Genome-Wide Identification and Expression Analysis of Isopentenyl transferase Family Genes during Development and Resistance to Abiotic Stresses in Tea Plant (Camellia sinensis)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification and Bioinformatics Analysis of Tea Plant CsIPT Genes
2.2. Multiple Sequence Alignment and Phylogenetic Analysis of CsIPT Gene Family
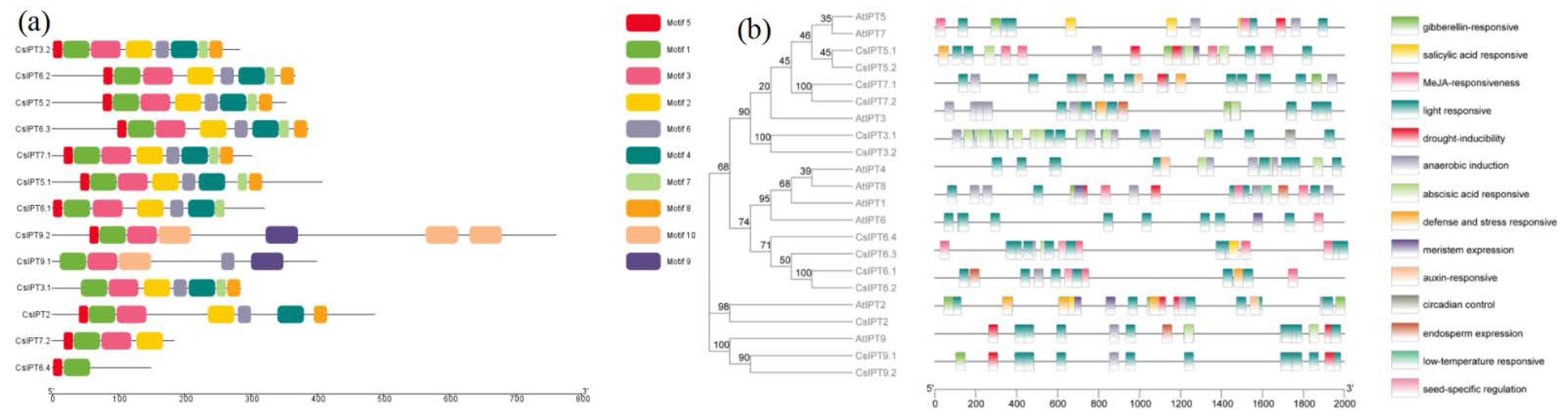
2.3. Gene Structure and Conserved Domain Analysis of CsIPT Genes
2.4. The cis-Acting Elements in the Promoter Regions of CsIPT Genes
2.5. Expression Pattern Analysis of CsIPT Family Numbers
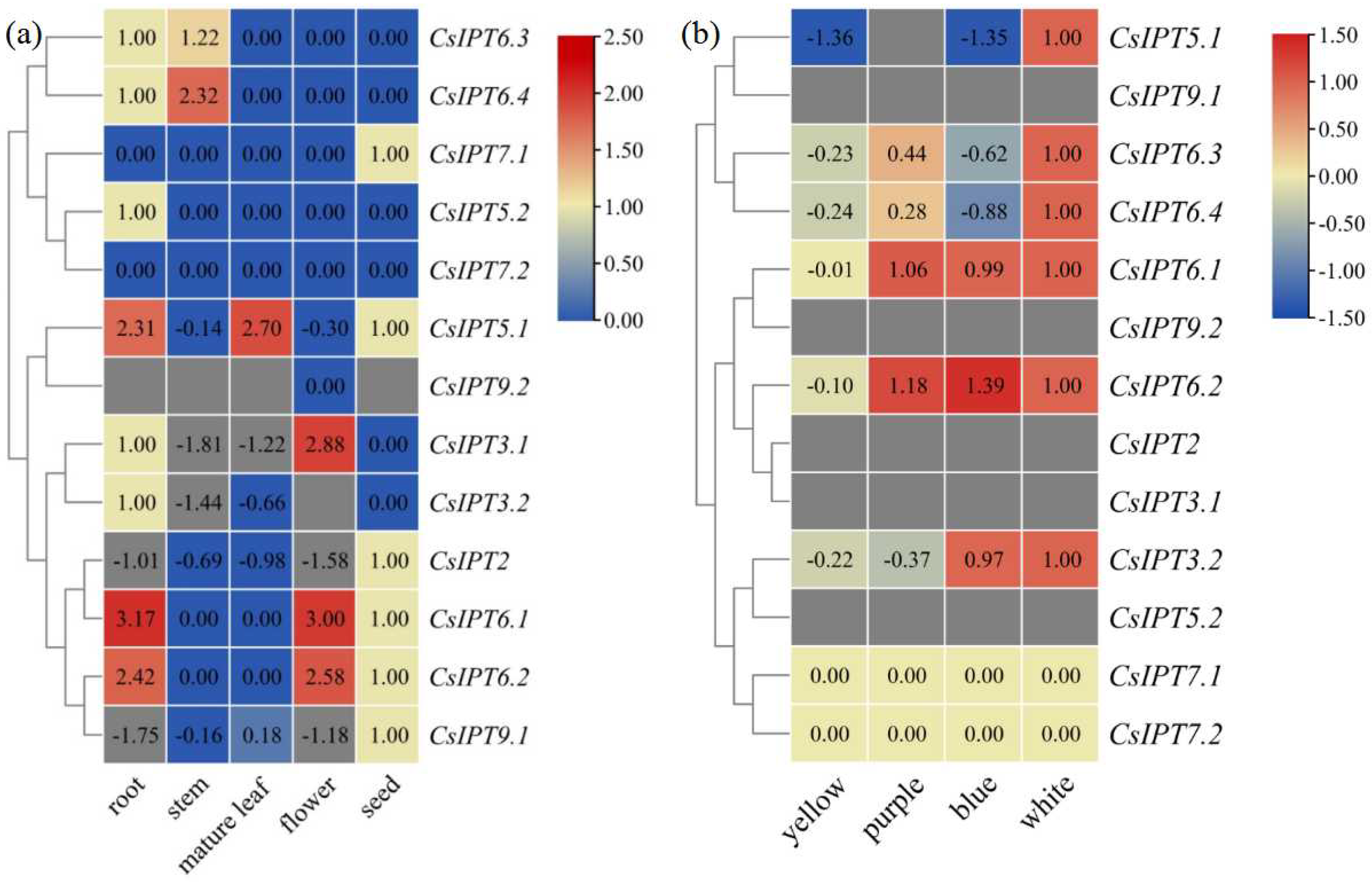
2.5.1. Expression Patterns of CsIPT Genes in Different Tissues and to Different Light Treatments
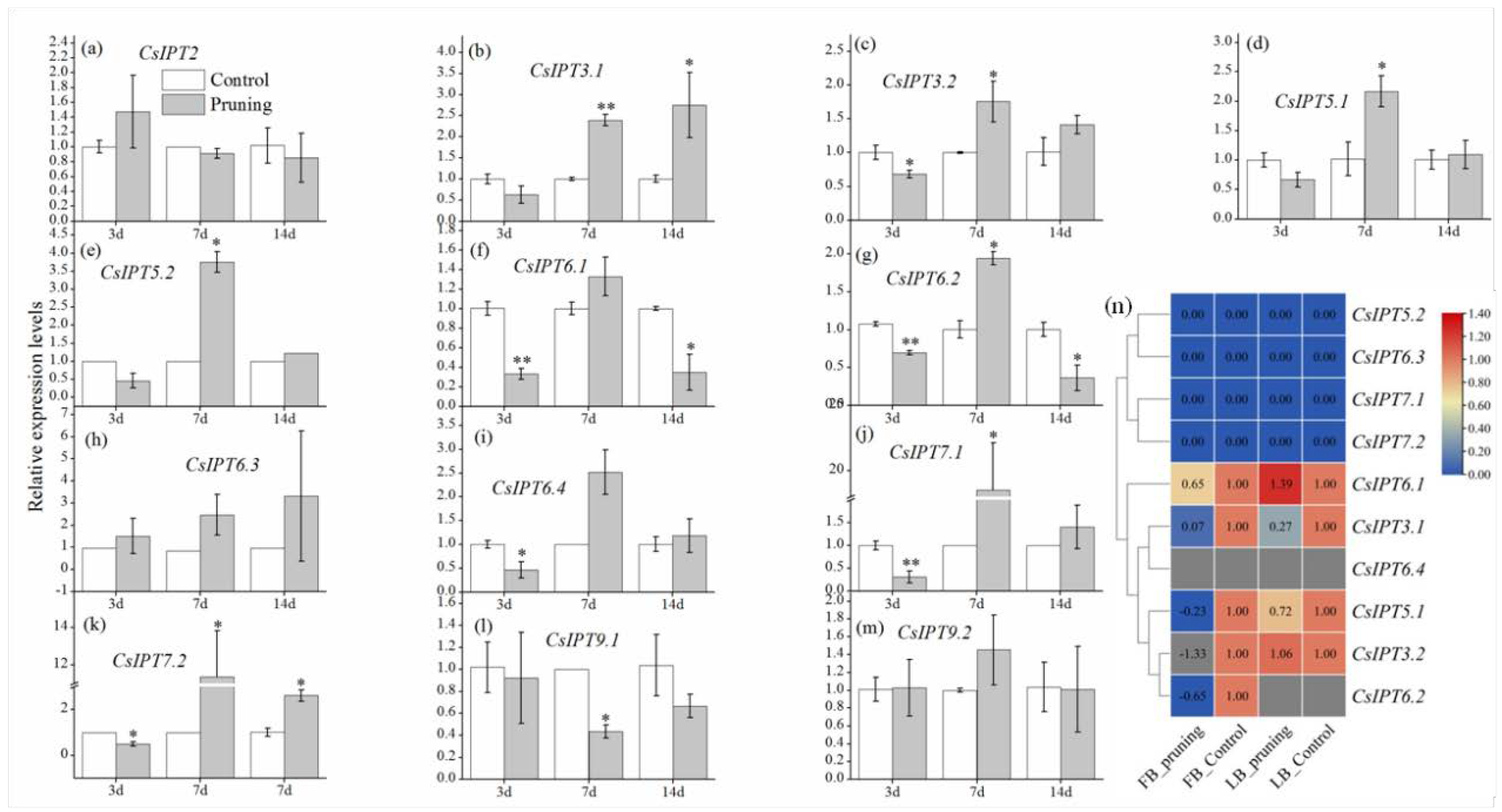
2.5.2. Involvement of CsIPT Genes in Tea Plant Development
The Regulatory Roles of CsIPT Genes in Tea Plant Growth and Development
CsIPT Numbers Synergistically Regulated the Growth of Leaf-Bud and Flower-Bud Induced by Pruning in Tea Plant
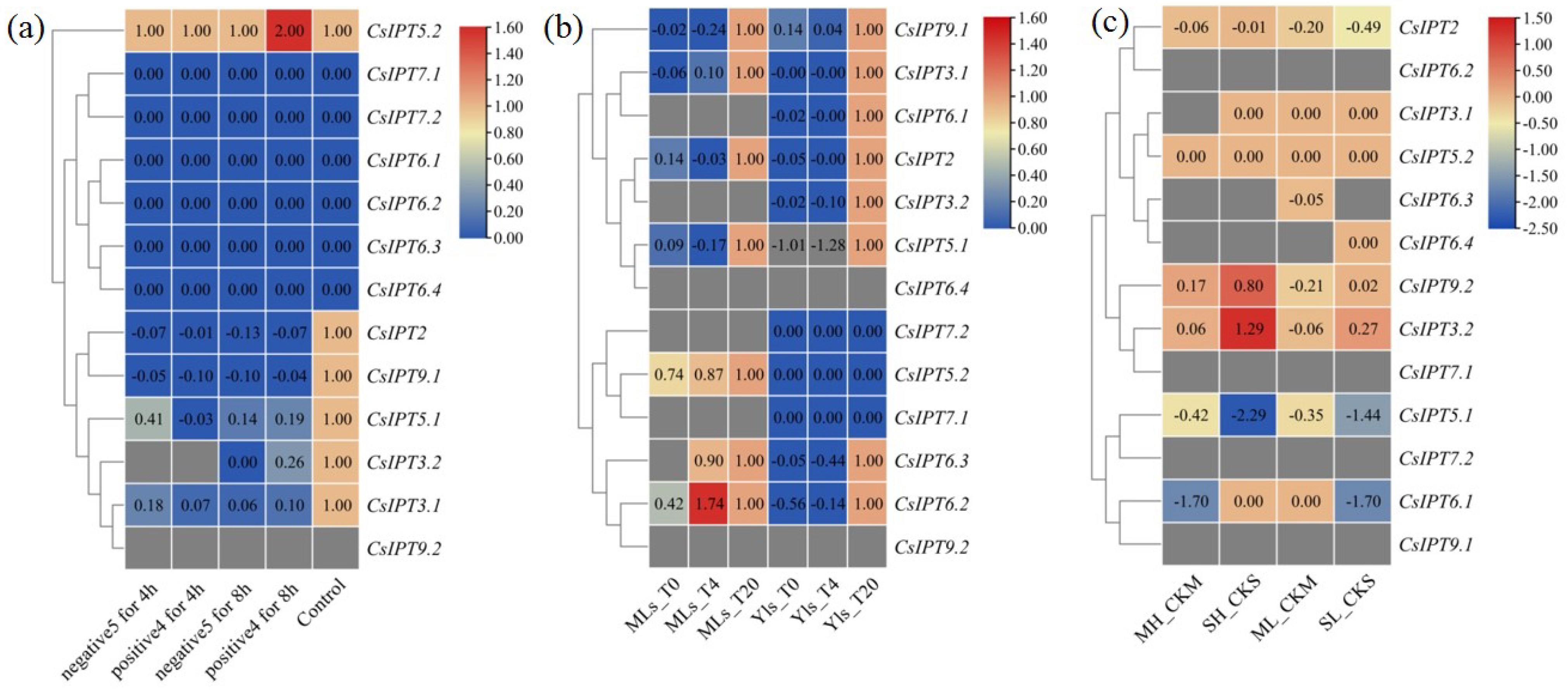
2.5.3. The Expression Patterns of CsIPT Genes in Response to Abiotic Stresses
The Involvement of CsIPT Family Members in Response to Temperature Stresses
The Involvement of CsIPT Family Members in Response to Drought and Salt Stresses
3. Materials and Methods
3.1. Identification of CsIPT Genes in Tea Plants
3.2. Multiple Sequence Alignment and Phylogenetic Analysis
3.3. Analysis of the Gene Structure and Conserved Motifs of CsIPT Genes
3.4. Physiological and Biochemical Property Analysis of CsIPT Proteins
3.5. Promoter Cis-Acting Regulatory Elements Analysis
3.6. Publicly Available Transcriptome Data Analysis
3.6.1. Analysis of CsIPT Expression Patterns in SRA Database during Tea Plant Development
3.6.2. Analysis of CsIPT Gene Expression in the SRA Database during Abiotic Stress
3.7. RNA-seq and qRT-PCR Detection
3.8. The Preparation and Observation of Paraffin Sections
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shoaib, M.; Yang, W.; Shan, Q.; Sajjad, M.; Zhang, A. Genome-wide identification and expression analysis of new cytokinin metabolic genes in bread wheat (Triricum aesrivum L.). PeerJ 2019, 7, e6300. [Google Scholar] [CrossRef] [PubMed]
- Nishii, K.; Wright, F.; Chen, Y.Y.; Moller, M. Tangled history of a multigene family: The evolution of ISOPENTENYLTRANSFERASE genes. PLoS ONE 2018, 13, e0201198. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, K.; Matsumoto-Kitano, M.; Kakimoto, T. Expression of cytokinin biosynthetic isopentenyl transferasegenes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004, 37, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Li, G.; Qi, S.; Liu, X.; Chen, X.; Ma, J.; Zhang, D.; Han, M. Identification and expression analysis of the IPT and CKX gene families during axillary bud outgrowth in apple (Males domestica Borkh.). Gene 2018, 651, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lin, S.; Liu, D.; Gan, L.; McAvoy, R.; Ding, J.; Li, Y. Evolution and roles of cytokinin genes in angiosperms 1: Do ancient IPTs play housekeeping while non-ancient IPTs play regulatory roles? Hortic. Res. 2020, 7, 28. [Google Scholar] [CrossRef]
- Immanen, J.; Nieminen, K.; Silva, H.D.; Rojas, F.R.; Meisel, L.A.; Silva, H.; Albert, V.A.; Hvidsten, T.R.; Helariutta, Y. Characterization of cytokinin signaling and homeostasis gene families in two hard wood tree species: Populus trichocarpa and Prunus persica. BMC Genom. 2013, 14, 885. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wei, Q.; Peng, F.; Yu, W.; Luo, J.; Zhao, Y. Identification and characterization of ATP/ADP isopentenyltransferases (ATP/ADP PpIPTs) genes in peach. J. Plant Growth Regul. 2019, 38, 416–430. [Google Scholar] [CrossRef]
- Ghosh, A.; Shah, M.N.A.; Jui, Z.S.; Saha, S.; Fariha, K.A.; Islam, T. Evolutionary variation and expression profiling of isopentenyl transferase gene family in Arabidopsis thaliana L. and Oryza sativa L. Plant Gene 2018, 15, 15–27. [Google Scholar] [CrossRef]
- Cai, L.; Zhang, L.; Fu, Q.; Xu, Z.F. Identification and expression analysis of cytokinin metabolic genes IPTs, CYP735A and CKXs in the biofuel plant Jatrophacurcas. PeerJ 2018, 6, e4812. [Google Scholar] [CrossRef]
- Liu, Z.; Lv, Y.; Zhang, M.; Liu, Y.; Kong, L.; Zou, M.; Lu, G.; Cao, J.; Yu, X. Identification, expression, and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 594. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.X.; Liu, Z.W.; Wu, Z.J.; Li, H.; Wang, W.L.; Cui, X.; Zhuang, J. Genome-wide identification andexpression analysis of GRAS family transcription factors in tea plant (Camellia sinensis). Sci. Rep. 2018, 8, 3949. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, P.; Gu, M.; Lin, X.; Hou, B.; Zheng, Y.; Sun, Y.; Jin, S.; Ye, N. R2R3-MYB transcription factor family in tea plant (Camellia sinensis): Genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics 2021, 113, 1565–1578. [Google Scholar] [CrossRef] [PubMed]
- Kakkar, R.K.; Nagar, P.K. Distribution and changes in endogenous polyamines during winter dormancy in tea [Camellia sinensis L. (O) Kuntze]. J. Plant Physiol. 1997, 151, 63–67. [Google Scholar] [CrossRef]
- Zhang, L.; Li, M.; Yan, P.; Fu, J.; Zhang, L.; Li, X.; Han, W. A novel adenylate isopentenyltransferase 5 regulates shoot branching via the ATTTA motif in Camellia sinensis. BMC Plant Biol. 2021, 21, 521. [Google Scholar] [CrossRef]
- Liu, S.C.; Jin, J.Q.; Ma, J.Q.; Yao, M.Z.; Ma, C.L.; Li, C.F.; Ding, Z.T.; Chen, L. Transcriptomic analysis of tea plant responding to drought stress and recovery. PLoS ONE 2016, 11, e0147306. [Google Scholar] [CrossRef]
- Li, N.; Yue, C.; Cao, H.; Qian, W.; Hao, X.; Wang, Y.; Wang, L.; Ding, C.; Wang, X.; Yang, Y. Transcriptome sequencing dissection of the mechanisms underlyingdifferential cold sensitivity in young and mature leaves of the tea plant (Camellia sinensis). J. Plant Physiol. 2018, 224–225, 144–155. [Google Scholar]
- Matsuo, S.; Kikuchi, K.; Fukuda, M.; Honda, I.; Imanishi, S. Roles and regulation of cytokinins in tomato fruit development. J. Exp. Bot. 2012, 63, 5569–5579. [Google Scholar] [CrossRef]
- Joshi, R.; Singla-Pareek, S.L.; Pareek, A. Engineering abiotic stress response in plants for biomass production. J. Biol. Chem. 2018, 293, 5035–5043. [Google Scholar] [CrossRef]
- Nishiyama, R.; Watanabe, Y.; Fujita, Y.; Le, D.T.; Kojima, M.; Werner, T.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Kakimoto, T.; et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 2011, 23, 2169–2183. [Google Scholar] [CrossRef]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Vankova, R.; Tanaka, M.; Seki, M.; Ham, L.H.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PLoS ONE 2012, 7, e42411. [Google Scholar]
- Xia, E.H.; Li, F.D.; Tong, W.; Li, P.H.; Wu, Q.; Zhao, H.J.; Ge, R.H.; Li, R.P.; Li, Y.Y.; Zhang, Z.Z.; et al. Tea Plant Information Archive (TPIA): A comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol. J. 2019, 17, 1938–1953. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Li, C.F.; Zhu, Y.; Yu, Y.; Zhao, Q.Y.; Wang, S.J.; Wang, X.C.; Yao, M.Z.; Luo, D.; Li, X.; Chen, L.; et al. Global transcriptome and gene regulation network for secondary metabolite biosynthesis of tea plant (Camellia sinensis). BMC Genom. 2015, 16, 560. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Li, L.; Hu, Y.; Zhou, C.; Wang, X.; Wang, L.; Zeng, J.; Yang, Y. Transcriptomic analysis of the effects of three different light treatments on the biosynthesis of characteristic compounds in the tea plant by RNA-Seq. Tree Genet. Genomes 2016, 12, 118. [Google Scholar] [CrossRef]
- Hao, X.; Yang, Y.; Yue, C.; Wang, L.; Horvath, D.P.; Wang, X. Comprehensive transcriptome analyses reveal differential gene expression profiles of Camellia sinensis axillary buds at para-, endo-, ecodormancy, and bud flush stages. Front. Plant Sci. 2017, 8, 553. [Google Scholar] [CrossRef]
- Liu, F.; Wang, Y.; Ding, Z.; Zhao, L.; Xiao, J.; Wang, L.; Ding, S. Transcriptomic analysis of flower development in tea (Camellia sinensis (L.)). Gene 2017, 631, 39–51. [Google Scholar] [CrossRef]
- Zheng, C.; Zhao, L.; Wang, Y.; Shen, J.; Zhang, Y.; Jia, S.; Li, Y.; Ding, Z. Integrated RNA-Seq and sRNA-Seq analysis identifies chilling and freezing responsive key molecular players and pathways in tea plant (Camellia sinensis). PLoS ONE 2015, 10, e0125031. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, D.; Zhou, L.; Zhang, X.; Liao, J.; Duan, Y.; Wen, B.; Ma, Y.; Wang, Y.; Fang, W.; et al. Transcriptomic and metabolomic profiling of Camellia sinensis L. cv. ‘Suchazao’ exposed to temperature stresses reveals modification in protein synthesis and photosynthetic and anthocyanin biosynthetic pathways. Tree Physiol. 2019, 39, 1583–1599. [Google Scholar] [CrossRef]
- Wan, S.; Wang, W.; Zhou, T.; Zhang, Y.; Chen, J.; Xiao, B.; Yang, Y.; Yu, Y. Transcriptomic analysis reveals the molecular mechanisms of Camellia sinensis in response to salt stress. Plant Growth Regul. 2018, 84, 481–492. [Google Scholar] [CrossRef]





| Gene | CDS Length (bp) | Protein | |||
|---|---|---|---|---|---|
| Name | Locus ID | Length (aa) | MW (kDa) | pI | |
| CsIPT2 | GWHPACFB004509 (CSS0044985.1) | 1464 | 55.42 | 6.09 | |
| CsIPT3.1 | GWHPACFB005294 | 858 | 285 | 32.68 | 8.81 |
| CsIPT3.2 | CSS0018660.1 | 852 | 283 | 31.76 | 7.10 |
| CsIPT5.1 | GWHPACFB026317 (CSS0005335.1) | 1227 | 408 | 45.17 | 8.56 |
| CsIPT5.2 | GWHPACFB017705 (CSS0010471.1) | 1062 | 353 | 39.66 | 5.77 |
| CsIPT6.1 | GWHPACFB021163 | 963 | 320 | 35.39 | 6.09 |
| CsIPT6.2 | CSS0006991.1 | 1104 | 367 | 41.69 | 8.99 |
| CsIPT6.3 | CSS0040585.1 | 1164 | 387 | 43.95 | 9.13 |
| CsIPT6.4 | GWHPACFB026680 | 447 | 149 | 15.90 | 6.21 |
| CsIPT7.1 | CSS0048745.1 | 909 | 302 | 34.31 | 6.08 |
| CsIPT7.2 | CSS0003027.1 | 555 | 184 | 20.63 | 5.11 |
| CsIPT9.1 | GWHPACFB024320 | 1383 | 400 | 45.73 | 6.25 |
| CsIPT9.2 | CSS0050211.1 | 2286 | 761 | 83.63 | 8.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Li, M.; Fu, J.; Huang, X.; Yan, P.; Ge, S.; Li, Z.; Bai, P.; Zhang, L.; Han, W.; et al. Genome-Wide Identification and Expression Analysis of Isopentenyl transferase Family Genes during Development and Resistance to Abiotic Stresses in Tea Plant (Camellia sinensis). Plants 2022, 11, 2243. https://doi.org/10.3390/plants11172243
Zhang L, Li M, Fu J, Huang X, Yan P, Ge S, Li Z, Bai P, Zhang L, Han W, et al. Genome-Wide Identification and Expression Analysis of Isopentenyl transferase Family Genes during Development and Resistance to Abiotic Stresses in Tea Plant (Camellia sinensis). Plants. 2022; 11(17):2243. https://doi.org/10.3390/plants11172243
Chicago/Turabian StyleZhang, Liping, Min Li, Jianyu Fu, Xiaoqin Huang, Peng Yan, Shibei Ge, Zhengzhen Li, Peixian Bai, Lan Zhang, Wenyan Han, and et al. 2022. "Genome-Wide Identification and Expression Analysis of Isopentenyl transferase Family Genes during Development and Resistance to Abiotic Stresses in Tea Plant (Camellia sinensis)" Plants 11, no. 17: 2243. https://doi.org/10.3390/plants11172243






