hey, S.J. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem. J. 2009, 419, 247–259. [Google Scholar] [CrossRef]
- Baena-González, E.; Hanson, J. Shaping plant development through the SnRK1–TOR metabolic regulators. Curr. Opin. Plant Biol. 2017, 35, 152–157. [Google Scholar] [CrossRef]
- Broeckx, T.; Hulsmans, S.; Rolland, F. The plant energy sensor: Evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot. 2016, 67, 6215–6252. [Google Scholar] [CrossRef]
- Robaglia, C.; Thomas, M.; Meyer, C. Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 2012, 15, 301–307. [Google Scholar] [CrossRef]
- Wu, Y.; Shi, L.; Li, L.; Fu, L.; Liu, Y.; Xiong, Y.; Sheen, J. Integration of nutrient, energy, light, and hormone signalling via TOR in plants. J. Exp. Bot. 2019, 70, 2227–2238. [Google Scholar] [CrossRef]
- Schepetilnikov, M.; Ryabova, L.A. Recent Discoveries on the Role of TOR (Target of Rapamycin) Signaling in Translation in Plants. Plant Physiol. 2018, 176, 1095–1105. [Google Scholar] [CrossRef]
- Caldana, C.; Martins, M.C.M.; Mubeen, U.; Urrea-Castellanos, R. The magic ‘hammer’ of TOR: The multiple faces of a single pathway in the metabolic regulation of plant growth and development. J. Exp. Bot. 2019, 70, 2217–2225. [Google Scholar] [CrossRef]
- Pu, Y.; Soto-Burgos, J.; Bassham, D.C. Regulation of autophagy through SnRK1 and TOR signaling pathways. Plant Signal. Behav. 2017, 12, e1395128. [Google Scholar] [CrossRef]
- Nukarinen, E.; Nägele, T.; Pedrotti, L.; Wurzinger, B.; Mair, A.; Landgraf, R.; Börnke, F.; Hanson, J.; Teige, M.; Baena-González, E.; et al. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 2016, 6, 31697. [Google Scholar] [CrossRef]
- Tomé, F.; Nã¤Gele, T.; Adamo, M.C.; Garg, A.; Marco-Llorca, C.; Nukarinen, E.; Pedrotti, L.; Peviani, A.; Simeunovic, A.; Tatkiewicz, A.; et al. The low energy signaling network. Front. Plant Sci. 2014, 5, 353. [Google Scholar] [CrossRef]
- Belda-Palazón, B.; Adamo, M.; Valerio, C.; Ferreira, L.J.; Confraria, A.; Reis-Barata, D.; Rodrigues, A.; Meyer, C.; Rodriguez, P.L.; Baena-González, E. A dual function of SnRK2 kinases in the regulation of SnRK1 and plant growth. Nat. Plants 2020, 6, 1345–1353. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 2015, 6, 84. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef]
- Fujii, H.; Zhu, J.-K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef]
- Kim, K.-N.; Lee, J.-S.; Han, H.; Choi, S.A.; Go, S.J.; Yoon, I.S. Isolation and characterization of a novel rice Ca2+-regulated protein kinase gene involved in responses to diverse signals including cold, light, cytokinins, sugars and salts. Plant Mol. Biol. 2003, 52, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 Protein Kinases, SRK2D/SnRK2. 2, SRK2E/SnRK2. 6/OST1 and SRK2I/SnRK2. 3, Involved in ABA Signaling are Essential for the Control of Seed Development and Dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Murata, M.; Minami, H.; Yamamoto, S.; Kagaya, Y.; Hobo, T.; Yamamoto, A.; Hattori, T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005, 44, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Y.; Li, S.-S.; Han, G.-Z. Insights into the Origin and Evolution of the Plant Hormone Signaling Machinery. Plant Physiol. 2015, 167, 872–886. [Google Scholar] [CrossRef] [PubMed]
- Lempiäinen, H.; Shore, D. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009, 21, 855–863. [Google Scholar] [CrossRef] [PubMed]
- De Block, M.; Van Lijsebettens, M. Energy efficiency and energy homeostasis as genetic and epigenetic components of plant performance and crop productivity. Curr. Opin. Plant Biol. 2011, 14, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Moin, M.; Bakshi, A.; Saha, A.; Kumar, M.U.; Reddy, A.R.; Rao, K.V.; Siddiq, E.A.; Kirti, P.B. Activation tagging in indica rice identifies ribosomal proteins as potential targets for manipulation of water-use efficiency and abiotic stress tolerance in plants: Activation Tagging Identifies Ribosomal Protein Genes. Plant Cell Environ. 2016, 39, 2440–2459. [Google Scholar] [CrossRef] [PubMed]
- Moin, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Expression Profiling of Ribosomal Protein Gene Family in Dehydration Stress Responses and Characterization of Transgenic Rice Plants Overexpressing RPL23A for Water-Use Efficiency and Tolerance to Drought and Salt Stresses. Front. Chem. 2017, 5, 97. [Google Scholar] [CrossRef]
- Saez-Vasquez, J.; Gallois, P.; Delseny, M. Accumulation and nuclear targeting of BnC24, a Brassica napus ribosomal protein corresponding to a mRNA accumulating in response to cold treatment. Plant Sci. 2000, 156, 35–46. [Google Scholar] [CrossRef]
- Moin, M.; Bakshi, A.; Saha, A.; Dutta, M.; Madhav, S.M.; Kirti, P.B. Rice Ribosomal Protein Large Subunit Genes and Their Spatio-temporal and Stress Regulation. Front. Plant Sci. 2016, 7, 1284. [Google Scholar] [CrossRef]
- Martinez-Seidel, F.; Suwanchaikasem, P.; Nie, S.; Leeming, M.G.; Firmino, A.A.P.; Williamson, N.A.; Kopka, J.; Roessner, U.; Boughton, B.A. Membrane-Enriched Proteomics Link Ribosome Accumulation and Proteome Reprogramming With Cold Acclimation in Barley Root Meristems. Front. Plant Sci. 2021, 12, 656683. [Google Scholar] [CrossRef]
- Martinez-Seidel, F.; Beine-Golovchuk, O.; Hsieh, Y.-C.; Eshraky, K.; Gorka, M.; Cheong, B.-E.; Jimenez-Posada, E.; Walther, D.; Skirycz, A.; Roessner, U.; et al. Spatially Enriched Paralog Rearrangements Argue Functionally Diverse Ribosomes Arise during Cold Acclimation in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 6160. [Google Scholar] [CrossRef]
- Garcia-Molina, A.; Kleine, T.; Schneider, K.; Mühlhaus, T.; Lehmann, M.; Leister, D. Translational Components Contribute to Acclimation Responses to High Light, Heat, and Cold in Arabidopsis. iScience 2020, 23, 101331. [Google Scholar] [CrossRef]
- Vandereyken, K.; Van Leene, J.; De Coninck, B.; Cammue, B.P.A. Hub Protein Controversy: Taking a Closer Look at Plant Stress Response Hubs. Front. Plant Sci. 2018, 9, 694. [Google Scholar] [CrossRef]
- Palm, D.; Streit, D.; Shanmugam, T.; Weis, B.L.; Ruprecht, M.; Simm, S.; Schleiff, E. Plant-specific ribosome biogenesis factors in Arabidopsis thaliana with essential function in rRNA processing. Nucleic Acids Res. 2019, 47, 1880–1895. [Google Scholar] [CrossRef] [PubMed]
- Weis, B.L.; Palm, D.; Missbach, S.; Bohnsack, M.T.; Schleiff, E. atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana. RNA 2015, 21, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Golovchuk, O.B.; Firmino, A.A.P.; Dąbrowska, A.; Schmidt, S.; Erban, A.; Walther, D.; Zuther, E.; Hincha, D.K.; Kopka, J. Plant Temperature Acclimation and Growth Rely on Cytosolic Ribosome Biogenesis Factor Homologs. Plant Physiol. 2018, 176, 2251–2276. [Google Scholar] [CrossRef]
- Cheong, B.E.; Beine-Golovchuk, O.; Gorka, M.; Ho, W.W.H.; Martinez-Seidel, F.; Firmino, A.A.P.; Skirycz, A.; Roessner, U.; Kopka, J. Arabidopsis REI-LIKE proteins activate ribosome biogenesis during cold acclimation. Sci. Rep. 2021, 11, 2410. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.; Dethloff, F.; Beine-Golovchuk, O.; Kopka, J. The REIL1 and REIL2 Proteins of Arabidopsis thaliana Are Required for Leaf Growth in the Cold. Plant Physiol. 2013, 163, 1623–1639. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Kong, X.; Huang, H.; Wu, W.; Park, J.; Yun, D.-J.; Lee, B.-H.; Shi, H.; Zhu, J.-K. STCH4/REIL2 Confers Cold Stress Tolerance in Arabidopsis by Promoting rRNA Processing and CBF Protein Translation. Cell Rep. 2020, 30, 229–242.e5. [Google Scholar] [CrossRef]
- Klingauf-Nerurkar, P.; Gillet, L.C.; Portugal-Calisto, D.; Oborská-Oplová, M.; Jäger, M.; Schubert, O.T.; Pisano, A.; Peña, C.; Rao, S.; Altvater, M.; et al. The GTPase Nog1 co-ordinates the assembly, maturation and quality control of distant ribosomal functional centers. eLife 2020, 9, e52474. [Google Scholar] [CrossRef]
- Guo, J.; Han, S.; Zhao, J.; Xin, C.; Zheng, X.; Liu, Y.; Wang, Y.; Qu, F. Essential role of NbNOG1 in ribosomal RNA processing: NOG1 and Ribosomal RNA Processing in Plants. J. Integr. Plant Biol. 2018, 60, 1018–1022. [Google Scholar] [CrossRef]
- Lee, S.; Senthil-Kumar, M.; Kang, M.; Rojas, C.M.; Tang, Y.; Oh, S.; Choudhury, S.R.; Lee, H.-K.; Ishiga, Y.; Allen, R.D.; et al. The small GTPase, nucleolar GTP-binding protein 1 (NOG1), has a novel role in plant innate immunity. Sci. Rep. 2017, 7, 9260. [Google Scholar] [CrossRef]
- Lee, S.; Rojas, C.M.; Oh, S.; Kang, M.; Choudhury, S.R.; Lee, H.-K.; Allen, R.D.; Pandey, S.; Mysore, K.S. Nucleolar GTP-Binding Protein 1-2 (NOG1-2) Interacts with Jasmonate-ZIMDomain Protein 9 (JAZ9) to Regulate Stomatal Aperture during Plant Immunity. Int. J. Mol. Sci. 2018, 19, 1922. [Google Scholar] [CrossRef] [PubMed]
- Pant, B.D.; Lee, S.; Lee, H.-K.; Krom, N.; Pant, P.; Jang, Y.; Mysore, K.S. Overexpression of Arabidopsis nucleolar GTP-binding 1 (NOG1) proteins confers drought tolerance in rice. Plant Physiol. 2022, 189, 988–1004. [Google Scholar] [CrossRef] [PubMed]
- Manzano, A.; Villacampa, A.; Sáez-Vásquez, J.; Kiss, J.Z.; Medina, F.J.; Herranz, R. The Importance of Earth Reference Controls in Spaceflight -Omics Research: Characterization of Nucleolin Mutants from the Seedling Growth Experiments. iScience 2020, 23, 101686. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.A. Bioregenerative life-support systems. Am. J. Clin. Nutr. 1994, 60, 820S–824S. [Google Scholar] [CrossRef] [PubMed]
- Sablok, G.; Powell, J.J.; Kazan, K. Emerging Roles and Landscape of Translating mRNAs in Plants. Front. Plant Sci. 2017, 8, 1443. [Google Scholar] [CrossRef] [PubMed]
- Chassé, H.; Boulben, S.; Costache, V.; Cormier, P.; Morales, J. Analysis of translation using polysome profiling. Nucleic Acids Res. 2017, 45, e15. [Google Scholar] [CrossRef]
- King, H.A.; Gerber, A.P. Translatome profiling: Methods for genome-scale analysis of mRNA translation. Brief. Funct. Genom. 2014, 15, 22–31. [Google Scholar] [CrossRef]
- Kage, U.; Powell, J.J.; Gardiner, D.M.; Kazan, K. Ribosome profiling in plants: What is not lost in translation? J. Exp. Bot. 2020, 71, 5323–5332. [Google Scholar] [CrossRef] [PubMed]
- Brar, G.A.; Weissman, J.S. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 2015, 16, 651–664. [Google Scholar] [CrossRef]
- Branco-Price, C.; Kawaguchi, R.; Ferreira, R.B.; Bailey-Serres, J. Genome-wide Analysis of Transcript Abundance and Translation in Arabidopsis Seedlings Subjected to Oxygen Deprivation. Ann. Bot. 2005, 96, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Branco-Price, C.; Kaiser, K.A.; Jang, C.J.H.; Larive, C.K.; Bailey-Serres, J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 2008, 56, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, R.; Girke, T.; Bray, E.A.; Bailey-Serres, J. Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 2004, 38, 823–839. [Google Scholar] [CrossRef]
- Yang, X.; Song, B.; Cui, J.; Wang, L.; Wang, S.; Luo, L.; Gao, L.; Mo, B.; Yu, Y.; Liu, L. Comparative ribosome profiling reveals distinct translational landscapes of salt-sensitive and -tolerant rice. BMC Genom. 2021, 22, 612. [Google Scholar] [CrossRef]
- Matsuura, H.; Ishibashi, Y.; Shinmyo, A.; Kanaya, S.; Kato, K. Genome-Wide Analyses of Early Translational Responses to Elevated Temperature and High Salinity in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 448–462. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, M.; Dong, Z. Preferential Ribosome Loading on the Stress-Upregulated mRNA Pool Shapes the Selective Translation under Stress Conditions. Plants 2021, 10, 304. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.; Barah, P.; Suarez-Rodriguez, M.C.; Bressendorff, S.; Friis, P.; Costantino, P.; Bones, A.M.; Nielsen, H.B.; Mundy, J. Transcriptome responses to combinations of stresses in Arabidopsis thaliana. Plant Physiol. 2013, 161, 1783–1794. [Google Scholar] [CrossRef]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Jewett, M.C.; Fritz, B.R.; Timmerman, L.E.; Church, G.M. In vitro integration of ribosomal RNA synthesis, ribosome assembly, and translation. Mol. Syst. Biol. 2013, 9, 678. [Google Scholar] [CrossRef] [PubMed]
- Fritz, B.R.; Jewett, M.C. The impact of transcriptional tuning on in vitro integrated rRNA transcription and ribosome construction. Nucleic Acids Res. 2014, 42, 6774–6785. [Google Scholar] [CrossRef] [PubMed]
- Hammerling, M.; Fritz, B.R.; Yoesep, D.J.; Kim, D.S.; Carlson, E.D.; Jewett, M.C. In vitro ribosome synthesis and evolution through ribosome display. Nat. Commun. 2020, 11, 1108. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dias-Fields, L.; Adamala, K.P. Engineering Ribosomes to Alleviate Abiotic Stress in Plants: A Perspective. Plants 2022, 11, 2097. https://doi.org/10.3390/plants11162097
Dias-Fields L, Adamala KP. Engineering Ribosomes to Alleviate Abiotic Stress in Plants: A Perspective. Plants. 2022; 11(16):2097. https://doi.org/10.3390/plants11162097
Chicago/Turabian StyleDias-Fields, Leticia, and Katarzyna P. Adamala. 2022. "Engineering Ribosomes to Alleviate Abiotic Stress in Plants: A Perspective" Plants 11, no. 16: 2097. https://doi.org/10.3390/plants11162097
APA StyleDias-Fields, L., & Adamala, K. P. (2022). Engineering Ribosomes to Alleviate Abiotic Stress in Plants: A Perspective. Plants, 11(16), 2097. https://doi.org/10.3390/plants11162097





