Proanthocyanidins Alleviate Cadmium Stress in Industrial Hemp (Cannabis sativa L.)
Abstract
:1. Introduction
2. Results and Analysis
2.1. Analysis of Physiological Indicators in Hemp Grown in Different Experimental Treatments
2.1.1. Effects of Different Treatment Conditions on the Growth of Hemp
2.1.2. Effects of Different Treatments on the Antioxidant Content of Hemp
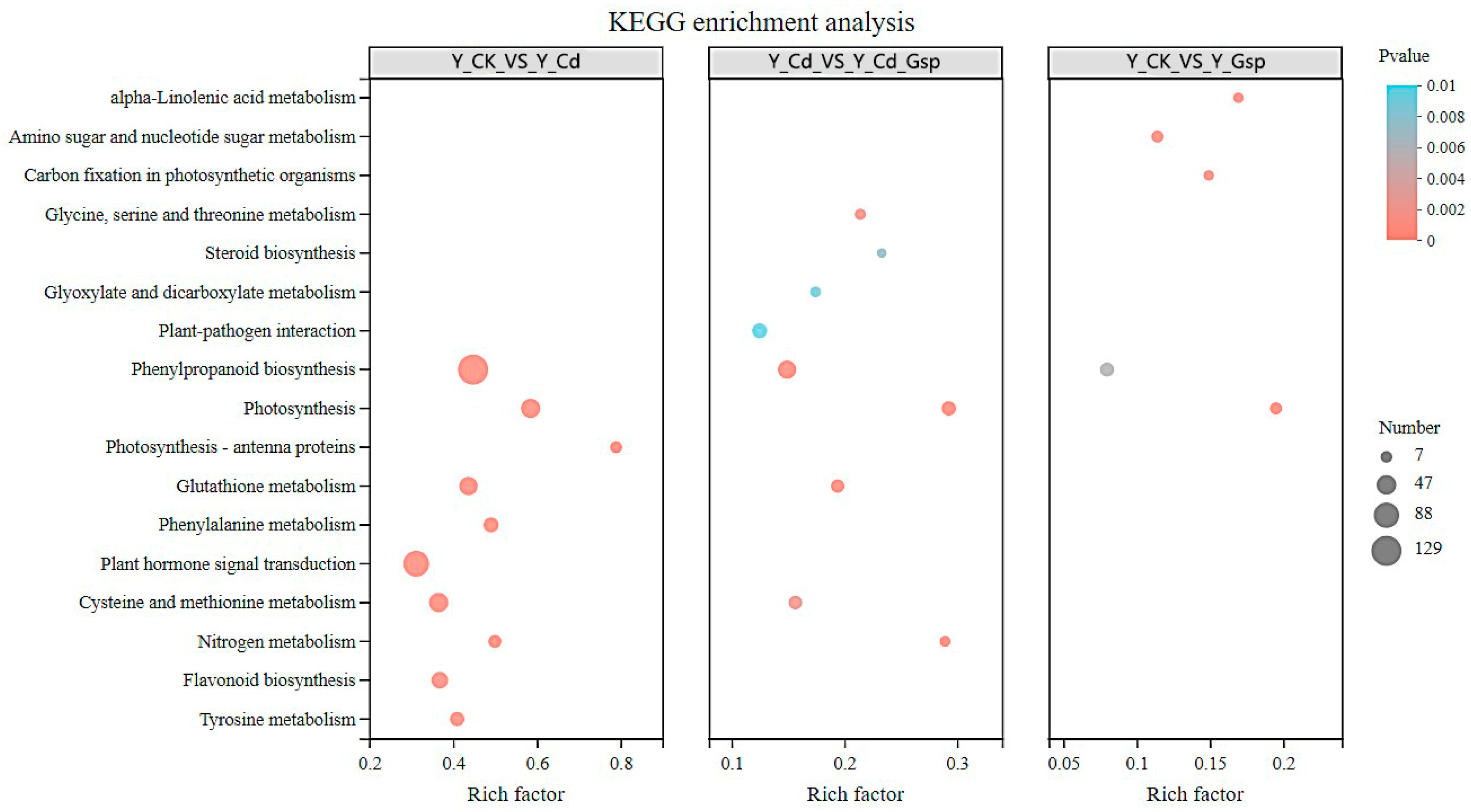
2.2. Multi-Omics Analysis of Hemp Grown in Different Treatment Settings
2.2.1. Transcriptome Results Analysis
2.2.2. Metabolome Results Analysis
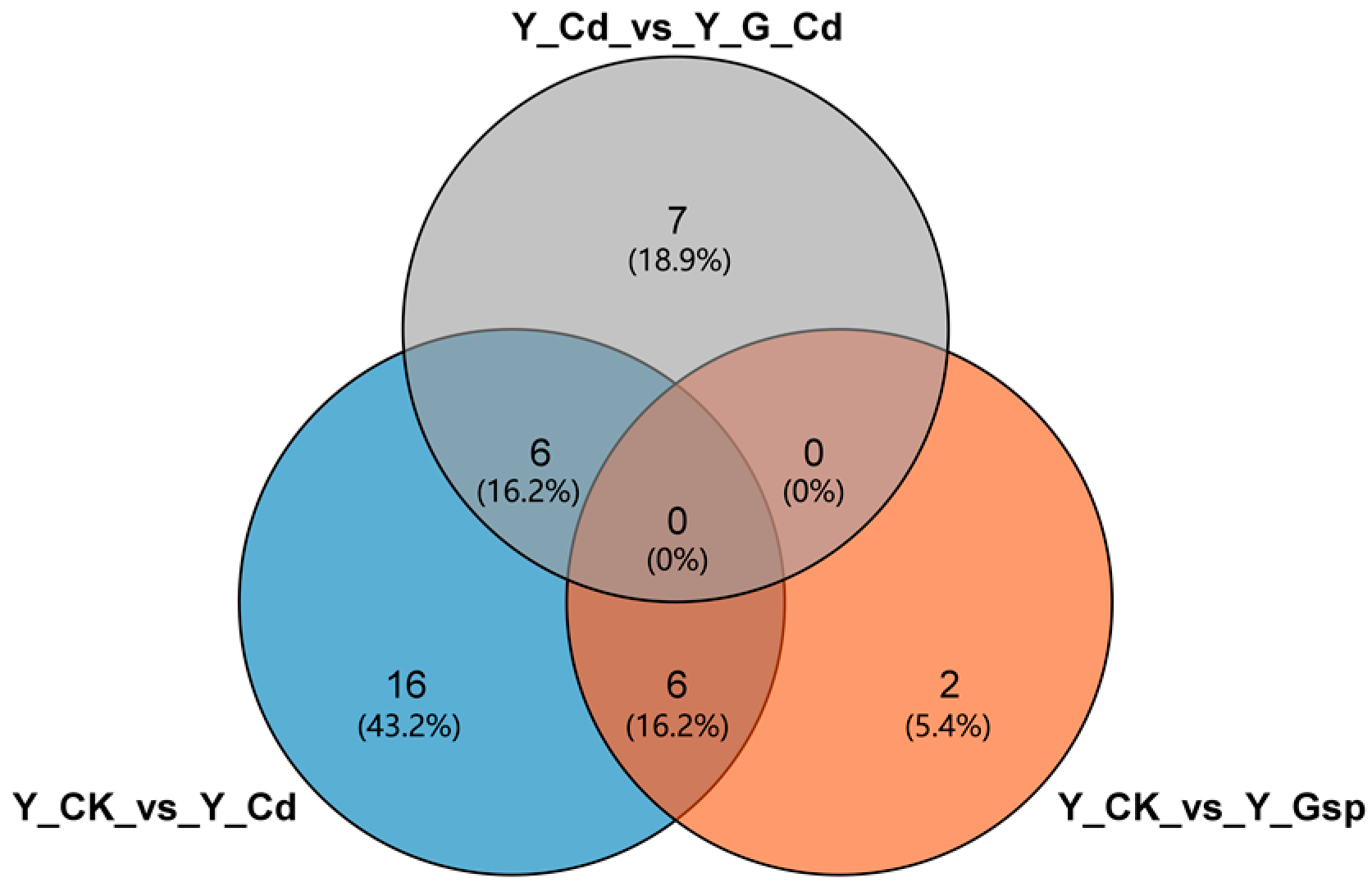
2.2.3. Combined Analysis of Transcriptome and Metabolome Results
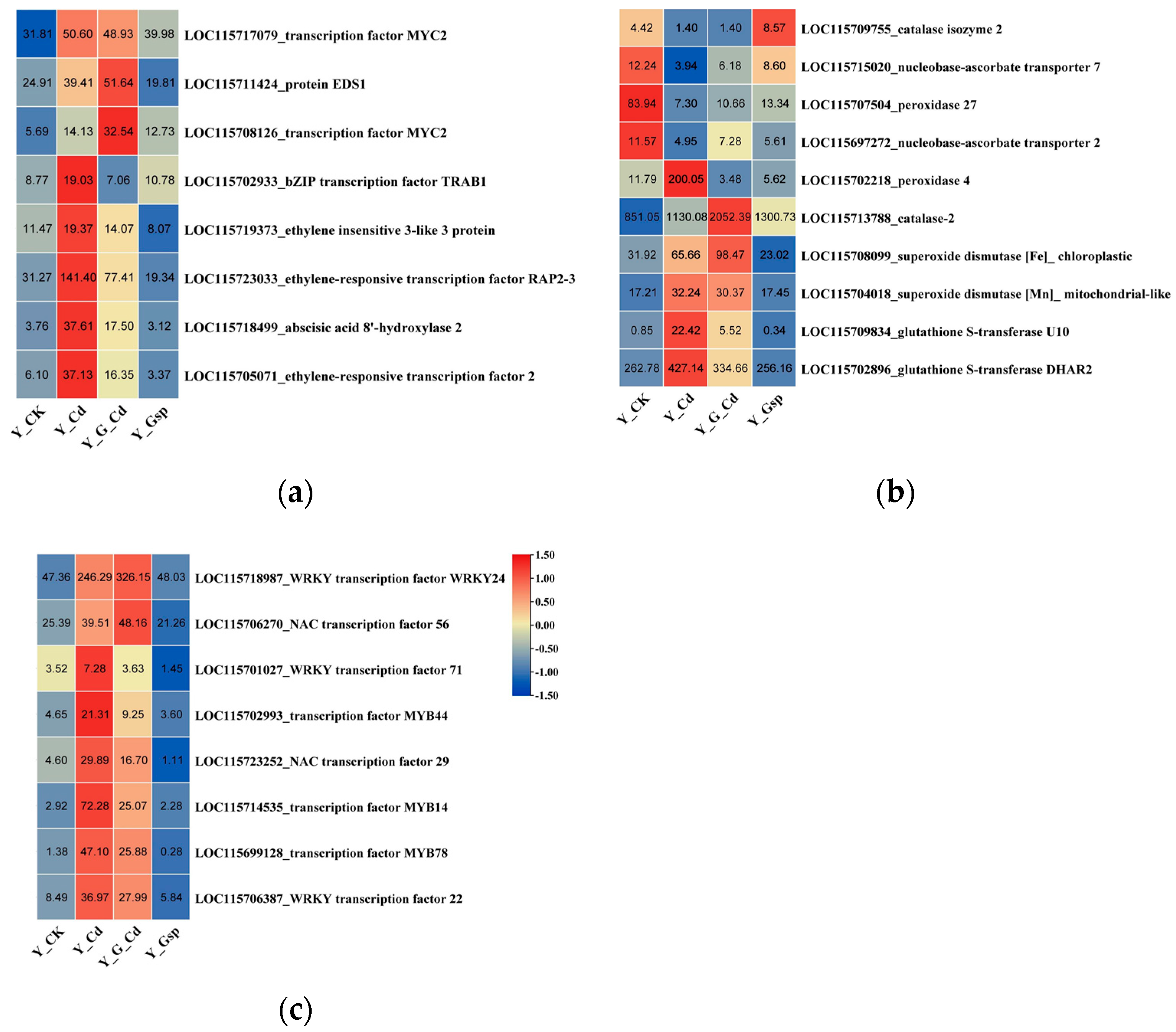
2.2.4. Validation of Differentially Expressed Genes by qRT-PCR
2.3. Cloning and Functional Analysis of Key Genes for Procyanidin Synthesis of Hemp
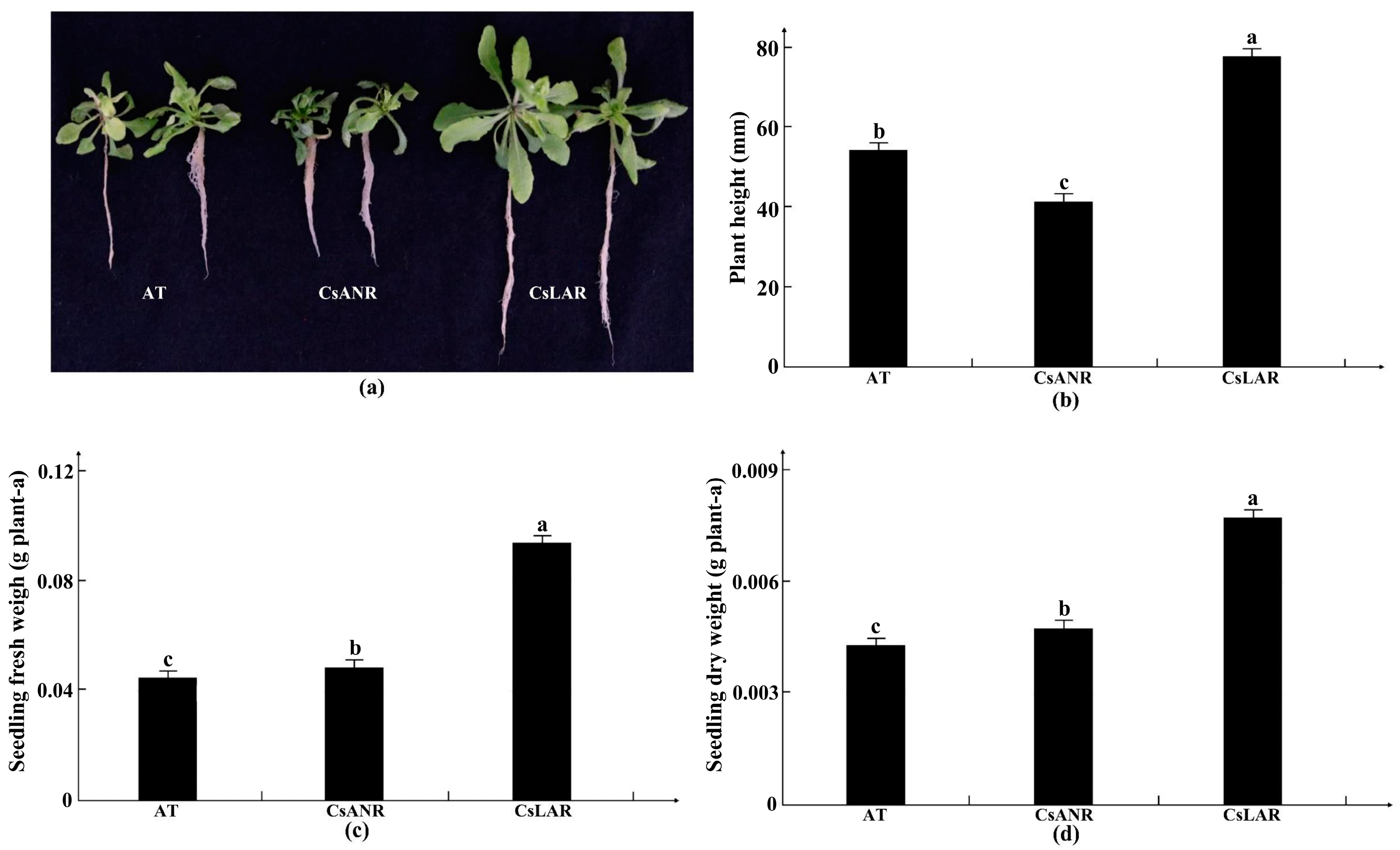
2.3.1. Construction of ANR and LAR Overexpression Lines of Hemp
2.3.2. Verification of Stress Tolerance in Transgenic Plants
2.3.3. Determination of Secondary Metabolites in Transgenic Plants
3. Discussion
3.1. Effects of Different Treatments on the Physiological Indicators of Industrial Hemp
3.2. Changes in Gene Expression and Metabolite Content of Hemp under Different Treatments
3.3. Construction and Functional Analysis of ANR and LAR Overexpression in Plants
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Stress Treatment and Determination of Antioxidant Substances
4.3. Transcriptome Analysis
4.4. Metabolome Analysis
4.5. The qRT-PCR Analysis
4.6. Vector Construction and Genetic Transformation
4.7. Measurement of Tolerance in Transgenic Plants
4.8. Measurement of Phytohormones in Transgenic Plants
4.9. Measurement of Anthocyanins in Transgenic Plants
4.10. Statistical Analysis and Presentation of Totals
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviations | Abbreviations Description |
| EDS1 | Protein enhanced disease susceptibility-1 |
| ANR | Anthocyanidin reductase |
| LAR | Leucoanthocyanidin reductase |
| AT | AT wild-type Arabidopsis |
| Cd2+ | Cadmium ions |
| ±SD | ± Standard deviation |
| GSH | Glutathione, r-glutamyl cysteingl, +glycine |
| SOD | Superoxide dismutase |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ABA | Abscisic acid |
| ETH | Ethylene |
| SA | Salicylic acid |
| JA | Jasmonic acid |
| POD | Pe-roxidase |
| ASA | Ascorbic acid |
| CAT | Catalase |
| OPLS-DA | Orthogonal partial least squares-discriminant analysis |
| FAD | Fatty acid desaturase |
| CBP | CREB-binding protein |
| CBT | Cannabitriol |
| LLP | Leptin-like peptide |
| PsbO | Photosystem II-oxygen-evolving enhancer protein 1 precursor |
| PsbP | Thylakoid lumenal subunit of photosystem II |
| FNR | Transcriptional activator |
| ATPC | ATP synthase |
| bZIP | Basic leucine zipper |
| NECD | N2 extracellular domain |
| PLA | Plasminogen activator |
| QRT-PCR | Real-Time Quantitative Reverse Transcription PCR |
| UPLC | High-performance liquid chromatography |
| YM1 | Yunnan hemp no. 1 |
References
- Bonini, S.A.; Premoli, M.; Tambaro, S.; Kumar, A.; Maccarinelli, G.; Memo, M.; Mastinu, A. Cannabis sativa: A comprehensive ethnopharmacological review of a medicinal plant with a long history. J. Ethnopharmacol. 2018, 227, 300–315. [Google Scholar] [CrossRef]
- Farinon, B.; Molinari, R.; Costantini, L.; Merendino, N. The seed of industrial hemp (Cannabis sativa L.): Nutritional Quality and Potential Functionality for Human Health and Nutrition. Nutrients 2020, 12, 1935. [Google Scholar] [CrossRef]
- Husain, R.; Weeden, H.; Bogush, D.; Deguchi, M.; Soliman, M.; Potlakayala, S.; Katam, R.; Goldman, S.; Rudrabhatla, S. Enhanced tolerance of industrial hemp (Cannabis sativa L.) plants on abandoned mine land soil leads to overexpression of cannabinoids. PLoS ONE 2019, 14, e0221570. [Google Scholar] [CrossRef]
- Xu, X.; Liu, C.; Zhao, X.; Li, R.; Deng, W. Involvement of an Antioxidant Defense System in the Adaptive Response to Cadmium in Maize Seedlings (Zea mays L.). Bull. Environ. Contam. Toxicol. 2014, 93, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Z.; Xu, S.; Zhang, L.; Hou, W.; Yu, N. Effect of Combined Pollution of Cd and B [a] P on Pho tosynthesis and Chlorophyll Fluorescence Characteristics of Wheat. Pol. J Env. Stud. 2015. [Google Scholar] [CrossRef]
- Lysenko, E.A.; Klaus, A.A.; Pshybytko, N.L.; Kusnetsov, V.V. Cadmium accumulation in chloroplasts and its impact on chloroplastic processes in barley and maize. Photosynth. Res. 2014, 125, 291–303. [Google Scholar] [CrossRef]
- Haider, F.U.; Liqun, C.; Coulter, J.A.; Cheema, S.A.; Wu, J.; Zhang, R.; Wenjun, M.; Farooq, M. Cadmium tox icity in plants: Impacts and remediation strategies. Ecotoxicol. Environ. Saf. 2021, 211, 111887. [Google Scholar] [CrossRef] [PubMed]
- Gallego, S.M.; Pena, L.B.; Barcia, R.A.; Azpilicueta, C.E.; Iannone, M.F.; Rosales, E.P.; Benavides, M.P. Unravelling cadmium toxicity and tolerance in plants: Insight into regulatory mechanisms. Environ. Exp. Bot. 2012, 83, 33–46. [Google Scholar] [CrossRef]
- Choppala, G.; Saifullah Bolan, N.; Bibi, S.; Iqbal, M.; Rengel, Z.; Kunhikrishnan, A.; Ashwath, N.; Ok, Y.S. Cellular Mechanisms in Higher Plants Governing Tolerance to Cadmium Toxicity. Crit. Rev. Plant Sci. 2014, 33, 374–391. [Google Scholar] [CrossRef]
- Pishchik, V.; Vorobyev, N.; Chernyaeva, I.; Timofeeva, S.; Kozhemyakov, A.; Alexeev, Y.; Lukin, S. Experimental and mathematical simulation of plant growth promoting rhizobacteria and plant interaction under cadmium stress. Plant Soil 2002, 243, 173–186. [Google Scholar] [CrossRef]
- Hussain, M.K.; Aziz, A.; Ditta, H.M.; Azhar, M.F.; El-Shehawi, A.M.; Hussain, S.; Mehboob, N.; Hussain, M.; Farooq, S. Foliar application of seed water extract of Nigella sativa im proved maize growth in cadmium-contaminated soil. PLoS ONE 2021, 16, e0254602. [Google Scholar] [CrossRef] [PubMed]
- Hassan, W.; Bashir, S.; Ali, F.; Ijaz, M.; Hussain, M.; David, J. Role of ACC-deaminase and/or nitrogen fixing rhizobacteria in growth promotion of wheat (Triticum aestivum L.) under cadmium pollution. Environ. Earth Sci. 2016, 75, 267. [Google Scholar] [CrossRef]
- Rauf, A.; Imran, M.; Abu-Izneid, T.; Patel, S.; Pan, X.; Naz, S.; Silva, A.S.; Saeed, F.; Suleria, H.A. Proanthocyanidins: A comprehensive review. Biomed. Pharmacother. 2019, 116, 108999. [Google Scholar] [CrossRef]
- Hosoda, K.; Sasahara, H.; Matsushita, K.; Tamura, Y.; Miyaji, M.; Matsuyama, H. Anthocyanin and proanthocyanidin contents, antioxidant activity, and in situ degradability of black and red rice grains. Asian-Australas. J. Anim. Sci. 2018, 31, 1213–1220. [Google Scholar] [CrossRef]
- Reuter, J.A.; Spacek, D.V.; Snyder, M.P. High-throughput sequencing technologies. Mol. Cell. 2015, 58, 586–597. [Google Scholar] [CrossRef]
- Qiang-long, Z.; Shi, L.; Peng, G.; Fei-shi, L. High-throughput Sequencing Technology and Its Application. J. Northeast Agric. Univ. 2014, 21, 84–96. [Google Scholar] [CrossRef]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics Technologies. PLoS Comput. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed]
- Buti, M.; Baldoni, E.; Formentin, E.; Milc, J.; Frugis, G.; Lo Schiavo, F.; Genga, A.; Francia, E. A Meta-Analysis of Comparative Transcriptomic Data Re veals a Set of Key Genes Involved in the Tolerance to Abiotic Stresses in Rice. Int. J. Mol. Sci. 2019, 20, 5662. [Google Scholar] [CrossRef] [PubMed]
- Nadella, K.D.; Marla, S.S.; Kumar, P.A. Metabolomics in agriculture. Omics A J. Integr. Biol. 2012, 16, 149. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechan isms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [Green Version]
- Hincha, D.K.; Espinoza, C.; Zuther, E. Transcriptomic and Metabolomic Approaches to the Analysis of Plant Freezing Tolerance and Cold Acclimation. Improv. Crop Resist. Abiotic 2012, 255–287. [Google Scholar] [CrossRef]
- Hirai, M.Y.; Yano, M.; Goodenowe, D.B.; Kanaya, S.; Kimura, T.; Awazuhara, M.; Arita, M.; Fujiwara, T.; Saito, K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 10205–10210. [Google Scholar] [CrossRef] [PubMed]
- Bogs, J.; Downey, M.O.; Harvey, J.S.; Ashton, A.R.; Tanner, G.J.; Robinson, S.P. Proanthocyanidin Synthesis and Expression of Genes Encoding Leucoanthocyanidin Reductase and Anthocyanidin Reductase in Developing Grape Berries and Grapevine Leaves. Plant Physiol. 2005, 139, 652–663. [Google Scholar] [CrossRef]
- Li, L.Z.; Tu, C.; Wu, L.H.; Peijnenburg, W.J.; Ebbs, S.; Luo, Y.M. Pathways of root uptake and membrane transport of Cd2+ in the zinc/cadmium hyperaccumulating plant Sedum plumbizincicola. Environ Toxicol Chem. 2017, 36, 1038–1046. [Google Scholar] [CrossRef] [PubMed]
- Ravaglia, D.; Espley, R.V.; Henry-Kirk, R.A.; Andreotti, C.; Ziosi, V.; Hellens, R.P.; Costa, G.; Allan, A.C. Tran scriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 2013, 13, 68. [Google Scholar] [CrossRef]
- Qin, Y.; Yu, H.; Cheng, S.; Liu, Z.; Yu, C.; Zhang, X.; Su, X.; Huang, J.; Shi, S.; Zou, Y.; et al. Ge nome-Wide Analysis of the WRKY Gene Family in Malus domestica and the Role of MdWRKY70L in Response to Drought and Salt Stresses. Genes 2022, 13, 1068. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, J.; Chen, H.; Jin, W.; Liang, Z. Characterization of NAC family genes in Salvia miltiorrhiza and NAC2 potentially involved in the biosynthesis of tanshinones. Phytochemistry 2021, 191, 112932. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, H.; Su, Z.; Khoo, C.; Gu, L. Identifying Cranberry Juice Consumers with Predictive OPLS-DA Models of Plasma Metabolome and Validation of Cranberry Juice Intake Biomarkers in a Double-Blinded, Randomized, Placebo-Controlled, Cross-Over Study. Mol. Nutr. Food Res. 2020, 64, 1901242. [Google Scholar] [CrossRef]
- Powers, R.; Worley, B. Multivariate Analysis in Metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Zhu, H.; Ai, H.; Cao, L.; Sui, R.; Ye, H.; Du, D.; Sun, J.; Yao, J.; Chen, K.; Chen, L. Transcriptome analysis providing novel insights for Cd-resistant tall fescue responses to Cd stress. Ecotoxicol. Environ. Saf. 2018, 160, 349–356. [Google Scholar] [CrossRef]
- Cui, H.; Qiu, J.; Zhou, Y.; Bhandari, D.D.; Zhao, C.; Bautor, J.; Parker, J.E. Antagonism of Transcription Factor MYC2 by EDS1/PAD4 Complexes Bolsters Salicylic Acid Defense in Arabidopsis Effector-Triggered Immunity. Mol. Plant 2018, 11, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Gobbato, E.; Kracher, B.; Qiu, J.; Bautor, J.; Parker, J.E. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 2017, 213, 1802–1817. [Google Scholar] [CrossRef] [PubMed]
- Janda, T.; Gondor, O.K.; Yordanova, R.; Szalai, G.; Pál, M. Salicylic acid and photosynthesis: Signalling and effects. Acta Physiol. Plant. 2014, 36, 2537–2546. [Google Scholar] [CrossRef]
- Yang, C.Q.; Fang, X.; Wu, X.M.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Transcriptional Regulation of Plant Secondary Metabolism. J. Integr. Plant Biol. 2012, 54, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Marques, D.N.; Carvalho, M.E.A.; Piotto, F.A.; Batagin-Piotto, K.D.; Nogueira, M.L.; Gaziola, S.A.; Azevedo, R.A. Antioxidant Defense Response in Plants to Cadmium Stress. In Cadmium Tolerance in Plants; Elsevier: Amsterdam, The Netherlands, 2019; pp. 423–461. [Google Scholar]
- Ozfidan-Konakci, C.; Yildiztugay, E.; Bahtiyar, M.; Kucukoduk, M. The humic acid-induced changes in the water status, chlorophyll fluorescence and antioxidant defense systems of wheat leaves with cadmium stress. Ecotoxicol. Environ. Saf. 2018, 155, 66–75. [Google Scholar] [CrossRef]
- Ahmad, P. Oxidative Damage to Plants: Antioxidant Networks and Signaling; Academic Press: Amsterdam, The Netherlands, 2014; ISBN 10: 0127999639. [Google Scholar]
- Zhen, S.; Shuai, H.; Xu, C.; Lv, G.; Zhu, X.; Zhang, Q.; Zhu, Q.; Núñez-Delgado, A.; Conde-Cid, M.; Zhou, Y.; et al. Foliar application of Zn reduces Cd accumulation in grains of late rice by regulating the antioxidant system, enhancing Cd chelation onto cell wall of leaves, and inhibiting Cd translocation in rice. Sci. Total Environ. 2021, 770, 145302. [Google Scholar] [CrossRef]
- Zhao, H.; Guan, J.; Liang, Q.; Zhang, X.; Hu, H.; Zhang, J. Effects of cadmium stress on growth and physiological characteristics of sassafras seedlings. Sci. Rep. 2021, 11, 9913. [Google Scholar] [CrossRef]
- Zhu, G.; Xiao, H.; Guo, Q.; Zhang, Z.; Zhao, J.; Yang, D. Effects of cadmium stress on growth and amino acid metabolism in two Compositae plants. Ecotoxicol. Environ. Saf. 2018, 158, 300–308. [Google Scholar] [CrossRef]
- Larkin, R.M. Tetrapyrrole Signaling in Plants. Front. Plant Sci. 2016, 7, 1586. [Google Scholar] [CrossRef]
- Solymosi, K.; Mysliwa-Kurdziel, B. Chlorophylls and their Derivatives Used in Food Industry and Medicine. Mini Rev. Med. Chem. 2017, 17, 1194–1222. [Google Scholar] [CrossRef]
- Ullah, C.; Tsai, C.J.; Unsicker, S.B.; Xue, L.; Reichelt, M.; Gershenzon, J.; Hammerbacher, A. Salicylic acid ac tivates poplar defense against the biotrophic rust fungus Melampsora larici-populina via increased biosynthesis of catechin and proanthocyanidins. New Phytol. 2019, 221, 960–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, D.; Huang, T.; Tian, B.; Zhan, J. Advances in Biosynthesis and Biological Functions of Proanthocya nidins in Horticultural Plants. Foods 2020, 9, 1774. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tian, J.; Yao, Y.Y.; Zhang, J.; Song, T.T.; Li, K.T.; Yao, Y.C. Identification of leucoanthocyanidin reduc tase and anthocyanidin reductase genes involved in proanthocyanidin biosynthesis in Malus crabapple plants. Plant Physiol. Biochem. 2019, 139, 141–151. [Google Scholar] [CrossRef]
- Luo, P.; Shen, Y.; Jin, S.; Huang, S.; Cheng, X.; Wang, Z.; Li, P.; Zhao, J.; Bao, M.; Ning, G. Overexpression of Rosa rugosa anthocyanidin reductase enhances tobacco tolerance to abiotic stress through increased ROS scavenging and modulation of ABA signaling. Plant Sci. 2016, 245, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.T.; Liu, J.; Zhou, S.M.; Xu, G.; Gao, Y.Q.; Liu, W.Y. UHPLC-QTOFMS-Based Metabolomic Analysis of the Hippocampus in Hypoxia Preconditioned Mouse. Front Physiol. 2019, 10, 1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, M.; Pan, L.; Liu, J.; Yang, X.; Tang, H.; Zhou, Y.; Huang, S.; Pan, G. Proanthocyanidins Alleviate Cadmium Stress in Industrial Hemp (Cannabis sativa L.). Plants 2022, 11, 2364. https://doi.org/10.3390/plants11182364
Yin M, Pan L, Liu J, Yang X, Tang H, Zhou Y, Huang S, Pan G. Proanthocyanidins Alleviate Cadmium Stress in Industrial Hemp (Cannabis sativa L.). Plants. 2022; 11(18):2364. https://doi.org/10.3390/plants11182364
Chicago/Turabian StyleYin, Ming, Langlang Pan, Junfei Liu, Xiaojuan Yang, Huijuan Tang, Yuxin Zhou, Siqi Huang, and Gen Pan. 2022. "Proanthocyanidins Alleviate Cadmium Stress in Industrial Hemp (Cannabis sativa L.)" Plants 11, no. 18: 2364. https://doi.org/10.3390/plants11182364
APA StyleYin, M., Pan, L., Liu, J., Yang, X., Tang, H., Zhou, Y., Huang, S., & Pan, G. (2022). Proanthocyanidins Alleviate Cadmium Stress in Industrial Hemp (Cannabis sativa L.). Plants, 11(18), 2364. https://doi.org/10.3390/plants11182364




