Co-Transcriptomic Analysis of the Maize–Western Corn Rootworm Interaction
Abstract
:1. Introduction
2. Results
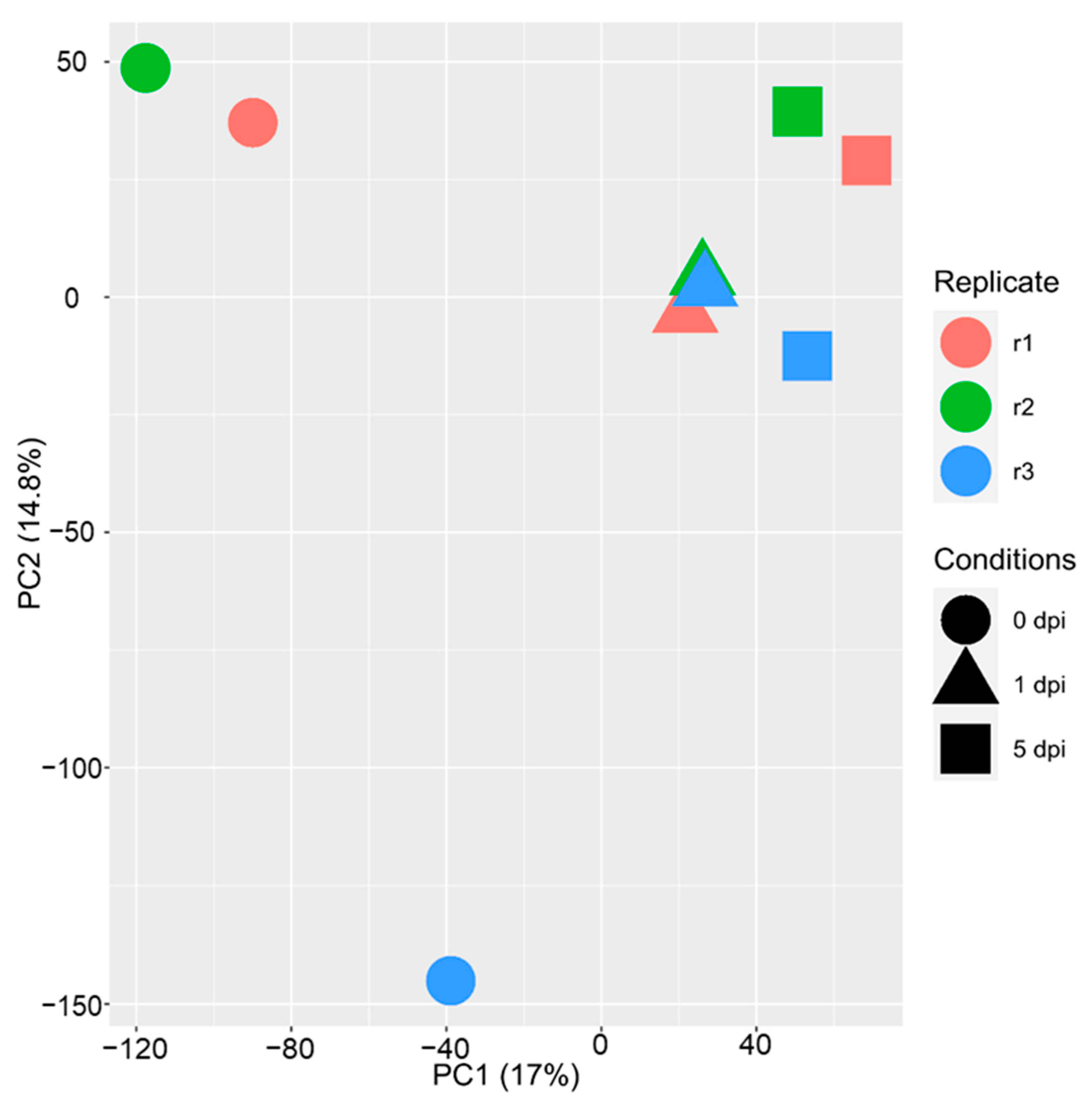
2.1. The Maize Transcriptomic Response Varies from Tissues and Time Points
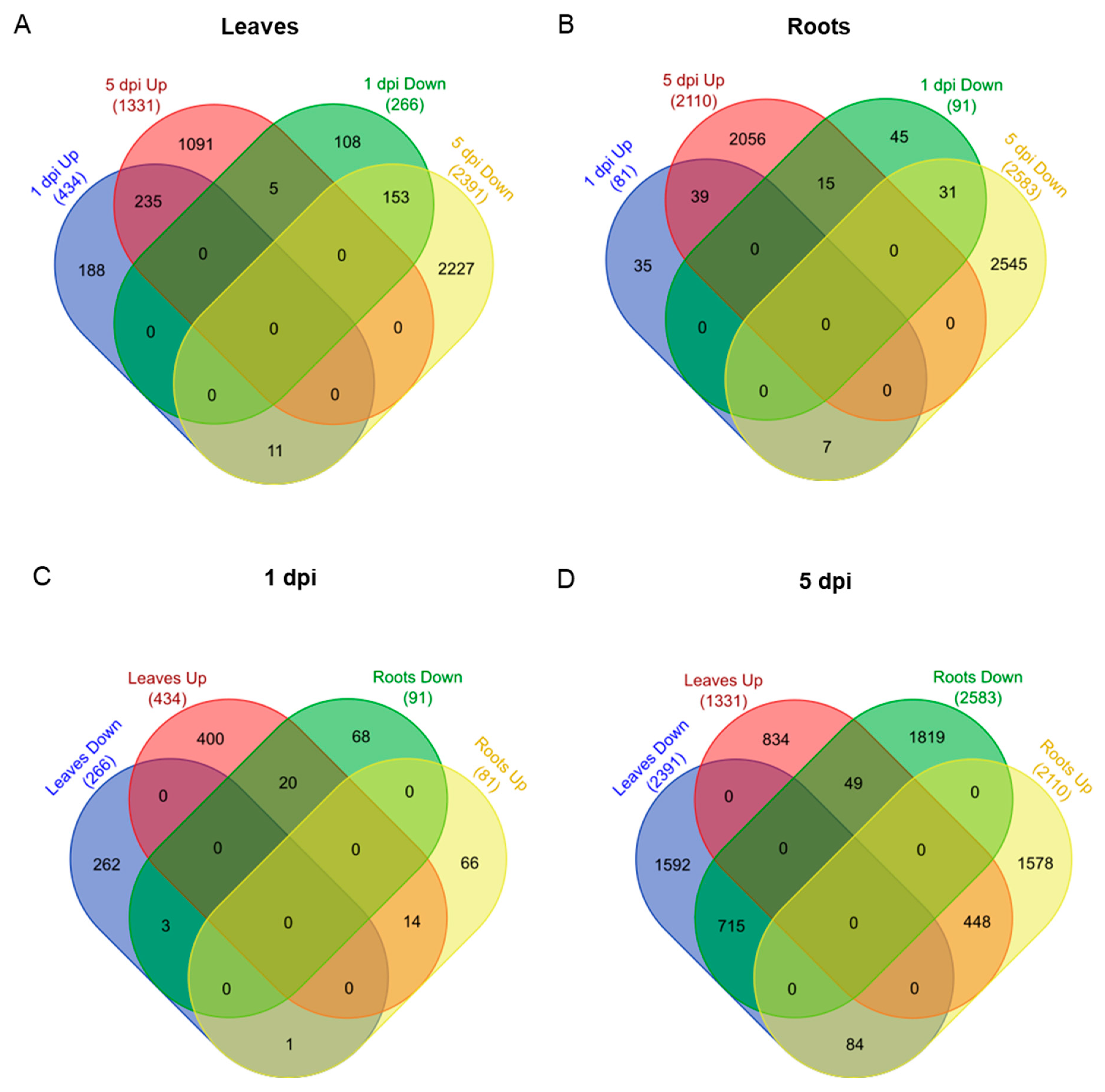
2.2. Differentially Expressed Genes Vary within Tissues and Have Minimal Overlap
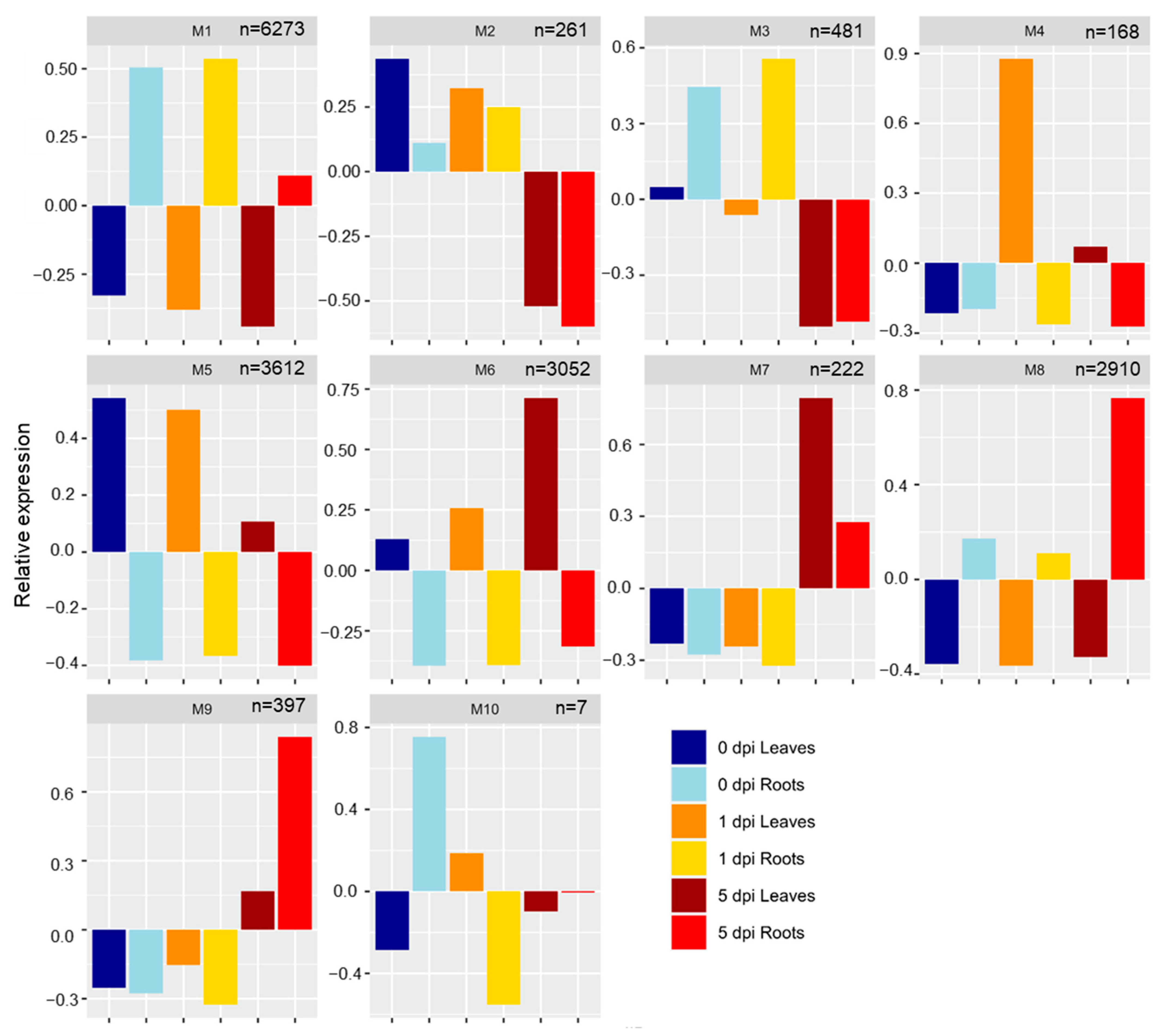
2.3. Genes Were Co-Expressed in Tissues and/or Time Points
2.3.1. Roots: M1, M3, M8
2.3.2. Leaves: M4, M5, M6
2.3.3. Time Points: M2, M7, M9, M10
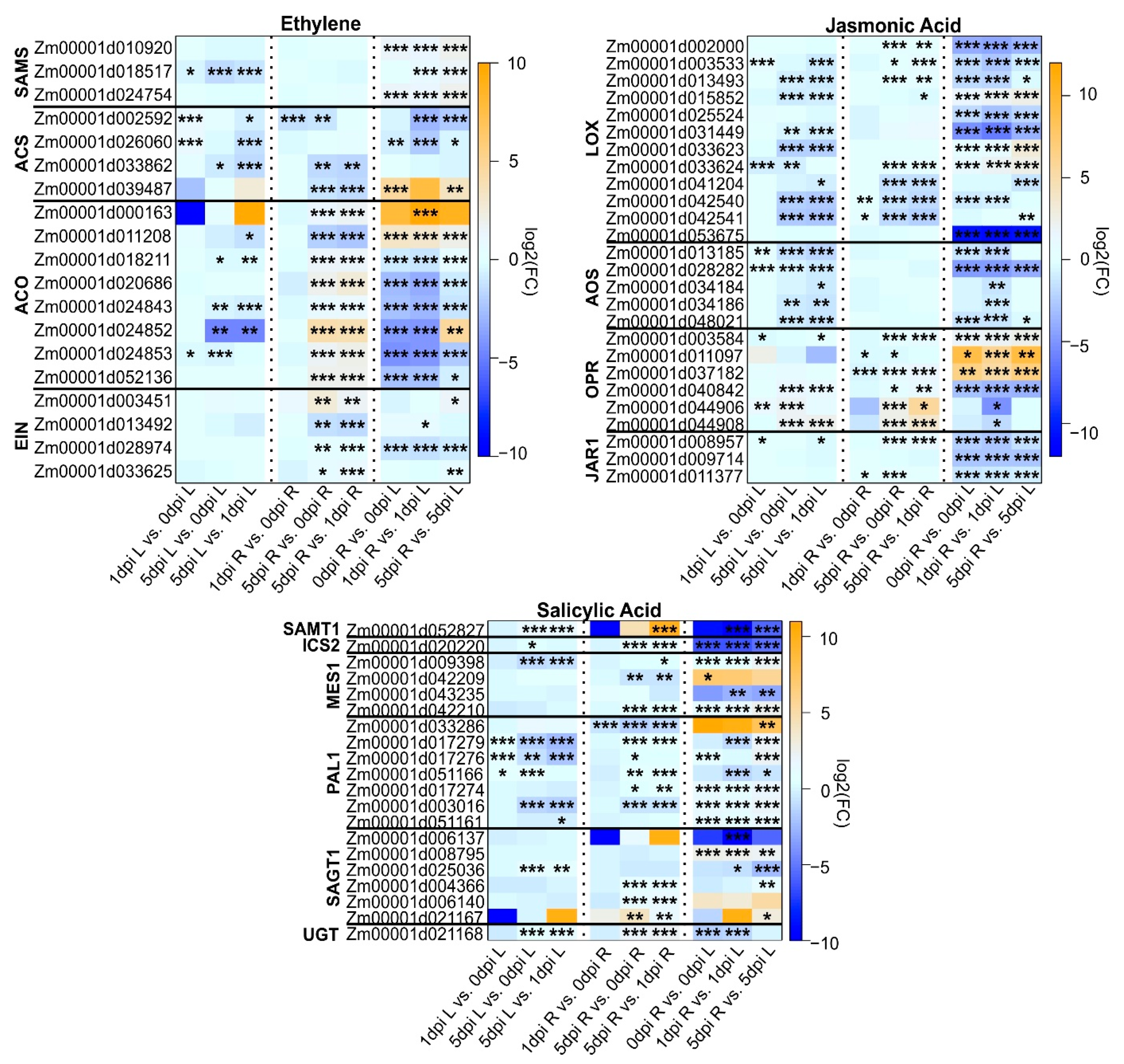
2.4. Modulations in Maize Hormone Pathways during WCR Infestation
2.4.1. Ethylene (ET)
2.4.2. Jasmonic Acid (JA)
2.4.3. Salicylic Acid (SA)
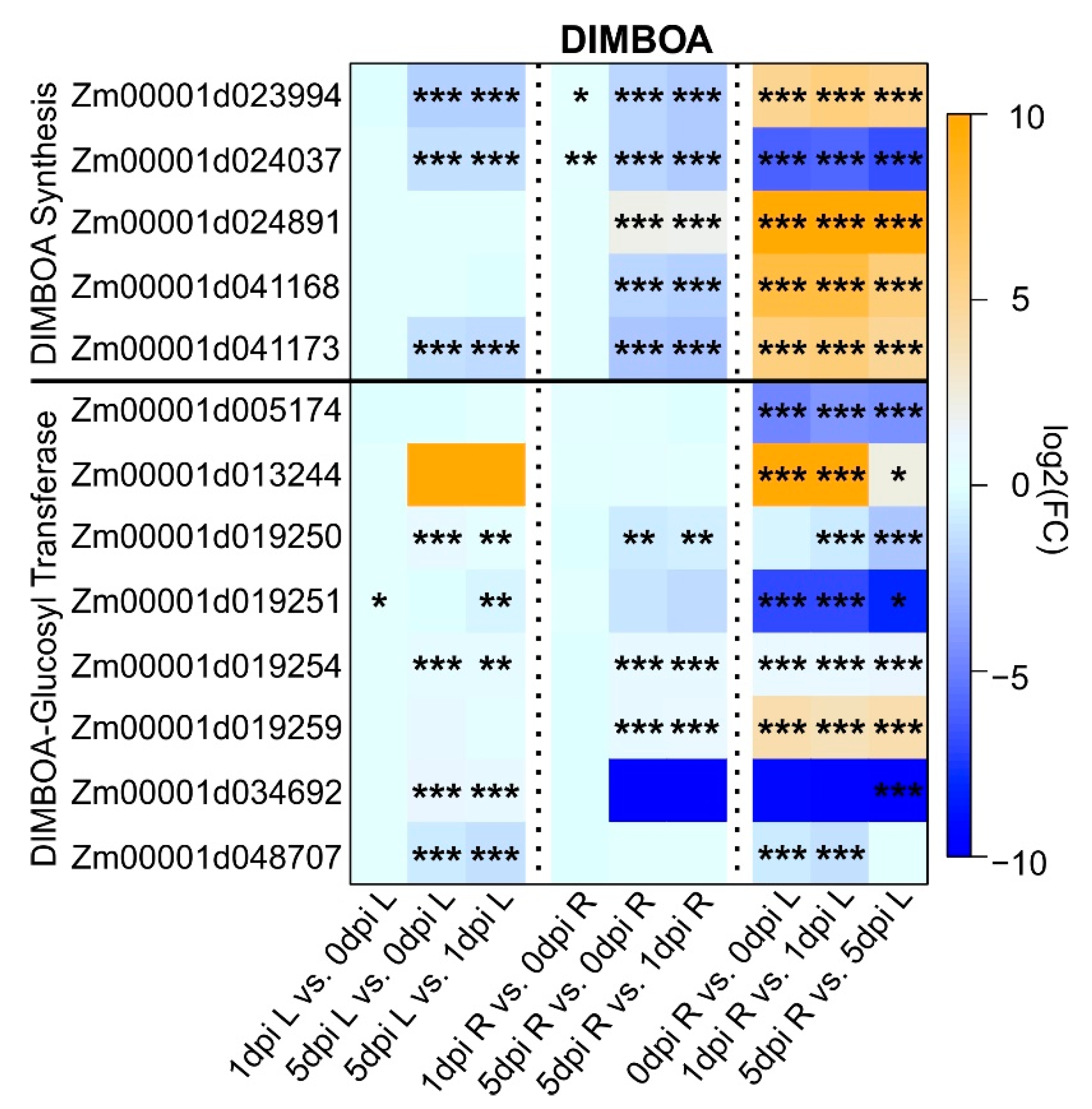
2.5. Maize DIMBOA Pathway Was Turned Off after Extended WCR Feeding
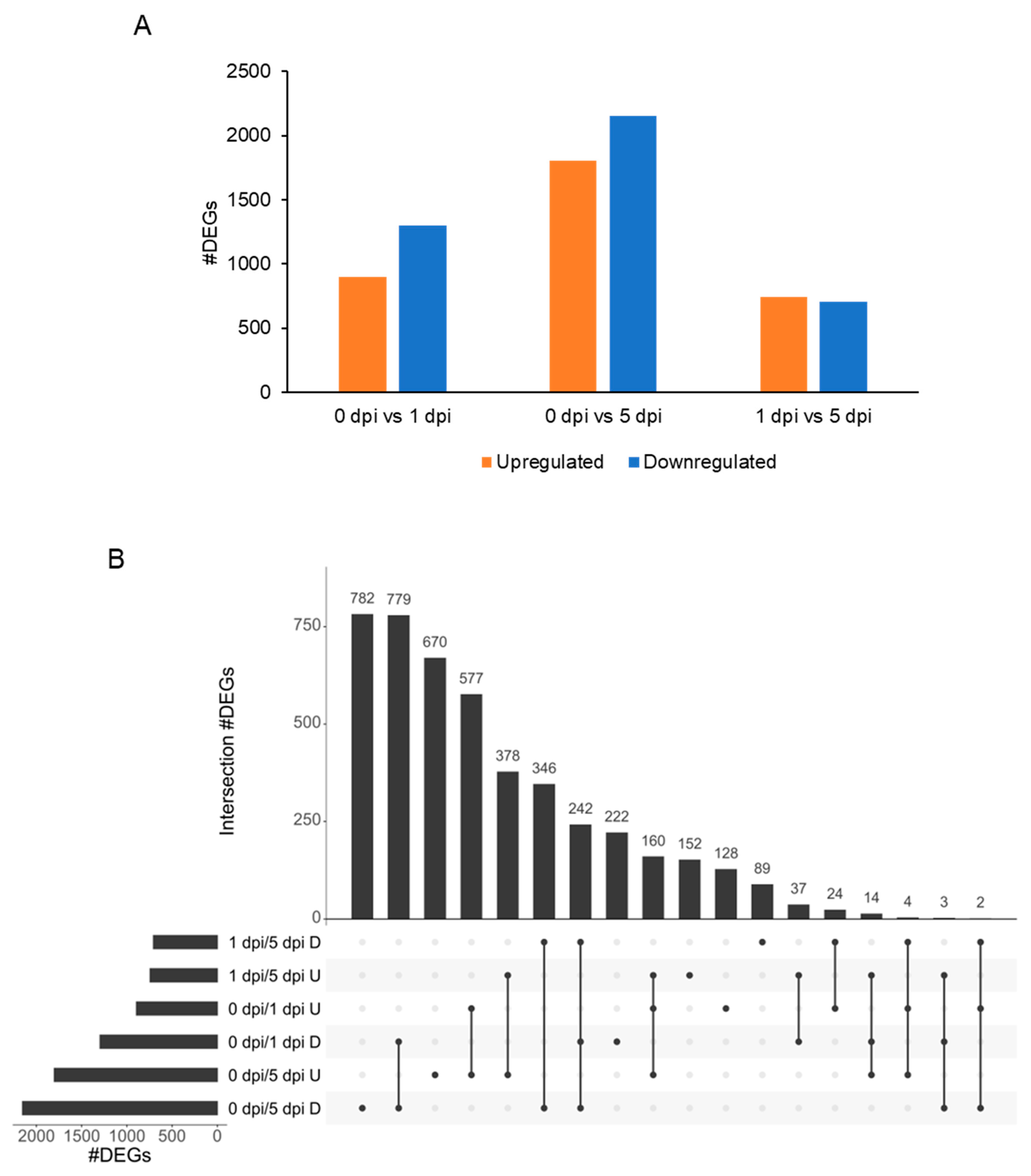
2.6. WCR Transcriptome Is Remodeled between 1 and 5 dpi
2.6.1. 0 h vs. 1 dpi
2.6.2. 0 h vs. 5 dpi
3. Discussion
4. Materials and Methods
4.1. Insect Growth
4.2. Plant Growth and WCR Infestations
4.3. RNA Extraction and RNA-Seq Libraries Construction and Sequencing
4.4. Analysis of RNA-Seq Libraries
4.5. Functional Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dun, Z.; Mitchell, P.D.; Agosti, M. Estimating Diabrotica virgifera virgifera Damage Functions with Field Trial Data: Applying an Unbalanced Nested Error Component Model. J. Appl. Entomol. 2010, 134, 409–419. [Google Scholar] [CrossRef]
- Tinsley, N.A.; Estes, R.E.; Gray, M.E. Validation of a Nested Error Component Model to Estimate Damage Caused by Corn Rootworm Larvae. J. Appl. Entomol. 2013, 137, 161–169. [Google Scholar] [CrossRef]
- Urías-López, M.A.; Meinke, L.J. Influence of Western Corn Rootworm (Coleoptera: Chrysomelidae) Larval Injury on Yield of Different Types of Maize. J. Econ. Entomol. 2001, 94, 106–111. [Google Scholar] [CrossRef]
- Branson, T. Larval Feeding Behavior and Host—Plant Resistance in Maize. In Methods for the Study of Pest Diabrotica; Springer: New York, NY, USA, 1986; pp. 159–182. [Google Scholar]
- Chiang, H.C. Bionomics of the Northern and Western Corn Rootworms. Annu. Rev. Entomol. 1973, 18, 47–72. [Google Scholar] [CrossRef]
- Riedell, W.E. Rootworm and Mechanical Damage Effects on Root Morphology and Water Relations in Maize. Crop Sci. 1990, 30, 628–631. [Google Scholar] [CrossRef]
- Riedell, W.; Kim, A. Anatomical Characterization of Western Corn Rootworm Damage in Adventitious Roots of Maize. J. Iowa Acad. Sci. 1990, 97, 15–17. [Google Scholar]
- Eshel, A.; Beeckman, T. Plant Roots: The Hidden Half, 4th ed.; CRC Press: Boca Raton, FL, USA, 2013; ISBN 978-1-4398-4649-0. [Google Scholar]
- Ball, H.J.; Weekman, G.T. Insecticide Resistance in the Adult Western Corn Rootworm in Nebraska. J. Econ. Entomol. 1962, 55, 439–441. [Google Scholar] [CrossRef]
- Meinke, L.J.; Siegfried, B.D.; Wright, R.J.; Chandler, L.D. Adult Susceptibility of Nebraska Western Corn Rootworm (Coleoptera: Chrysomelidae) Populations to Selected Insecticides. J. Econ. Entomol. 1998, 91, 594–600. [Google Scholar] [CrossRef]
- Parimi, S.; Meinke, L.J.; Wade French, B.; Chandler, L.D.; Siegfried, B.D. Stability and Persistence of Aldrin and Methyl-Parathion Resistance in Western Corn Rootworm Populations (Coleoptera: Chrysomelidae). Crop Prot. 2006, 25, 269–274. [Google Scholar] [CrossRef]
- Pereira, A.E.; Wang, H.; Zukoff, S.N.; Meinke, L.J.; French, B.W.; Siegfried, B.D. Evidence of Field-Evolved Resistance to Bifenthrin in Western Corn Rootworm (Diabrotica virgifera virgifera LeConte) Populations in Western Nebraska and Kansas. PLoS ONE 2015, 10, e0142299. [Google Scholar] [CrossRef]
- Zhu, K.Y.; Wilde, G.E.; Higgins, R.A.; Sloderbeck, P.E.; Buschman, L.L.; Shufran, R.A.; Whitworth, R.J.; Starkey, S.R.; He, F. Evidence of Evolving Carbaryl Resistance in Western Corn Rootworm (Coleoptera: Chrysomelidae) in Areawide-Managed Cornfields in North Central Kansas. J. Econ. Entomol. 2001, 94, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Levine, E.; Spencer, J.L.; Isard, S.A.; Onstad, D.W.; Gray, M.E. Adaptation of the Western Corn Rootworm to Crop Rotation: Evolution of a New Strain in Response to a Management Practice. Am. Entomol. 2002, 48, 94–107. [Google Scholar] [CrossRef]
- Gassmann, A.J.; Petzold-Maxwell, J.L.; Keweshan, R.S.; Dunbar, M.W. Field-Evolved Resistance to Bt Maize by Western Corn Rootworm. PLoS ONE 2011, 6, e22629. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, A.J.; Shrestha, R.B.; Jakka, S.R.K.; Dunbar, M.W.; Clifton, E.H.; Paolino, A.R.; Ingber, D.A.; French, B.W.; Masloski, K.E.; Dounda, J.W.; et al. Evidence of Resistance to Cry34/35Ab1 Corn by Western Corn Rootworm (Coleoptera: Chrysomelidae): Root Injury in the Field and Larval Survival in Plant-Based Bioassays. J. Econ. Entomol. 2016, 109, 1872–1880. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, A.J.; Shrestha, R.B.; Kropf, A.L.; Clair, C.R.S.; Brenizer, B.D. Field-Evolved Resistance by Western Corn Rootworm to Cry34/35Ab1 and Other Bacillus thuringiensis Traits in Transgenic Maize. Pest Manag. Sci. 2020, 76, 268–276. [Google Scholar] [CrossRef]
- Ludwick, D.C.; Meihls, L.N.; Ostlie, K.R.; Potter, B.D.; French, L.; Hibbard, B.E. Minnesota Field Population of Western Corn Rootworm (Coleoptera: Chrysomelidae) Shows Incomplete Resistance to Cry34Ab1/Cry35Ab1 and Cry3Bb1. J. Appl. Entomol. 2017, 141, 28–40. [Google Scholar] [CrossRef]
- Zukoff, S.N.; Ostlie, K.R.; Potter, B.; Meihls, L.N.; Zukoff, A.L.; French, L.; Ellersieck, M.R.; Wade French, B.; Hibbard, B.E. Multiple Assays Indicate Varying Levels of Cross Resistance in Cry3Bb1-Selected Field Populations of the Western Corn Rootworm to MCry3A, ECry3.1Ab, and Cry34/35Ab1. J. Econ. Entomol. 2016, 109, 1387–1398. [Google Scholar] [CrossRef]
- Branson, T.F.; Sutter, G.R.; Fisher, J.R. Comparison of a Tolerant and a Susceptible Maize Inbred Under Artificial Infestations of Diabrotica virgifera virgifera: Yield and Adult Emergence. Environ. Entomol. 1982, 11, 371–372. [Google Scholar] [CrossRef]
- Meihls, L.N.; Kaur, H.; Jander, G. Natural Variation in Maize Defense against Insect Herbivores. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 269–283. [Google Scholar] [CrossRef]
- Van Dam, N.M. Belowground Herbivory and Plant Defenses. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 373–391. [Google Scholar] [CrossRef]
- War, A.R.; Taggar, G.K.; Hussain, B.; Taggar, M.S.; Nair, R.M.; Sharma, H.C. Plant Defence against Herbivory and Insect Adaptations. Aob Plants 2018, 10, ply037. [Google Scholar] [CrossRef]
- Levin, D.A. The Role of Trichomes in Plant Defense. Q. Rev. Biol. 1973, 48, 3–15. [Google Scholar] [CrossRef]
- Tian, D.; Tooker, J.; Peiffer, M.; Chung, S.H.; Felton, G.W. Role of Trichomes in Defense against Herbivores: Comparison of Herbivore Response to Woolly and Hairless Trichome Mutants in Tomato (Solanum lycopersicum). Planta 2012, 236, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Williams, W.P.; Davis, F.M. Registration of Mp704 Germplasm Line of Maize (Reg. No. GP116). Crop Sci. 1982, 22, 1269–1270. [Google Scholar] [CrossRef]
- Williams, W.P.; Davis, F.M.; Windham, G.L. Registration of Mp708 Germplasm Line of Maize. Crop Sci. 1990, 30, 757. [Google Scholar] [CrossRef]
- Williams, P.W.; Buckley, P.M.; Davis, F.M. Larval Growth and Behavior of the Fall Armyworm (Lepidoptera: Noctuidae) on Callus Initiated from Susceptible and Resistant Corn Hybrids. J. Econ. Entomol. 1985, 78, 951–954. [Google Scholar] [CrossRef]
- Erb, M.; Meldau, S.; Howe, G.A. Role of Phytohormones in Insect-Specific Plant Reactions. Trends Plant Sci. 2012, 17, 250–259. [Google Scholar] [CrossRef]
- Al-Zahrani, W.; Bafeel, S.O.; El-Zohri, M. Jasmonates Mediate Plant Defense Responses to Spodoptera exigua Herbivory in Tomato and Maize Foliage. Plant Signal Behav. 2020, 15, 1746898. [Google Scholar] [CrossRef]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of Jasmonate and Salicylate Signal Crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef]
- Chung, S.H.; Rosa, C.; Scully, E.D.; Peiffer, M.; Tooker, J.F.; Hoover, K.; Luthe, D.S.; Felton, G.W. Herbivore Exploits Orally Secreted Bacteria to Suppress Plant Defenses. Proc. Natl. Acad. Sci. USA 2013, 110, 15728–15733. [Google Scholar] [CrossRef]
- Harfouche, A.L.; Shivaji, R.; Stocker, R.; Williams, P.W.; Luthe, D.S. Ethylene Signaling Mediates a Maize Defense Response to Insect Herbivory. MPMI 2006, 19, 189–199. [Google Scholar] [CrossRef] [Green Version]
- Ankala, A.; Luthe, D.S.; Williams, W.P.; Wilkinson, J.R. Integration of Ethylene and Jasmonic Acid Signaling Pathways in the Expression of Maize Defense Protein Mir1-CP. Mol. Plant Microbe Interact. 2009, 22, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Louis, J.; Basu, S.; Varsani, S.; Castano-Duque, L.; Jiang, V.; Williams, W.P.; Felton, G.W.; Luthe, D.S. Ethylene Contributes to Maize Insect Resistance1-Mediated Maize Defense against the Phloem Sap-Sucking Corn Leaf Aphid. Plant Physiol. 2015, 169, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Pechan, T.; Ye, L.; Chang, Y.; Mitra, A.; Lin, L.; Davis, F.M.; Williams, W.P.; Luthe, D.S. A Unique 33-KD Cysteine Proteinase Accumulates in Response to Larval Feeding in Maize Genotypes Resistant to Fall Armyworm and Other Lepidoptera. Plant Cell 2000, 12, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Gill, T.A.; Sandoya, G.; Williams, P.; Luthe, D.S. Belowground Resistance to Western Corn Rootworm in Lepidopteran-Resistant Maize Genotypes. J. Econ. Entomol. 2011, 104, 299–307. [Google Scholar] [CrossRef]
- Castano-Duque, L.; Loades, K.W.; Tooker, J.F.; Brown, K.M.; Paul Williams, W.; Luthe, D.S. A Maize Inbred Exhibits Resistance Against Western Corn Rootworm, Diabrotica virgifera virgifera. J. Chem. Ecol. 2017, 43, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Varsani, S.; Basu, S.; Williams, W.P.; Felton, G.W.; Luthe, D.S.; Louis, J. Intraplant Communication in Maize Contributes to Defense against Insects. Plant Signal. Behav. 2016, 11, e1212800. [Google Scholar] [CrossRef]
- Varsani, S.; Grover, S.; Zhou, S.; Koch, K.G.; Huang, P.-C.; Kolomiets, M.V.; Williams, W.P.; Heng-Moss, T.; Sarath, G.; Luthe, D.S.; et al. 12-Oxo-Phytodienoic Acid Acts as a Regulator of Maize Defense against Corn Leaf Aphid. Plant Physiol. 2019, 179, 1402–1415. [Google Scholar] [CrossRef]
- Pingault, L.; Basu, S.; Zogli, P.; Williams, W.P.; Palmer, N.; Sarath, G.; Louis, J. Aboveground Herbivory Influences Belowground Defense Responses in Maize. Front. Ecol. Evol. 2021, 9, 804. [Google Scholar] [CrossRef]
- Mohan, S.; Ma, P.W.K.; Williams, W.P.; Luthe, D.S. A Naturally Occurring Plant Cysteine Protease Possesses Remarkable Toxicity against Insect Pests and Synergizes Bacillus thuringiensis Toxin. PLoS ONE 2008, 3, e1786. [Google Scholar] [CrossRef]
- Paddock, K.J.; Robert, C.A.M.; Erb, M.; Hibbard, B.E. Western Corn Rootworm, Plant and Microbe Interactions: A Review and Prospects for New Management Tools. Insects 2021, 12, 171. [Google Scholar] [CrossRef]
- Castano-Duque, L.; Luthe, D.S. Protein Networks Reveal Organ-Specific Defense Strategies in Maize in Response to an Aboveground Herbivore. Arthropod-Plant Interact. 2018, 12, 147–175. [Google Scholar] [CrossRef]
- Pechan, T.; Cohen, A.; Williams, W.P.; Luthe, D.S. Insect Feeding Mobilizes a Unique Plant Defense Protease That Disrupts the Peritrophic Matrix of Caterpillars. Proc. Natl. Acad. Sci. USA 2002, 99, 13319–13323. [Google Scholar] [CrossRef]
- Nguyen, D.; Rieu, I.; Mariani, C.; van Dam, N.M. How Plants Handle Multiple Stresses: Hormonal Interactions Underlying Responses to Abiotic Stress and Insect Herbivory. Plant Mol. Biol. 2016, 91, 727–740. [Google Scholar] [CrossRef]
- Wang, K.L.-C.; Li, H.; Ecker, J.R. Ethylene Biosynthesis and Signaling Networks. Plant Cell 2002, 14, s131–s151. [Google Scholar] [CrossRef]
- Li, W.; Ma, M.; Feng, Y.; Li, H.; Wang, Y.; Ma, Y.; Li, M.; An, F.; Guo, H. EIN2-Directed Translational Regulation of Ethylene Signaling in Arabidopsis. Cell 2015, 163, 670–683. [Google Scholar] [CrossRef]
- Ogunola, O.F.; Hawkins, L.K.; Mylroie, E.; Kolomiets, M.V.; Borrego, E.; Tang, J.D.; Williams, W.P.; Warburton, M.L. Characterization of the Maize Lipoxygenase Gene Family in Relation to Aflatoxin Accumulation Resistance. PLoS ONE 2017, 12, e0181265. [Google Scholar] [CrossRef]
- Woldemariam, M.G.; Ahern, K.; Jander, G.; Tzin, V. A Role for 9-Lipoxygenases in Maize Defense against Insect Herbivory. Plant Signal Behav. 2018, 13, e1422462. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.J. Enzymes Involved in Jasmonic Acid Biosynthesis. Physiol. Plant 1997, 100, 653–663. [Google Scholar] [CrossRef]
- Stenzel, I.; Hause, B.; Maucher, H.; Pitzschke, A.; Miersch, O.; Ziegler, J.; Ryan, C.A.; Wasternack, C. Allene Oxide Cyclase Dependence of the Wound Response and Vascular Bundle-Specific Generation of Jasmonates in Tomato—Amplification in Wound Signalling. Plant J. 2003, 33, 577–589. [Google Scholar] [CrossRef]
- Staswick, P.E.; Tiryaki, I. The Oxylipin Signal Jasmonic Acid Is Activated by an Enzyme That Conjugates It to Isoleucine in Arabidopsis. Plant Cell 2004, 16, 2117–2127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.J.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 Reveals the Versatile Functions of Jasmonic Acid in Maize Development and Defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef] [PubMed]
- Shivaji, R.; Camas, A.; Ankala, A.; Engelberth, J.; Tumlinson, J.H.; Williams, W.P.; Wilkinson, J.R.; Luthe, D.S. Plants on Constant Alert: Elevated Levels of Jasmonic Acid and Jasmonate-Induced Transcripts in Caterpillar-Resistant Maize. J. Chem. Ecol. 2010, 36, 179–191. [Google Scholar] [CrossRef]
- Dempsey, D.A.; Vlot, A.C.; Wildermuth, M.C.; Klessig, D.F. Salicylic Acid Biosynthesis and Metabolism. Arab. Book 2011, 9, e0156. [Google Scholar] [CrossRef]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic Acid Biosynthesis in Plants. Front. Plant Sci. 2020, 11, 338. [Google Scholar] [CrossRef] [PubMed]
- Dutartre, L.; Hilliou, F.; Feyereisen, R. Phylogenomics of the Benzoxazinoid Biosynthetic Pathway of Poaceae: Gene Duplications and Origin of the Bx Cluster. BMC Evol. Biol. 2012, 12, 64. [Google Scholar] [CrossRef]
- Niemeyer, H.M. Hydroxamic Acids (4-Hydroxy-1,4-Benzoxazin-3-Ones), Defence Chemicals in the Gramineae. Phytochemistry 1988, 27, 3349–3358. [Google Scholar] [CrossRef]
- Sicker, D.; Frey, M.; Schulz, M.; Gierl, A. Role of Natural Benzoxazinones in the Survival Strategy of Plants. Int. Rev. Cytol. 2000, 198, 319–346. [Google Scholar] [CrossRef]
- Jonczyk, R.; Schmidt, H.; Osterrieder, A.; Fiesselmann, A.; Schullehner, K.; Haslbeck, M.; Sicker, D.; Hofmann, D.; Yalpani, N.; Simmons, C.; et al. Elucidation of the Final Reactions of DIMBOA-Glucoside Biosynthesis in Maize: Characterization of Bx6 and Bx7. Plant Physiol. 2008, 146, 1053–1063. [Google Scholar] [CrossRef]
- Woodhouse, M.R.; Cannon, E.K.; Portwood, J.L.; Harper, L.C.; Gardiner, J.M.; Schaeffer, M.L.; Andorf, C.M. A Pan-Genomic Approach to Genome Databases Using Maize as a Model System. BMC Plant Biol. 2021, 21, 385. [Google Scholar] [CrossRef]
- Erb, M.; Flors, V.; Karlen, D.; de Lange, E.; Planchamp, C.; D’Alessandro, M.; Turlings, T.C.J.; Ton, J. Signal Signature of Aboveground-Induced Resistance upon Belowground Herbivory in Maize. Plant J. 2009, 59, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, I.T.; Schmelz, E.A.; Ohnmeiss, T.E. Wound-Induced Changes in Root and Shoot Jasmonic Acid Pools Correlate with Induced Nicotine Synthesis In Nicotiana sylvestris Spegazzini and Comes. J. Chem. Ecol. 1994, 20, 2139–2157. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.M.; Marchi-Werle, L.; Hunt, T.E.; Heng-Moss, T.M.; Louis, J. Abscisic and Jasmonic Acids Contribute to Soybean Tolerance to the Soybean Aphid (Aphis glycines Matsumura). Sci. Rep. 2018, 8, 15148. [Google Scholar] [CrossRef]
- Liu, L.; Sonbol, F.-M.; Huot, B.; Gu, Y.; Withers, J.; Mwimba, M.; Yao, J.; He, S.Y.; Dong, X. Salicylic Acid Receptors Activate Jasmonic Acid Signalling through a Non-Canonical Pathway to Promote Effector-Triggered Immunity. Nat. Commun. 2016, 7, 13099. [Google Scholar] [CrossRef]
- Ye, W.; Bustos-Segura, C.; Degen, T.; Erb, M.; Turlings, T.C.J. Belowground and Aboveground Herbivory Differentially Affect the Transcriptome in Roots and Shoots of Maize. Plant Direct 2022, 6, e426. [Google Scholar] [CrossRef]
- Robert, C.A.M.; Veyrat, N.; Glauser, G.; Marti, G.; Doyen, G.R.; Villard, N.; Gaillard, M.D.P.; Köllner, T.G.; Giron, D.; Body, M.; et al. A Specialist Root Herbivore Exploits Defensive Metabolites to Locate Nutritious Tissues. Ecol. Lett. 2012, 15, 55–64. [Google Scholar] [CrossRef]
- Robert, C.A.; Zhang, X.; Machado, R.A.; Schirmer, S.; Lori, M.; Mateo, P.; Erb, M.; Gershenzon, J. Sequestration and Activation of Plant Toxins Protect the Western Corn Rootworm from Enemies at Multiple Trophic Levels. eLife 2017, 6, e29307. [Google Scholar] [CrossRef] [PubMed]
- Schnepf, H.E.; Lee, S.; Dojillo, J.; Burmeister, P.; Fencil, K.; Morera, L.; Nygaard, L.; Narva, K.E.; Wolt, J.D. Characterization of Cry34/Cry35 Binary Insecticidal Proteins from Diverse Bacillus thuringiensis Strain Collections. Appl. Environ. Microbiol. 2005, 71, 1765–1774. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.T.; Stockhoff, B.A.; Stamp, L.; Schnepf, H.E.; Schwab, G.E.; Knuth, M.; Russell, J.; Cardineau, G.A.; Narva, K.E. Novel Bacillus Thuringiensis Binary Insecticidal Crystal Proteins Active on Western Corn Rootworm, Diabrotica virgifera virgifera LeConte. Appl. Environ. Microbiol. 2002, 68, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Eyun, S.-I.; Arora, K.; Tan, S.Y.; Gandra, P.; Moriyama, E.; Khajuria, C.; Jurzenski, J.; Li, H.; Donahue, M.; et al. Patterns of Gene Expression in Western Corn Rootworm (Diabrotica virgifera virgifera) Neonates, Challenged with Cry34Ab1, Cry35Ab1 and Cry34/35Ab1, Based on Next-Generation Sequencing. Toxins 2017, 9, 124. [Google Scholar] [CrossRef] [PubMed]
- Sparks, M.E.; Blackburn, M.B.; Kuhar, D.; Gundersen-Rindal, D.E. Transcriptome of the Lymantria dispar (Gypsy Moth) Larval Midgut in Response to Infection by Bacillus thuringiensis. PLoS ONE 2013, 8, e61190. [Google Scholar] [CrossRef] [Green Version]
- Lei, Y.; Zhu, X.; Xie, W.; Wu, Q.; Wang, S.; Guo, Z.; Xu, B.; Li, X.; Zhou, X.; Zhang, Y. Midgut Transcriptome Response to a Cry Toxin in the Diamondback Moth, Plutella xylostella (Lepidoptera: Plutellidae). Gene 2014, 533, 180–187. [Google Scholar] [CrossRef]
- Ritchie, J.T.; Singh, U.; Godwin, D.C.; Bowen, W.T. Cereal Growth, Development and Yield. In Understanding Options for Agricultural Production; Systems Approaches for Sustainable Agricultural, Development; Tsuji, G.Y., Hoogenboom, G., Thornton, P.K., Eds.; Springer: Dordrecht, The Netherlands, 1998; pp. 79–98. ISBN 978-94-017-3624-4. [Google Scholar]
- Ankala, A.; Kelley, R.Y.; Rowe, D.E.; Williams, W.P.; Luthe, D.S. Foliar Herbivory Triggers Local and Long Distance Defense Responses in Maize. Plant Sci. 2013, 199, 103–112. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 30 May 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate Alignment of Transcriptomes in the Presence of Insertions, Deletions and Gene Fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Pingault, L.; Varsani, S.; Palmer, N.; Ray, S.; Williams, W.P.; Luthe, D.S.; Ali, J.G.; Sarath, G.; Louis, J. Transcriptomic and Volatile Signatures Associated with Maize Defense against Corn Leaf Aphid. BMC Plant Biol. 2021, 21, 138. [Google Scholar] [CrossRef]
- Lin, W.-D.; Chen, Y.-C.; Ho, J.; Hsiao, C. GOBU: Toward an Integration Interface for Biological Objects. J. Inf. Sci. Eng. 2006, 22, 19–29. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pingault, L.; Basu, S.; Vellichirammal, N.N.; Williams, W.P.; Sarath, G.; Louis, J. Co-Transcriptomic Analysis of the Maize–Western Corn Rootworm Interaction. Plants 2022, 11, 2335. https://doi.org/10.3390/plants11182335
Pingault L, Basu S, Vellichirammal NN, Williams WP, Sarath G, Louis J. Co-Transcriptomic Analysis of the Maize–Western Corn Rootworm Interaction. Plants. 2022; 11(18):2335. https://doi.org/10.3390/plants11182335
Chicago/Turabian StylePingault, Lise, Saumik Basu, Neetha N. Vellichirammal, William Paul Williams, Gautam Sarath, and Joe Louis. 2022. "Co-Transcriptomic Analysis of the Maize–Western Corn Rootworm Interaction" Plants 11, no. 18: 2335. https://doi.org/10.3390/plants11182335





