Pathogen Adaptation to American (Rpv3-1) and Eurasian (Rpv29) Grapevine Loci Conferring Resistance to Downy Mildew
Abstract
1. Introduction
2. Results
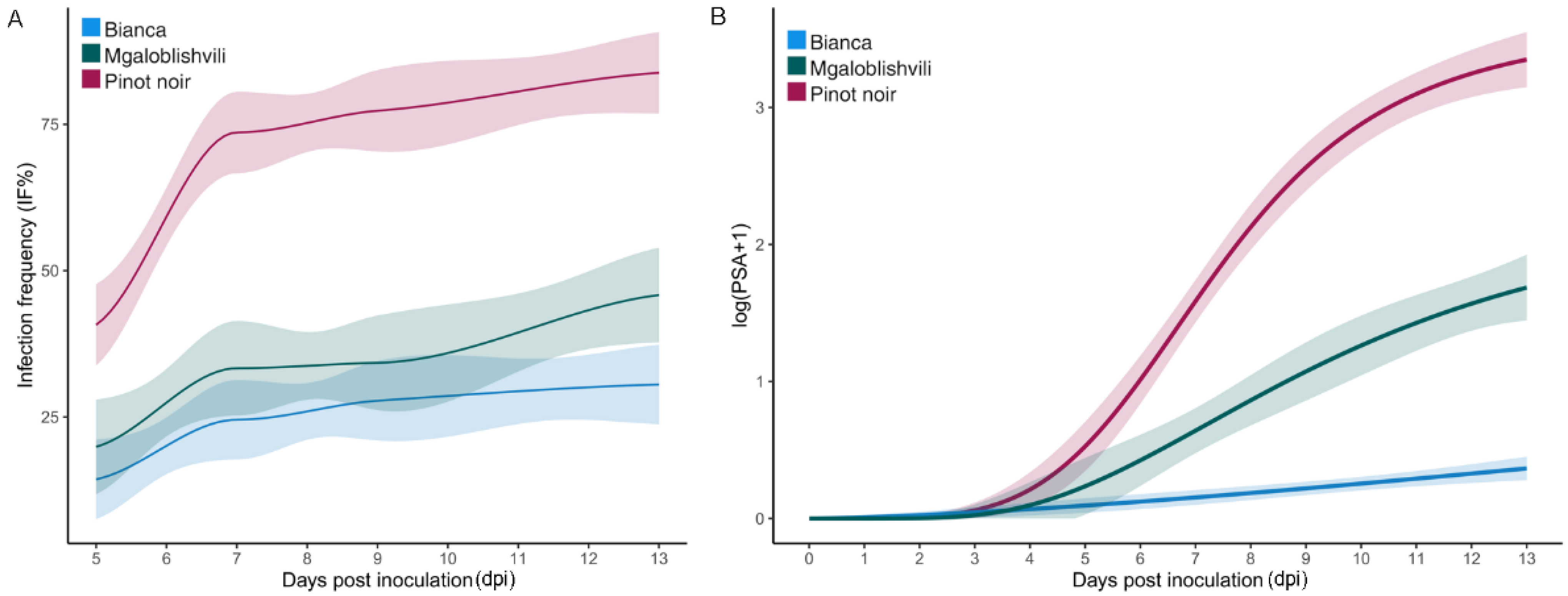
2.1. Infection Parameters of the Isolates
2.2. Identification of Potentially Adapted Isolates
2.3. Infection Parameters of the Isolates Divided into Pathogenicity Classes
2.4. Oospore Production and Viability
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. P. viticola Isolates
4.3. Experimental Inoculation and Pathogenicity Evaluation
4.4. Oospore Production and Viability
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McGovern, P.; Jalabadze, M.; Batiuk, S.; Callahan, M.P.; Smith, K.E.; Hall, G.R.; Kvavadze, E.; Maghradze, D.; Rusishvili, N.; Bouby, L.; et al. Early Neolithic Wine of Georgia in the South Caucasus. Proc. Natl. Acad. Sci. USA 2017, 114, E10309–E10318. [Google Scholar] [CrossRef] [PubMed]
- Bois, B.; Zito, S.; Calonnec, A.; Ollat, N. Climate vs. Grapevine Pests and Diseases Worldwide: The First Results of a Global Survey. OENO One 2017, 51, 133–139. [Google Scholar] [CrossRef]
- Gessler, C.; Pertot, I.; Perazzolli, M. Plasmopara viticola: A Review of Knowledge on Downy Mildew of Grapevine and Effective Disease Management. Phytopathol. Mediterr. 2011, 50, 3–44. [Google Scholar] [CrossRef]
- Toffolatti, S.L.; Russo, G.; Campia, P.; Bianco, P.A.; Borsa, P.; Coatti, M.; Torriani, S.F.F.; Sierotzki, H. A Time-Course Investigation of Resistance to the Carboxylic Acid Amide Mandipropamid in Field Populations of Plasmopara viticola Treated with Anti-Resistance Strategies. Pest Manag. Sci. 2018, 74, 2822–2834. [Google Scholar] [CrossRef] [PubMed]
- Burruano, S. The Life-Cycle of Plasmopara viticola, Cause of Downy Mildew of Vine. Mycologist 2000, 14, 179–182. [Google Scholar] [CrossRef]
- Fröbel, S.; Zyprian, E. Colonization of Different Grapevine Tissues by Plasmopara viticola—A Histological Study. Front. Plant Sci. 2019, 10, 951. [Google Scholar] [CrossRef] [PubMed]
- Gindro, K.; Pezet, R.; Viret, O. Histological Study of the Responses of Two Vitis vinifera Cultivars (Resistant and Susceptible) to Plasmopara viticola Infections. Plant Physiol. Biochem. 2003, 41, 846–853. [Google Scholar] [CrossRef]
- Massi, F.; Torriani, S.F.F.; Borghi, L.; Toffolatti, S.L. Fungicide Resistance Evolution and Detection in Plant Pathogens: Plasmopara viticola as a Case Study. Microorganisms 2021, 9, 119. [Google Scholar] [CrossRef]
- Eibach, R.; Töpfer, R. Traditional Grapevine Breeding Techniques. In Grapevine Breeding Programs for the Wine Industry; Reynolds, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 3–22. [Google Scholar] [CrossRef]
- Vezzulli, S.; Gramaje, D.; Tello, J.; Gambino, G.; Bettinelli, P.; Pirrello, C.; Schwandner, A.; Barba, P.; Angelini, E.; Anfora, G.; et al. Genomic Designing for Biotic Stress Resistant Grapevine. In Genomic Designing for Biotic Stress Resistant Fruit Crops; Chittaranjan, K., Ed.; Springer Nature: Cham, Switzerland, 2022; pp. 87–255. ISBN 9783030918026. [Google Scholar]
- Di Gaspero, G.; Copetti, D.; Coleman, C.; Castellarin, S.D.; Eibach, R.; Kozma, P.; Lacombe, T.; Gambetta, G.; Zvyagin, A.; Cindrić, P.; et al. Selective Sweep at the Rpv3 Locus during Grapevine Breeding for Downy Mildew Resistance. Theor. Appl. Genet. 2012, 124, 277–286. [Google Scholar] [CrossRef]
- Zini, E.; Dolzani, C.; Stefanini, M.; Gratl, V.; Bettinelli, P.; Nicolini, D.; Betta, G.; Dorigatti, C.; Velasco, R.; Letschka, T.; et al. R-Loci Arrangement Versus Downy and Powdery Mildew Resistance Level: A Vitis Hybrid Survey. Int. J. Mol. Sci. 2019, 20, 3526. [Google Scholar] [CrossRef]
- Chitarrini, G.; Riccadonna, S.; Zulini, L.; Vecchione, A.; Stefanini, M.; Larger, S.; Pindo, M.; Cestaro, A.; Franceschi, P.; Magris, G.; et al. Two-Omics Data Revealed Commonalities and Differences between Rpv12- and Rpv3-Mediated Resistance in Grapevine. Sci. Rep. 2020, 10, 12193. [Google Scholar] [CrossRef]
- Toffolatti, S.L.; Venturini, G.; Maffi, D.; Vercesi, A. Phenotypic and Histochemical Traits of the Interaction between Plasmopara viticola and Resistant or Susceptible Grapevine Varieties. BMC Plant Biol. 2012, 12, 124. [Google Scholar] [CrossRef]
- Bellin, D.; Peressotti, E.; Merdinoglu, D.; Wiedemann-Merdinoglu, S.; Adam-Blondon, A.F.; Cipriani, G.; Morgante, M.; Testolin, R.; Di Gaspero, G. Resistance to Plasmopara viticola in Grapevine “Bianca” Is Controlled by a Major Dominant Gene Causing Localised Necrosis at the Infection Site. Theor. Appl. Genet. 2009, 120, 163–176. [Google Scholar] [CrossRef]
- Toffolatti, S.; Maddalena, G.; Salomoni; Maghradze; Bianco, P.A.P.; Failla, O.; Salomoni, D.; Maghradze, D.; Bianco, P.A.P.; Failla, O. Evidence of Resistance to the Downy Mildew Agent Plasmopara viticola in the Georgian Vitis Vinifera Germplasm. Vitis 2016, 55, 121–128. [Google Scholar] [CrossRef]
- Toffolatti, S.L.; De Lorenzis, G.; Costa, A.; Maddalena, G.; Passera, A.; Bonza, M.C.; Pindo, M.; Stefani, E.; Cestaro, A.; Casati, P.; et al. Unique Resistance Traits against Downy Mildew from the Center of Origin of Grapevine (Vitis vinifera). Sci. Rep. 2018, 8, 12523. [Google Scholar] [CrossRef]
- Bourguet, D.; Delmotte, F.; Franck, P.; Guillemaud, T.; Reboud, X.; Vacher, C.; Walker, A.S. Combining Selective Pressures to Enhance the Durability of Disease Resistance Genes. Front. Plant Sci. 2016, 7, 1916. [Google Scholar] [CrossRef]
- Peressotti, E.; Wiedemann-Merdinoglu, S.; Delmotte, F.; Bellin, D.; Di Gaspero, G.; Testolin, R.; Merdinoglu, D.; Mestre, P. Breakdown of Resistance to Grapevine Downy Mildew upon Limited Deployment of a Resistant Variety. BMC Plant Biol. 2010, 10, 147. [Google Scholar] [CrossRef]
- Eisenmann, B.; Czemmel, S.; Ziegler, T.; Buchholz, G.; Kortekamp, A.; Trapp, O.; Rausch, T.; Dry, I.; Bogs, J. Rpv3-1 Mediated Resistance to Grapevine Downy Mildew Is Associated with Specific Host Transcriptional Responses and the Accumulation of Stilbenes. BMC Plant Biol. 2019, 19, 343. [Google Scholar] [CrossRef]
- Wingerter, C.; Eisenmann, B.; Weber, P.; Dry, I.; Bogs, J. Grapevine Rpv3-, Rpv10- and Rpv12-Mediated Defense Responses against Plasmopara viticola and the Impact of Their Deployment on Fungicide Use in Viticulture. BMC Plant Biol. 2021, 21, 470. [Google Scholar] [CrossRef]
- Delmotte, F.; Mestre, P.; Schneider, C.; Kassemeyer, H.H.; Kozma, P.; Richart-Cervera, S.; Rouxel, M.; Delière, L. Rapid and Multiregional Adaptation to Host Partial Resistance in a Plant Pathogenic Oomycete: Evidence from European Populations of Plasmopara viticola, the Causal Agent of Grapevine Downy Mildew. Infect. Genet. Evol. 2014, 27, 500–508. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the Plant Immune System from Dissection to Deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Maddalena, G.; Bitsadze, N.; Maghradze, D.; Bianco, P.A.; Failla, O.; Toffolatti, S.L.; De Lorenzis, G. Rpv29, Rpv30 and Rpv31: Three Novel Genomic Loci Associated With Resistance to Plasmopara viticola in Vitis Vinifera. Front. Plant Sci. 2020, 11, 562432. [Google Scholar] [CrossRef]
- Toffolatti, S.L.; De Lorenzis, G.; Brilli, M.; Moser, M.; Shariati, V.; Tavakol, E.; Maddalena, G.; Passera, A.; Casati, P.; Pindo, M.; et al. Novel Aspects on the Interaction Between Grapevine and Plasmopara viticola: Dual-RNA-Seq Analysis Highlights Gene Expression Dynamics in the Pathogen and the Plant During the Battle For Infection. Genes 2020, 11, 261. [Google Scholar] [CrossRef]
- McRoberts, N.; Hughes, G.; Madden, L.V. The Theoretical Basis and Practical Application of Relationships between Different Disease Intensity Measurements in Plants. Ann. Appl. Biol. 2003, 142, 191–211. [Google Scholar] [CrossRef]
- Bove, F.; Rossi, V. Components of Partial Resistance to Plasmopara viticola Enable Complete Phenotypic Characterization of Grapevine Varieties. Sci. Rep. 2020, 10, 585. [Google Scholar] [CrossRef]
- Possamai, T.; Wiedemann-Merdinoglu, S. Phenotyping for QTL Identification: A Case Study of Resistance to Plasmopara viticola and Erysiphe necator in Grapevine. Front. Plant Sci. 2022, 13, 930954. [Google Scholar] [CrossRef]
- Fontaine, M.C.; Labbé, F.; Dussert, Y.; Delière, L.; Richart-cervera, S.; Giraud, T.; Delmotte, F. Europe as a Bridgehead in the Worldwide Invasion History of Grapevine Downy Mildew, Plasmopara viticola. Curr. Biol. 2021, 31, 2155–2166. [Google Scholar] [CrossRef]
- Ricciardi, V.; Marcianò, D.; Sargolzaei, M.; Maddalena, G.; Maghradze, D.; Tirelli, A.; Casati, P.; Bianco, P.A.; Failla, O.; Fracassetti, D.; et al. From Plant Resistance Response to the Discovery of Antimicrobial Compounds: The Role of Volatile Organic Compounds (VOCs) in Grapevine Downy Mildew Infection. Plant Physiol. Biochem. 2021, 160, 294–305. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Rustioni, L.; Cola, G.; Ricciardi, V.; Bianco, P.A.; Maghradze, D.; Failla, O.; Quaglino, F.; Toffolatti, S.L.; De Lorenzis, G. Georgian Grapevine Cultivars: Ancient Biodiversity for Future Viticulture. Front. Plant Sci. 2021, 12, 630122. [Google Scholar] [CrossRef]
- Brilli, M.; Asquini, E.; Moser, M.; Bianchedi, P.L.; Perazzolli, M.; Si-Ammour, A. A Multi-Omics Study of the Grapevine-Downy Mildew (Plasmopara viticola) Pathosystem Unveils a Complex Protein Coding-A Nd Noncoding-Based Arms Race during Infection. Sci. Rep. 2018, 8, 757. [Google Scholar] [CrossRef]
- Xiang, J.; Li, X.; Wu, J.; Yin, L.; Zhang, Y.; Lu, J. Studying the Mechanism of Plasmopara viticola RxLR Effectors on Suppressing Plant Immunity. Front. Microbiol. 2016, 7, 709. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Chen, S.; Liu, J.; Fu, P.; Wu, W.; Song, S.; Gao, Y.; Ye, W.; Lu, J. Plasmopara viticola Effector PvRXLR111 Stabilizes VvWRKY40 to Promote Virulence. Mol. Plant Pathol. 2021, 22, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Vercesi, A.; Vavassori, A.; Faoro, F.; Bisiach, M. Effect of Azoxystrobin on the Oospores of Plasmopara viticola. In Advances in Downy Mildew Research; Spencer-Phillips, P.T.N., Gisi, U., Lebeda, A., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 195–199. ISBN 0306479141. [Google Scholar]
- Maddalena, G.; Russo, G.; Toffolatti, S.L. The Study of the Germination Dynamics of Plasmopara viticola Oospores Highlights the Presence of Phenotypic Synchrony With the Host. Front. Microbiol. 2021, 12, 698586. [Google Scholar] [CrossRef] [PubMed]
- Toffolatti, S.L.; Venturini, G.; Campia, P.; Cirio, L.; Bellotto, D.; Vercesi, A. Sensitivity to Cymoxanil in Italian Populations of Plasmopara viticola Oospores. Pest Manag. Sci. 2015, 71, 1182–1188. [Google Scholar] [CrossRef]
- Vercesi, A.; Toffolatti, S.L.; Zocchi, G.; Guglielmann, R.; Ironi, L. A New Approach to Modelling the Dynamics of Oospore Germination in Plasmopara viticola. Eur. J. Plant Pathol. 2010, 128, 113–126. [Google Scholar] [CrossRef]
- Delbac, L.; Delière, L.; Schneider, C.; Delmotte, F. Evidence for Sexual Reproduction and Fertile Oospore Production by Plasmopara viticola on the Leaves of Partially Resistant Grapevine Cultivars. Acta Hortic. 2019, 1248, 607–619. [Google Scholar] [CrossRef]
- Wong, F.P.; Wilcox, W.F. Distribution of Baseline Sensitivities to Azoxystrobin among Isolates of Plasmopara viticola. Plant Dis. 2000, 84, 275–281. [Google Scholar] [CrossRef]
- Hernández, I.; Gutiérrez, S.; Ceballos, S.; Palacios, F.; Toffolatti, S.L.; Maddalena, G.; Diago, M.P.; Tardaguila, J. Assessment of Downy Mildew in Grapevine Using Computer Vision and Fuzzy Logic. Development and Validation of a New Method. OENO One 2022, 56, 41–53. [Google Scholar] [CrossRef]
- Précigout, P.A.; Claessen, D.; Makowski, D.; Robert, C. Does the Latent Period of Leaf Fungal Pathogens Reflect Their Trophic Type? A Meta-Analysis of Biotrophs, Hemibiotrophs, and Necrotrophs. Phytopathology 2020, 110, 345–361. [Google Scholar] [CrossRef]
- Scherer, E.; Gisi, U. Characterization of Genotype and Mating Type in European Isolates of Plasmopara viticola. J. Phytopathol. 2006, 154, 489–495. [Google Scholar] [CrossRef]
- Wong, F.P.; Burr, H.N.; Wilcox, W.F. Heterothallism in Plasmopara viticola. Plant Pathol. 2001, 50, 427–432. [Google Scholar] [CrossRef]
- Dussert, Y.; Legrand, L.; Mazet, I.; Couture, C.; Piron, M.-C.; Serre, R.-F.; Bouchez, O.; Mestre, P.; Toffolatti, S.L.; Giraud, T.; et al. Identification of the First Oomycete Mating-Type Locus Sequence in the Grapevine Downy Mildew Pathogen, Plasmopara viticola. Curr. Biol. 2020, 30, 3897–3907. [Google Scholar] [CrossRef]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef]
- Bates, D.; Watts, D. Nonlinear Regression Analysis and Its Applications; Wiley: New York, NY, USA, 1988; ISBN 0471816434. [Google Scholar]
- Berger, R.D. The Analysis of Effects of Control Measures on the Development of Epidemics. In Experimental Techniques in Plant Disease Epidemiology; Kranz, J., Rotem, J., Eds.; Springer: Berlin/Heidelberg, Germany, 1988; pp. 137–151. ISBN 978-3-642-95536-5. [Google Scholar]
- Green, K.K.; Stenberg, J.A.; Lankinen, Å. Making Sense of Integrated Pest Management (IPM) in the Light of Evolution. Evol. Appl. 2020, 13, 1791–1805. [Google Scholar] [CrossRef]




| Grapevine cv | Variable | dpi | ||||
|---|---|---|---|---|---|---|
| PC | 5 | 7 | 9 | 13 | ||
| Pinot noir | IF (%) ± SD | 1 | 30 ± 30 a | 89 ± 30 b | 98 ± 8 c | 100 ± 0 c |
| 2 | 63 ± 38 a | 92 ± 15 ab | 92 ± 15 bc | 92 ± 15 abc | ||
| 3 | 33 ± 32 a | 56 ± 33 a | 53 ± 32 a | 67 ± 25 a | ||
| 4 | 44 ± 34 a | 69 ± 29 ab | 70 ± 26 ab | 80 ± 26 ab | ||
| all isolates | 41 ± 37 | 74 ± 31 | 77 ± 27 | 84 ± 24 | ||
| PSA (%) ± SD | 1 | 0.5 ± 0.5 a | 4 ± 4 a | 13 ± 13a | 30 ± 30 a | |
| 2 | 2 ± 2 a | 8 ± 5 a | 26 ± 14 a | 48 ± 31 a | ||
| 3 | 1.4 ± 1.4 a | 6 ± 6 a | 16 ± 10 a | 47 ± 30 a | ||
| 4 | 2 ± 2 a | 7 ± 7 a | 19 ± 15 a | 38 ± 20 a | ||
| all isolates | 1 ± 1 | 6 ± 6 | 18 ± 14 | 39 ± 27 | ||
| Mgaloblishvili | IF (%) ± SD | 1 | - | - | - | - |
| 2 | - | - | - | - | ||
| 3 | 25 ± 25 a | 50 ± 27 a | 53 ± 26 a | 67 ± 32 a | ||
| 4 | 34 ± 32 a | 55 ± 29 a | 56 ± 27 a | 76 ± 25 a | ||
| all isolates | 20 ± 20 | 33 ± 33 | 34 ± 34 | 46 ± 42 | ||
| PSA (%) ± SD | 1 | - | - | - | - | |
| 2 | - | - | - | - | ||
| 3 | 0.4 ± 0.4 a | 3 ± 3 a | 5 ± 5 a | 15 ± 11 a | ||
| 4 | 0.7 ± 0.7 a | 3 ± 3 a | 7 ± 7 a | 23 ± 16 a | ||
| all isolates | 0.4 ± 0.4 | 2 ± 2 | 4 ± 4 | 13 ± 13 | ||
| Bianca | IF (%) ± SD | 1 | - | - | - | - |
| 2 | 29 ± 27 a | 42 ± 30 a | 50 ± 31 a | 54 ± 25 a | ||
| 3 | - | - | - | - | ||
| 4 | 24 ± 22 a | 43 ± 26 a | 48 ± 26 a | 54 ± 26 a | ||
| all isolates | 14 ± 14 | 25 ± 25 | 28 ± 28 | 31 ± 31 | ||
| PSA (%) ± SD | 1 | - | - | - | - | |
| 2 | 0.5 ± 0.5 a | 0.5 ± 0.5 a | 1 ± 1a | 1.7 ± 1.7 a | ||
| 3 | - | - | - | - | ||
| 4 | 0.1 ± 0.1 a | 0.4 ± 0.4 a | 0.8 ± 0.8 a | 1.1 ± 1.1 a | ||
| all isolates | 0.1 ± 0.1 | 0.3 ± 0.3 | 0.5 ± 0.5 | 0.7 ± 0.7 | ||
| Grapevine Cultivar | Model Parameters | ||
|---|---|---|---|
| b | D | e (dpi) | |
| Pinot noir | −4.53 a | 3.61 (38) b | 7.39 a |
| Mgaloblishvili | −3.53 ab | 2.13 (9.4) a | 8.95 a |
| Bianca | −1.50 b | 3.26 * | 51.42 * |
| PSA Range | Grapevine Variety | ||
|---|---|---|---|
| Pinot Noir | Bianca | Mgaloblishvili | |
| 0 | 0 | 32 (44%) | 27 (38%) |
| 0.1–5% | 7 (10%) | 38 (53%) | 7 (10%) |
| 5.1–10% | 4 (5%) | 2 (3%) | 7 (10%) |
| 10.1–25% | 16 (22%) | 0 | 13 (18%) |
| 25.1–50% | 23 (32%) | 0 | 16 (22%) |
| 50.1–75% | 14 (20%) | 0 | 2 (2%) |
| 75.1–100% | 8 (11%) | 0 | 0 |
| Pathogenicity Class | Isolates | Number and Percentage (in Brackets) of Isolates |
|---|---|---|
| 1 | BET4-BET6-CASB1-CASB16-CASB5-CASB9-CAST10-REF.F12-REF13-REF1A-REF1E-REF9-SAC9-ZEN9A-ZIN2-ZOX11.2.2-ZOX16.1.1-ZOX16.29-ZOX20.2.9 | 19 (26%) |
| 2 | CASB12-CASB28-CASB3-SAC2-SAC6-ZIN4-ZOX11.1.2.4-ZOX6.19 | 8 (11%) |
| 3 | BEB6-BET3-CASB30-CASB31-CASB51-CAST12-CAST8-REF12-REF8-ZIN7-ZOX14.2.3-ZOX16.2.1 | 12 (17%) |
| 4 | BEB3-BET17-CASB11-CASB13-CASB14-CASB15-CASB18-CASB23-CASB36-CASB4-CASB6-CAST1-CAST3-CAST6-CAST7-CAST9-REF11-REF14-REF3A-REF3C-SAC3-SAC4-VILLO6-ZEN3-ZIN1-ZIN6-ZIN8-ZOX16.1.8-ZOX16.2.6-ZOX20.11-ZOX20.2.2-ZOX6.1.1.1-ZOX6.1.8 | 33 (46%) |
| Mating Pairs | OD (oospores/cm2) | OV (%) | ||||
|---|---|---|---|---|---|---|
| Pinot Noir | Bianca | Mgaloblishvili | Pinot Noir | Bianca | Mgaloblishvili | |
| CAST1xCAST7 | 7.1 ± 2 | 6.3 ± 2 | 4.9 ± 1,4 | 67 ± 15 | 80 ± 26 | 70 ± 10 |
| CASB11xCAST1 | 6 ± 1.7 | 5.9 ± 1.6 | 5.2 ± 0.9 | 77 ± 15 | 83 ± 15 | 77 ± 15 |
| CASB11xCAST7 | 8.2 ± 1.9 b | 4.6 ± 0.4 a | 5.6 ± 0.6 a | 87 ± 15 | 93 ± 12 | 87 ± 15 |
| Vineyard N. | Vineyard Location | Region | Name of the Isolates | Number of Isolates per Vineyard |
|---|---|---|---|---|
| 1 | Santa Maria della Versa (PV) | Lombardy (NW) | BEB3, BEB6 | 2 |
| 2 | BET3-BET4-BET6-BET17 | 4 | ||
| 3 | Casarsa della Delizia (PN) | Friuli-Venezia Giulia (NE) | CASB1-CASB3-CASB4-CASB5-CASB6-CASB9-CASB11-CASB12-CASB13-CASB14-CASB15-CASB16-CASB18-CASB23-CASB28-CASB30-CASB31-CASB36-CASB51 | 19 |
| 4 | CAST1-CAST3-CAST6-CAST7-CAST8-CAST9-CAST10-CAST12 | 8 | ||
| 5 | Refrontolo (TV) | Veneto (NE) | REF1A-REF1E-REF3A-REF3C-REF8-REF9-REF11-REF.F12-REF13-REF14-REF12 | 11 |
| 6 | Fognano di Brisighella (RA) | Emilia Romagna (NE) | SAC2-SAC3-SAC4-SAC6-SAC9 | 5 |
| 7 | Villorba (TV) | Veneto (NE) | VILLO6 | 1 |
| 8 | Zenson di Piave (TV) | Veneto (NE) | ZEN9A, ZEN3 | 2 |
| 9 | Casarsa della Delizia (PN) | Friuli-Venezia Giulia (NE) | ZIN1-ZIN2-ZIN4-ZIN6-ZIN7-ZIN8 | 6 |
| 10 | Valeggio sul Mincio (VR) | Veneto (NE) | ZOX6.1.8-ZOX6.1.1.1-ZOX6.19 | 3 |
| 11 | Roverè della Luna (TN) | Trentino-Alto Adige (NE) | ZOX11.1.2.4-ZOX11.2.2 | 2 |
| 12 | Valdobbiadene, f. San Giovanni (TV) | Veneto (NE) | ZOX14.2.3 | 1 |
| 13 | Vidor di Valdobbiadene (TV) | Veneto (NE) | ZOX16.2.6-ZOX16.2.1-ZOX16.1.1-ZOX16.1.8-ZOX16.29 | 5 |
| 14 | Valdobbiadene (TV) | Veneto (NE) | ZOX20.2.2-ZOX20.2.9-ZOX20.11 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marone Fassolo, E.; Lecchi, B.; Marcianò, D.; Maddalena, G.; Toffolatti, S.L. Pathogen Adaptation to American (Rpv3-1) and Eurasian (Rpv29) Grapevine Loci Conferring Resistance to Downy Mildew. Plants 2022, 11, 2619. https://doi.org/10.3390/plants11192619
Marone Fassolo E, Lecchi B, Marcianò D, Maddalena G, Toffolatti SL. Pathogen Adaptation to American (Rpv3-1) and Eurasian (Rpv29) Grapevine Loci Conferring Resistance to Downy Mildew. Plants. 2022; 11(19):2619. https://doi.org/10.3390/plants11192619
Chicago/Turabian StyleMarone Fassolo, Elena, Beatrice Lecchi, Demetrio Marcianò, Giuliana Maddalena, and Silvia Laura Toffolatti. 2022. "Pathogen Adaptation to American (Rpv3-1) and Eurasian (Rpv29) Grapevine Loci Conferring Resistance to Downy Mildew" Plants 11, no. 19: 2619. https://doi.org/10.3390/plants11192619
APA StyleMarone Fassolo, E., Lecchi, B., Marcianò, D., Maddalena, G., & Toffolatti, S. L. (2022). Pathogen Adaptation to American (Rpv3-1) and Eurasian (Rpv29) Grapevine Loci Conferring Resistance to Downy Mildew. Plants, 11(19), 2619. https://doi.org/10.3390/plants11192619








