Biological Assay-Guided Fractionation and Mass Spectrometry-Based Metabolite Profiling of Annona muricata L. Cytotoxic Compounds against Lung Cancer A549 Cell Line
Abstract
:1. Introduction
2. Results
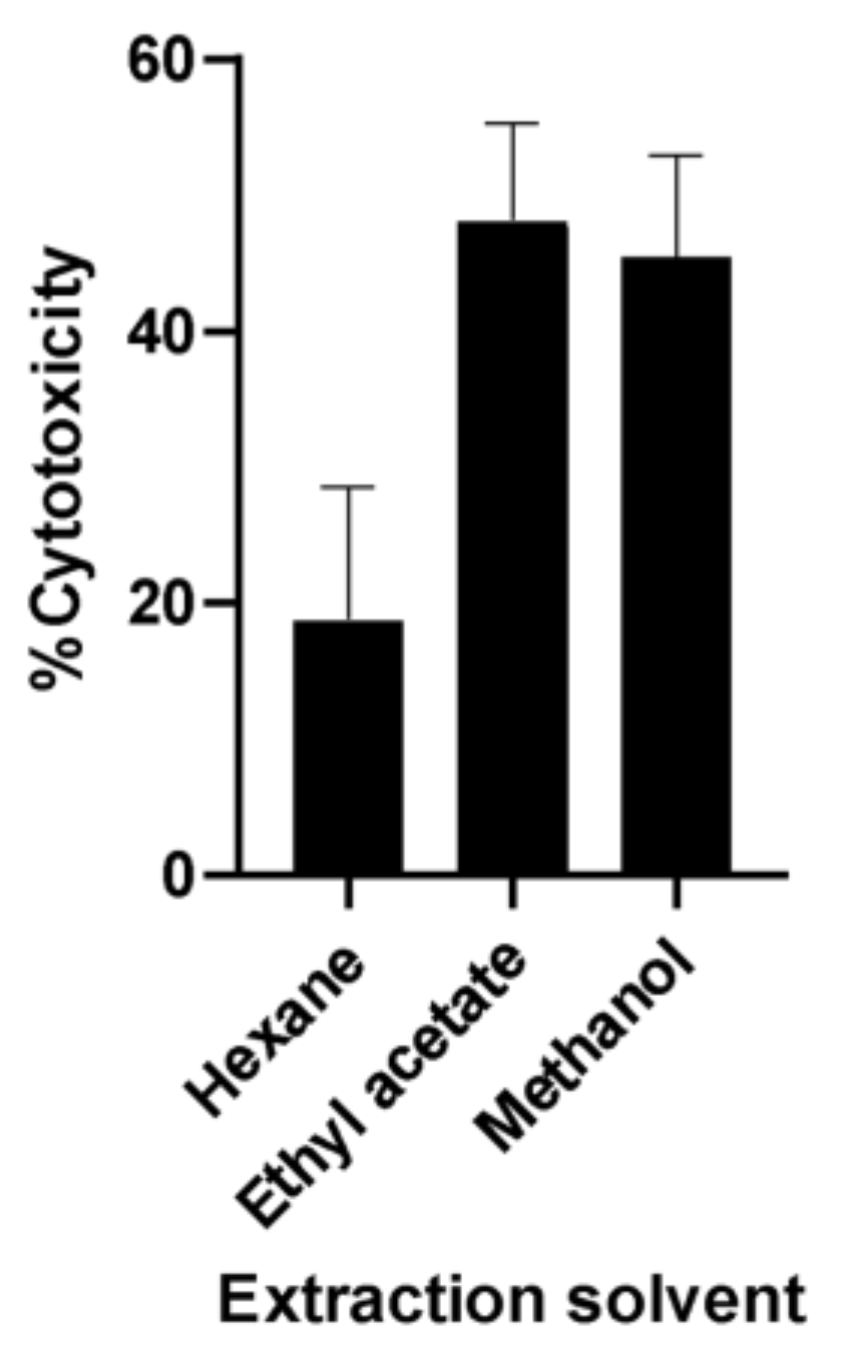
2.1. Bioassay-Guided Fractionation of A. muricata Ethyl Acetate Leaf Extract
2.2. Metabolomics Profiling of A. muricata Ethyl Acetate Leaf Extract
2.3. Molecular Networking of Identified Compounds in A. muricata Ethyl Acetate Leaf Extract
3. Discussion
4. Materials and Methods
4.1. Sample Collection, Preparation, and Extraction
4.2. Bioassay-Guided Fractionation
4.3. Cell Culture and Bioassay Screening
4.4. LC-MS/MS Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Oser, M.G.; Niederst, M.J.; Sequist, L.V.; Engelman, J.A. Transformation from Non-Small-Cell Lung Cancer to Small-Cell Lung Cancer: Molecular Drivers and Cells of Origin. Lancet Oncol. 2015, 16, e165–e172. [Google Scholar] [CrossRef]
- Lemjabbar, H.; Basbaum, C. Platelet-Activating Factor Receptor and ADAM10 Mediate Responses to Staphylococcus Aureus in Epithelial Cells. Nat. Med. 2002, 8, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Chen, X. Drug Resistance and Combating Drug Resistance in Cancer. Cancer Drug Resist. 2019, 2, 141–160. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.A.; Perez de Souza, L.; Serag, A.; Fernie, A.R.; Farag, M.A.; Ezzat, S.M.; Alseekh, S. Metabolomics in the Context of Plant Natural Products Research: From Sample Preparation to Metabolite Analysis. Metabolites 2020, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Molyneux, R.J.; Colegate, S.M. Bioactive Natural Products: Detection, Isolation, and Structural Determination, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Coria-Téllez, A.V.; Montalvo-Gónzalez, E.; Yahia, E.M.; Obledo-Vázquez, E.N. Annona muricata: A Comprehensive Review on Its Traditional Medicinal Uses, Phytochemicals, Pharmacological Activities, Mechanisms of Action and Toxicity. Arab. J. Chem. 2018, 11, 662–691. [Google Scholar] [CrossRef]
- Wahab, S.M.A.; Jantan, I.; Haque, A.; Arshad, L. Exploring the Leaves of Annona muricata L. as a Source of Potential Anti-Inflammatory and Anticancer Agents. Front. Pharmacol. 2018, 9, 661. [Google Scholar] [CrossRef]
- Prasad, S.K.; Varsha, V.; Devananda, D. Anti-Cancer Properties of Annona muricata (L.): A Review. Med. Plants Int. J. Phytomed. Relat. Ind. 2019, 11, 123. [Google Scholar] [CrossRef]
- Zhu, T.; Bao, X.; Chen, M.; Lin, R.; Zhuyan, J.; Zhen, T.; Xing, K.; Zhou, W.; Zhu, S. Mechanisms and Future of Non-Small Cell Lung Cancer Metastasis. Front. Oncol. 2020, 10, 585284. [Google Scholar] [CrossRef]
- Perez de Souza, L.; Alseekh, S.; Naake, T.; Fernie, A. Mass Spectrometry-Based Untargeted Plant Metabolomics. Curr. Protoc. Plant Biol. 2019, 4, e20100. [Google Scholar] [CrossRef]
- Yang, Y.-Y.; Wu, Z.-Y.; Zhang, H.; Yin, S.-J.; Xia, F.-B.; Zhang, Q.; Wan, J.-B.; Gao, J.-L.; Yang, F.-Q. LC–MS-Based Multivariate Statistical Analysis for the Screening of Potential Thrombin/Factor Xa Inhibitors from Radix salvia Miltiorrhiza. Chin. Med. 2020, 15, 38. [Google Scholar] [CrossRef]
- Rubakhin, S.S.; Romanova, E.V.; Nemes, P.; Sweedler, J.V. Profiling Metabolites and Peptides in Single Cells. Nat. Methods 2011, 8, S20–S29. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2012, 1, 92–107. [Google Scholar] [CrossRef]
- Chen, K.; Preuß, A.; Hackbarth, S.; Wacker, M.; Langer, K.; Röder, B. Novel Photosensitizer-Protein Nanoparticles for Photodynamic Therapy: Photophysical Characterization and in Vitro Investigations. J. Photochem. Photobiol. B 2009, 96, 66–74. [Google Scholar] [CrossRef]
- Son, J.; Yang, S.M.; Yi, G.; Roh, Y.J.; Park, H.; Park, J.M.; Choi, M.-G.; Koo, H. Folate-Modified PLGA Nanoparticles for Tumor-Targeted Delivery of Pheophorbide isn Vivo. Biochem. Biophys. Res. Commun. 2018, 498, 523–528. [Google Scholar] [CrossRef]
- Jonker, J.W.; Buitelaar, M.; Wagenaar, E.; van der Valk, M.A.; Scheffer, G.L.; Scheper, R.J.; Plosch, T.; Kuipers, F.; Elferink, R.P.J.O.; Rosing, H.; et al. Nonlinear Partial Differential Equations and Applications: The Breast Cancer Resistance Protein Protects against a Major Chlorophyll-Derived Dietary Phototoxin and Protoporphyria. Proc. Natl. Acad. Sci. USA 2002, 99, 15649–15654. [Google Scholar] [CrossRef]
- Robey, R.W.; Steadman, K.; Polgar, O.; Morisaki, K.; Blayney, M.; Mistry, P.; Bates, S.E. Pheophorbide a Is a Specific Probe for ABCG2 Function and Inhibition. Cancer Res. 2004, 64, 1242–1246. [Google Scholar] [CrossRef]
- Raheem, D.J.; Tawfike, A.F.; Abdelmohsen, U.R.; Edrada-Ebel, R.; Fitzsimmons-Thoss, V. Application of Metabolomics and Molecular Networking in Investigating the Chemical Profile and Antitrypanosomal Activity of British Bluebells (Hyacinthoides non-scripta). Sci. Rep. 2019, 9, 2547. [Google Scholar] [CrossRef]
- Zhong, G.-X.; Li, P.; Zeng, L.-J.; Guan, J.; Li, D.-Q.; Li, S.-P. Chemical Characteristics of Salvia miltiorrhiza (Danshen) Collected from Different Locations in China. J. Agric. Food Chem. 2009, 57, 6879–6887. [Google Scholar] [CrossRef]
- Yang, H.; Li, X.; Li, X.; Yu, H.; Shen, Z. Identification of Lipopeptide Isoforms by MALDI-TOF-MS/MS Based on the Simultaneous Purification of Iturin, Fengycin, and Surfactin by RP-HPLC. Anal. Bioanal. Chem. 2015, 407, 2529–2542. [Google Scholar] [CrossRef]
- Salsabila, I.; Nugraheni, N.; Ahlina, F.; Haryanti, S.; Meiyanto, E. Synergistic Cotreatment Potential of Soursop (Annona muricata L.) Leaves Extract with Doxorubicin on 4T1 Cells with Antisenescence and Anti-reactive-oxygen-species Properties. Iran. J. Pharm. Sci. 2021, 20, 57–67. [Google Scholar] [CrossRef]
- Mroczek, T.; Dymek, A.; Widelski, J.; Wojtanowski, K.K. The Bioassay-Guided Fractionation and Identification of Potent Acetylcholinesterase Inhibitors from Narcissus c.v. ‘Hawera’ Using Optimized Vacuum Liquid Chromatography, High Resolution Mass Spectrometry and Bioautography. Metabolites 2020, 10, 395. [Google Scholar] [CrossRef]
- Rady, I.; Bloch, M.B.; Chamcheu, R.-C.N.; Banang Mbeumi, S.; Anwar, M.R.; Mohamed, H.; Babatunde, A.S.; Kuiate, J.-R.; Noubissi, F.K.; El Sayed, K.A.; et al. Anticancer Properties of Graviola (Annona muricata): A Comprehensive Mechanistic Review. Oxidative Med. Cell. Longev. 2018, 2018, 1826170. [Google Scholar] [CrossRef]
- Moghadamtousi, S.Z.; Karimian, H.; Rouhollahi, E.; Paydar, M.; Fadaeinasab, M.; Kadir, H.A. Annona Muricata Leaves Induce G1 Cell Cycle Arrest and Apoptosis through Mitochondria-Mediated Pathway in Human HCT-116 and HT-29 Colon Cancer Cells. J. Ethnopharmacol. 2014, 156, 277–289. [Google Scholar] [CrossRef]
- Moghadamtousi, S.Z.; Fadaeinasab, M.; Nikzad, S.; Mohan, G.; Ali, H.; Kadir, H.A. Annona muricata (Annonaceae): A Review of Its Traditional Uses, Isolated Acetogenins and Biological Activities. Int. J. Mol. Sci. 2015, 16, 15625–15658. [Google Scholar] [CrossRef]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-Based Metabolomics. Mol. BioSyst. 2012, 8, 470–481. [Google Scholar] [CrossRef]
- Castrosanto, M.; Alvarez, M.R.; Salamanez, K.; Nacario, R.; Completo, G.C. Barnyard Grass [Echinochloa Crus-galli (L.) Beauv] Leaves Extract against Tomato Pests. J. Sci. Food Agric. 2021, 101, 6289–6299. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; Vandergheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-Independent MS/MS Deconvolution for Comprehensive Metabolome Analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.-F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F. Reproducible Molecular Networking of Untargeted Mass Spectrometry Data Using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- Ricart, E.; Pupin, M.; Müller, M.; Lisacek, F. Automatic Annotation and Dereplication of Tandem Mass Spectra of Peptidic Natural Products. Anal. Chem. 2020, 92, 15862–15871. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.; Ong, C.W.; Wandy, J.; Ernst, M.; Ridder, L.; van der Hooft, J.J.J. Deciphering Complex Metabolite Mixtures by Unsupervised and Supervised Substructure Discovery and Semi-Automated Annotation from MS/MS Spectra. Faraday Discuss. 2019, 218, 284–302. [Google Scholar] [CrossRef]
- Da Silva, R.R.; Wang, M.; Nothias, L.-F.; van der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating Annotations of Molecular Networks Using in Silico Fragmentation. PLoS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.-F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef] [Green Version]




| Alignment ID | m/z | rt | Formula | Ontology |
|---|---|---|---|---|
| 13271 | 633.43335 | 19.473 | NA | NA |
| 12388 | 617.43817 | 20.733 | NA | NA |
| 12357 | 617.17627 | 20.72 | NA | NA |
| 12376 | 617.30951 | 20.734 | NA | NA |
| 3078 | 335.17929 | 19.474 | C21H28O2 | Androgens and derivatives |
| 13297 | 633.61481 | 19.472 | NA | NA |
| 12369 | 617.27972 | 20.737 | NA | NA |
| 11145 | 591.42114 | 18.254 | C40H56O2 | Xanthophylls |
| 12433 | 618.26306 | 20.723 | C22H47N5O21S2 | 4,6-disubstituted 2-deoxystreptamines |
| 12422 | 617.61731 | 20.752 | NA | NA |
| 11920 | 609.11182 | 20.41 | C28H23BCl2F4N2O4 | NA |
| 11996 | 610.47235 | 16.595 | C35H63NO7 | Macrolides and analogues |
| 11931 | 609.26654 | 20.402 | C32H42O10 | Limonoids |
| 12363 | 617.23511 | 20.714 | C31H35FN4O7 | Dipeptides |
| 13254 | 633.33405 | 19.48 | C31H50N2O10 | Peptides |
| 10417 | 573.40094 | 18.334 | NA | NA |
| 13144 | 631.40973 | 19.491 | NA | NA |
| 11923 | 609.13605 | 20.392 | NA | NA |
| 9844 | 557.41901 | 19.811 | NA | NA |
| 11985 | 610.30011 | 18.351 | C35H39N5O5 | Ergotamines, dihydroergotamines, and derivatives |
| Alignment ID | m/z | rt | Formula | Ontology |
|---|---|---|---|---|
| 15623 | 659.44159 | 20.911 | NA | NA |
| 14263 | 633.43433 | 20.944 | NA | NA |
| 3653 | 336.20688 | 17.861 | C21H25N3O | 4-benzylpiperidines |
| 17871 | 703.41687 | 19.107 | NA | NA |
| 3618 | 335.20947 | 17.844 | C18H32O4 | NA |
| 14942 | 645.50934 | 22.086 | NA | NA |
| 3722 | 338.1687 | 18.385 | NA | NA |
| 1551 | 239.23094 | 17.983 | NA | NA |
| 19912 | 772.49536 | 20.734 | NA | NA |
| 14387 | 635.42743 | 20.948 | NA | NA |
| 14941 | 645.50061 | 22.144 | NA | NA |
| 17456 | 692.46521 | 20.672 | NA | NA |
| 5453 | 395.37494 | 20.871 | NA | NA |
| 19882 | 771.52161 | 21.33 | NA | NA |
| 10807 | 567.42865 | 22.217 | NA | NA |
| 21854 | 871.30621 | 20.854 | NA | NA |
| 8693 | 505.40161 | 19.697 | NA | NA |
| 6588 | 431.19675 | 20.974 | C22H32O7 | NA |
| 3683 | 337.1861 | 18.402 | NA | NA |
| 17677 | 699.5072 | 22.895 | NA | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salac, E.L.O.; Alvarez, M.R.; Gaurana, R.S.; Grijaldo, S.J.B.; Serrano, L.M.; Juan, F.d.; Abogado, R.; Padolina Jr., I.; Deniega, F.M.; Delica, K.; et al. Biological Assay-Guided Fractionation and Mass Spectrometry-Based Metabolite Profiling of Annona muricata L. Cytotoxic Compounds against Lung Cancer A549 Cell Line. Plants 2022, 11, 2380. https://doi.org/10.3390/plants11182380
Salac ELO, Alvarez MR, Gaurana RS, Grijaldo SJB, Serrano LM, Juan Fd, Abogado R, Padolina Jr. I, Deniega FM, Delica K, et al. Biological Assay-Guided Fractionation and Mass Spectrometry-Based Metabolite Profiling of Annona muricata L. Cytotoxic Compounds against Lung Cancer A549 Cell Line. Plants. 2022; 11(18):2380. https://doi.org/10.3390/plants11182380
Chicago/Turabian StyleSalac, Edcyl Lee O., Michael Russelle Alvarez, Rnie Shayne Gaurana, Sheryl Joyce B. Grijaldo, Luster Mae Serrano, Florence de Juan, Rowell Abogado, Isagani Padolina Jr., Froila Marie Deniega, Kimberly Delica, and et al. 2022. "Biological Assay-Guided Fractionation and Mass Spectrometry-Based Metabolite Profiling of Annona muricata L. Cytotoxic Compounds against Lung Cancer A549 Cell Line" Plants 11, no. 18: 2380. https://doi.org/10.3390/plants11182380
APA StyleSalac, E. L. O., Alvarez, M. R., Gaurana, R. S., Grijaldo, S. J. B., Serrano, L. M., Juan, F. d., Abogado, R., Padolina Jr., I., Deniega, F. M., Delica, K., Fernandez, K., Lebrilla, C. B., Manalo, M. N., Heralde III, F. M., Completo, G. C. J., & Nacario, R. C. (2022). Biological Assay-Guided Fractionation and Mass Spectrometry-Based Metabolite Profiling of Annona muricata L. Cytotoxic Compounds against Lung Cancer A549 Cell Line. Plants, 11(18), 2380. https://doi.org/10.3390/plants11182380







