Study on the Role of Phytohormones in Resistance to Watermelon Fusarium Wilt
Abstract
:1. Introduction
2. Results
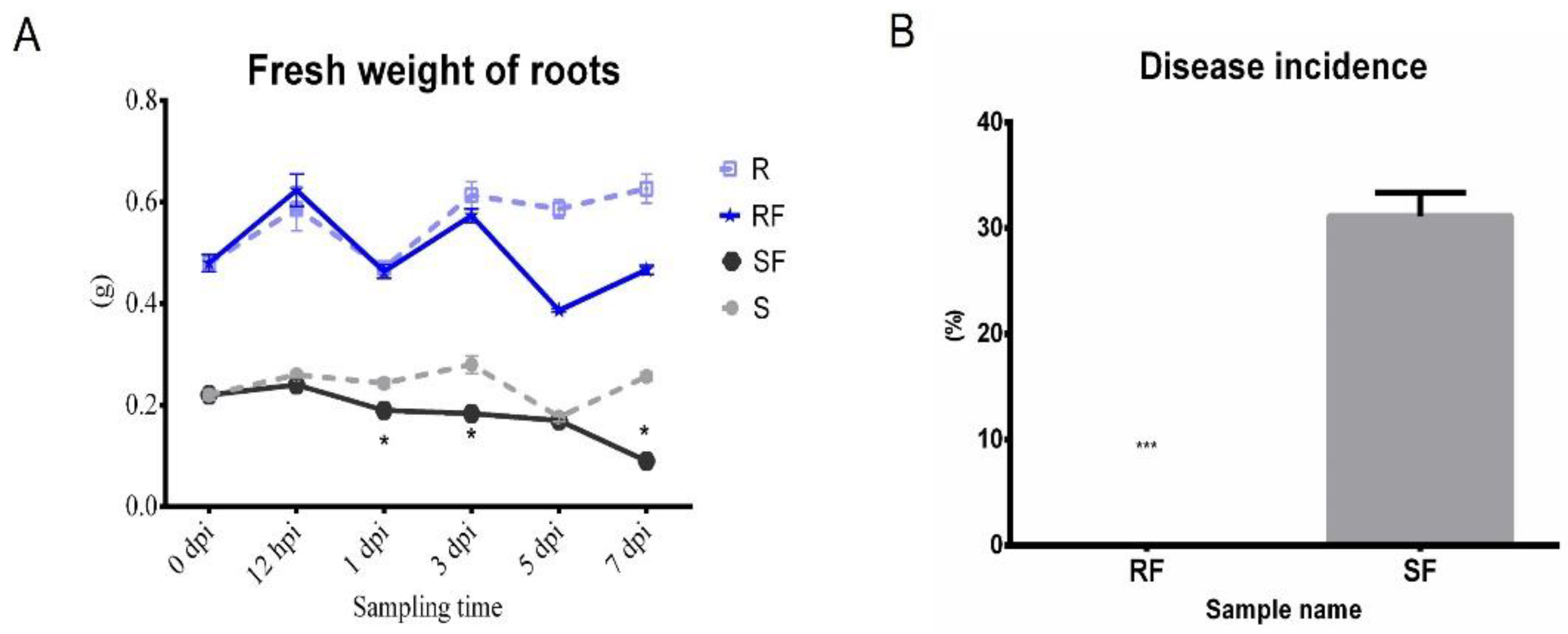
2.1. Comparison Analysis of Phenotype and Fusarium Wilt Disease Occurrence in Resistant and Susceptible Watermelon after FON Inoculation
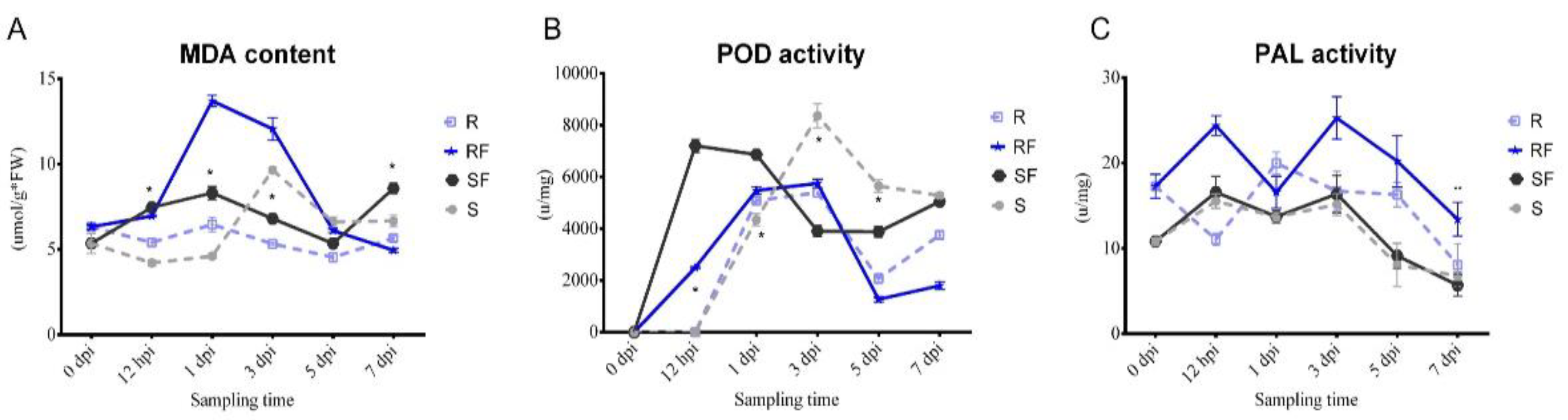
2.2. Dynamic Changes in MDA Content and PAL, POD Enzyme Activities after FON Inoculation
2.3. Comparison Analysis of SA, JA, and ABA Contents at 7 dpi
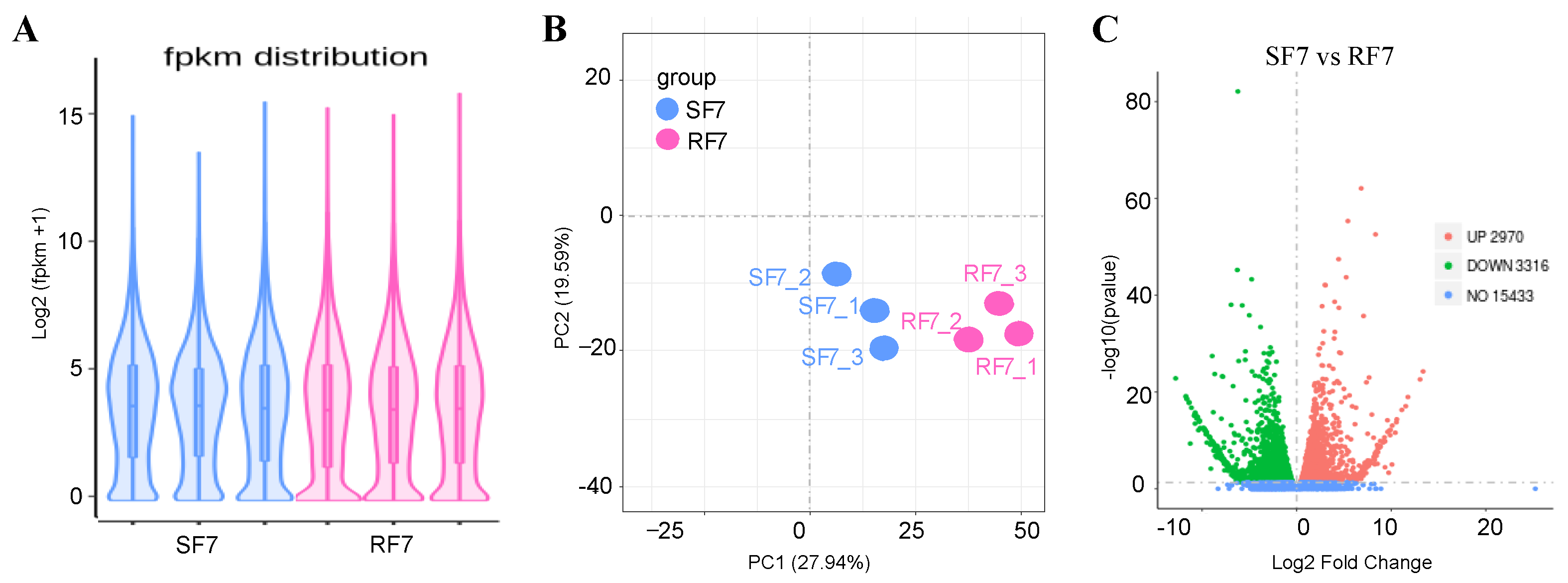
2.4. Comparison Analysis of Transcriptome Differences at 7 dpi
2.5. Functional Annotation of Genes Expressed
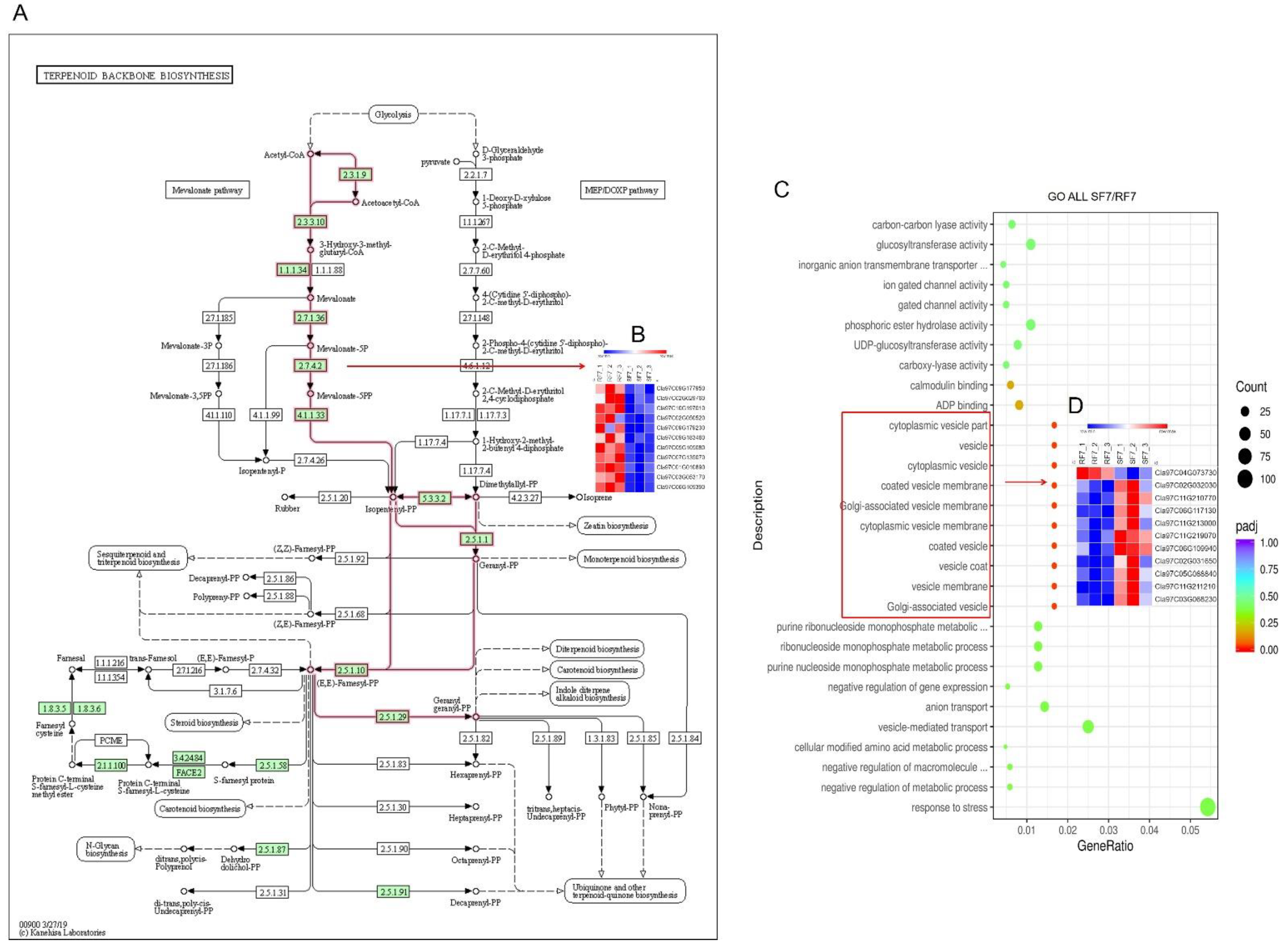
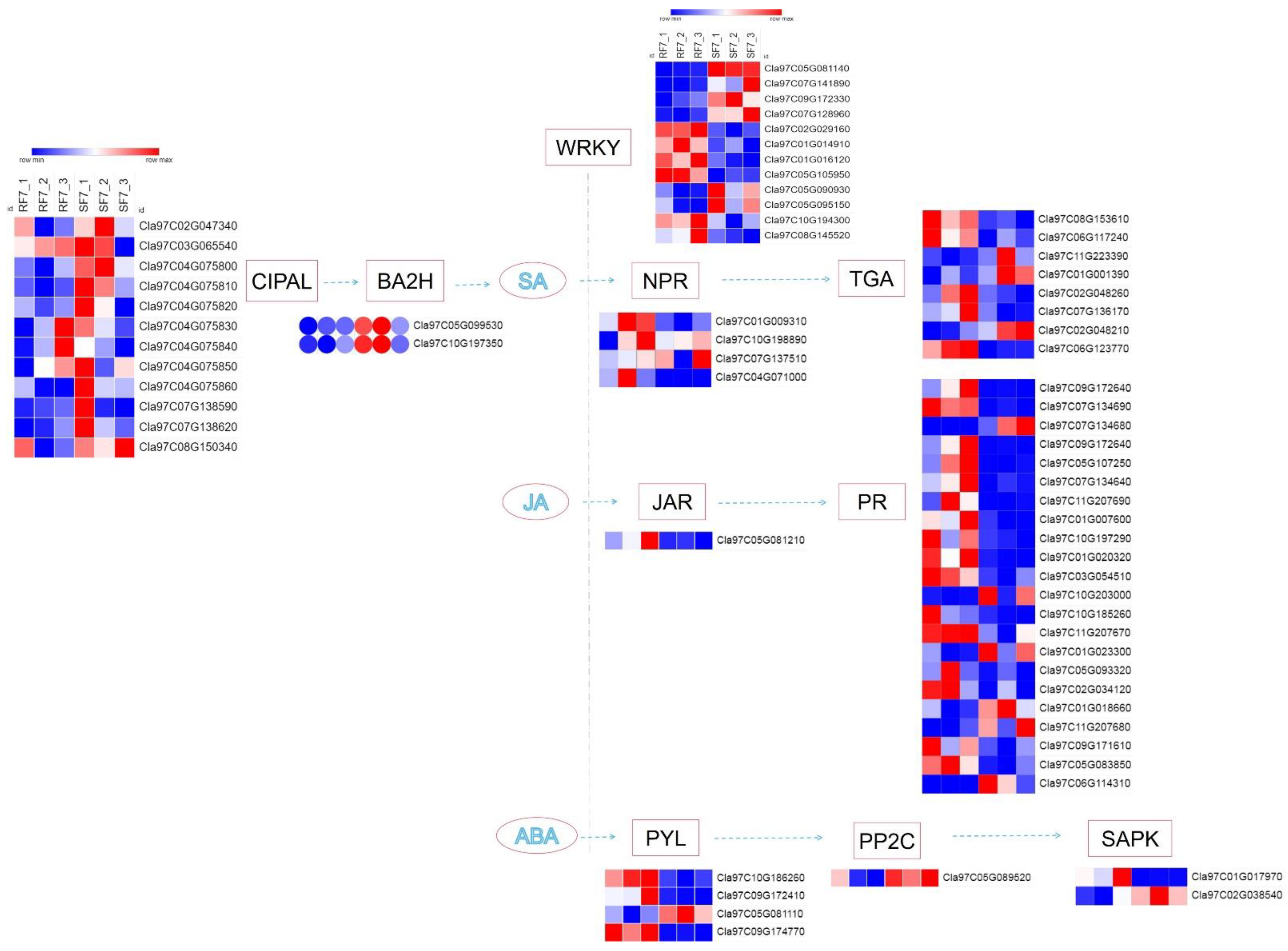
2.6. Analysis of Hormone-Related DEGs
2.7. Bioinformatics Analysis of Candidate Genes
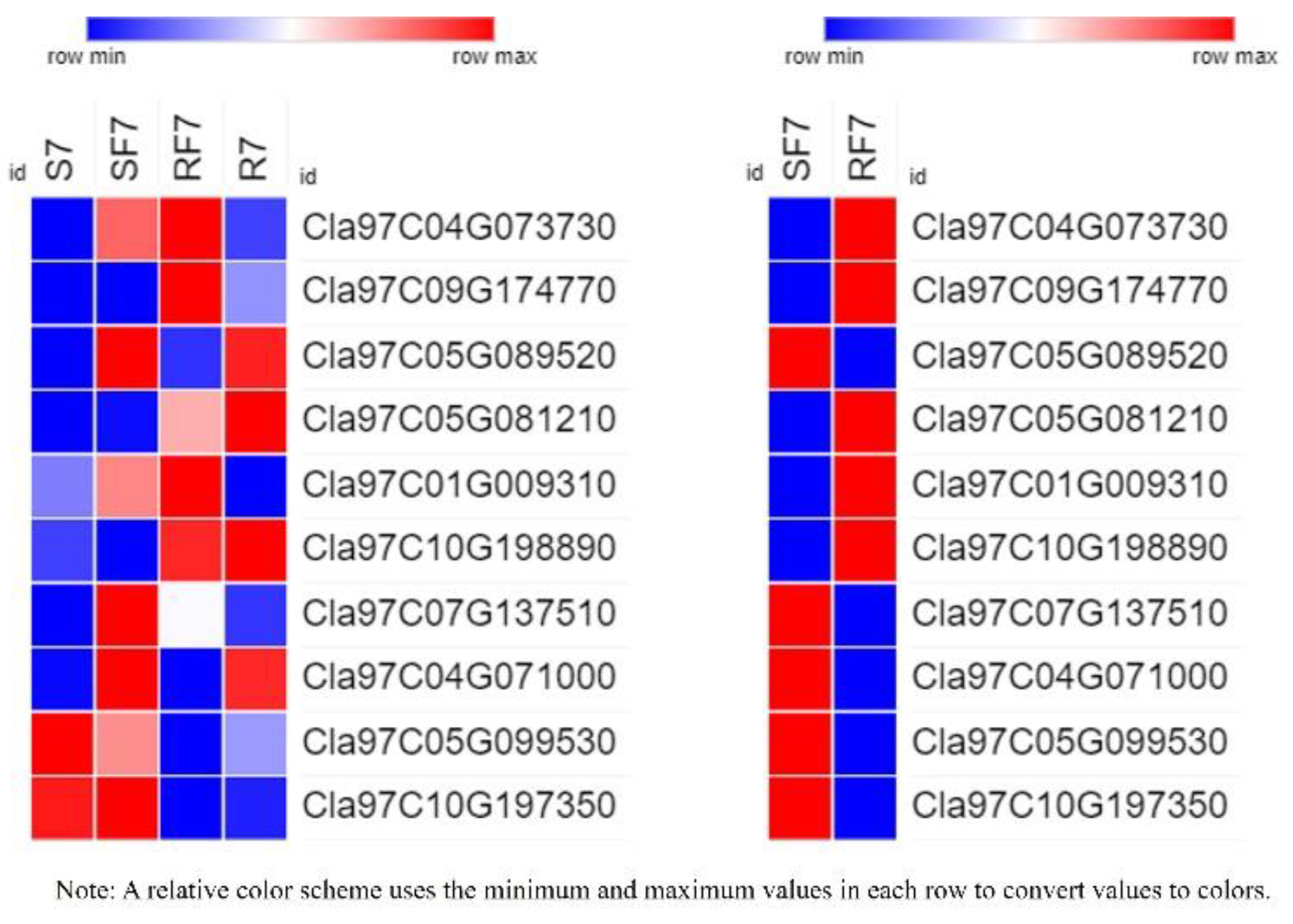
2.8. Expression Verification of 10 DEGs
2.9. Prediction Analysis of Phytohormones cis-Acting Regulatory Elements of 9 DEGs
3. Discussion
4. Materials and Methods
4.1. Experimental Site Description and Sampling
4.2. Determination of the Physiological and Biochemical Indexes in Watermelon Plant
4.3. Establishment of SA, ABA, and JA Determination System in Watermelon Root
4.4. RNA-Seq Sample Collection and Preparation
4.5. Quantification of Gene Expression Level and Differential Expression Analysis
4.6. Enrichment and Expression Level Analysis of Differential Genes
4.7. Quantitative Detection of Candidate Genes by RT-qPCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Everts, K.L.; Himmelstein, J.C. Fusarium wilt of watermelon: Towards sustainable management of a re-emerging plant disease. Crop Prot. 2015, 73, 93–99. [Google Scholar] [CrossRef]
- Li, C.X.; Fu, X.P.; Zhou, X.G.; Liu, S.W.; Xia, Y.; Li, N.H.; Zhang, X.X.; Wu, F.Z. Treatment With Wheat Root Exudates and Soil Microorganisms From Wheat/Watermelon Companion Cropping Can Induce Watermelon Disease Resistance Against Fusarium oxysporum f. sp. niveum. Plant Dis. 2019, 103, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Xiao, J.; Zhang, Y.; Wei, L.; Liang, Z. Dazomet application suppressed watermelon wilt by the altered soil microbial community. Sci. Rep. 2020, 10, 21668. [Google Scholar] [CrossRef] [PubMed]
- Everts, K.; Egel, D.S.; Langston, D.; Zhou, X.G. Chemical management of Fusarium wilt of watermelon. Crop. Prot. 2014, 66, 114–119. [Google Scholar] [CrossRef]
- Zhu, F.Y.; Li, J.G.; Zhang, Y.; Xiao, J.L.; Liang, Z.H. Watermelon rhizosphere soil bacterial diversity affects the occurrence of Fusarium wilt. Chin. Agric. Sci. Bull. 2018, 34, 69–76. (In Chinese) [Google Scholar] [CrossRef]
- Zhu, F.Y.; Tian, C.; Zhang, Y.; Xiao, J.L.; Wei, L.; Liang, Z.H. Effects of different fertilization treatments on soil microbial community structure and the occurrence of watermelon wilt. Chin. J. Biol. Control. 2018, 34, 589–597. (In Chinese) [Google Scholar] [CrossRef]
- Boughalleb, N.; Mhamdi, M.; El Assadi, B.; El Bourgi, Z.; Tarchoun, N.; Romdhani, M.S. Resistance Evaluation of Grafted Watermelon (Citrullus lanatus L.) Against Fusarium wilt and Fusarium Crown and Root Rot. Asian J. Plant Pathol. 2008, 2, 24–29. [Google Scholar] [CrossRef]
- Pandey, A.; Khan, M.K.; Isik, R.; Turkmen, O.; Acar, R.; Seymen, M.; Hakki, E.E. Genetic diversity and population structure of watermelon (Citrullus sp.) genotypes. 3 Biotech 2019, 9, 210. [Google Scholar] [CrossRef]
- Wang, Z.; Jia, C.; Li, J.; Huang, S.; Xu, B.; Jin, Z. Activation of salicylic acid metabolism and signal transduction can enhance resistance to Fusarium wilt in banana (Musa acuminata L. AAA group, cv. Cavendish). Funct. Integr. Genom. 2015, 15, 47–62. [Google Scholar] [CrossRef]
- Lv, H.; Cao, H.; Nawaz, M.A.; Sohail, H.; Huang, Y.; Cheng, F.; Kong, Q.; Bie, Z. Wheat Intercropping Enhances the Resistance of Watermelon to Fusarium wilt. Front. Plant Sci. 2018, 9, 696. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Del Rio, T.G.; Jones, C.D.; Tringe, S.G.; et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef] [Green Version]
- Yoo, H.; Greene, G.H.; Yuan, M.; Xu, G.; Burton, D.; Liu, L.; Marqués, J.; Dong, X. Translational Regulation of Metabolic Dynamics during Effector-Triggered Immunity. Mol. Plant 2020, 13, 88–98. [Google Scholar] [CrossRef] [Green Version]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of Salicylic Acid for the Induction of Systemic Acquired Resistance. Science 1993, 261, 754–756. [Google Scholar] [CrossRef]
- Qi, G.; Chen, J.; Chang, M.; Chen, H.; Hall, K.; Korin, J.; Liu, F.; Wang, D.; Fu, Z.Q. Pandemonium Breaks Out: Disruption of Salicylic Acid-Mediated Defense by Plant Pathogens. Mol. Plant 2018, 11, 1427–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuda, K.; Sato, M.; Stoddard, T.; Glazebrook, J.; Katagiri, F. Network Properties of Robust Immunity in Plants. PLoS Genet. 2009, 5, e1000772. [Google Scholar] [CrossRef] [Green Version]
- Van Verk, M.C.; Bol, J.F.; Linthorst, H.J. Prospecting for Genes involved in transcriptional regulation of plant defenses, a bioinformatics approach. BMC Plant Biol. 2011, 11, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, F.Y.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant–pathogen interactions. J. Plant Res. 2011, 124, 489–499. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.; Gao, P.; Liu, S.; Zhu, Z.; Amanullah, S.; Davis, A.R.; Luan, F. Comparative transcriptome analysis of two contrasting watermelon genotypes during fruit development and ripening. BMC Genom. 2017, 18, 3. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Wang, Z.; Wu, F. Companion cropping with wheat increases resistance to Fusarium wilt in watermelon and the roles of root exudates in watermelon root growth. Physiol. Mol. Plant Pathol. 2015, 90, 12–20. [Google Scholar] [CrossRef]
- Cheng, F.; Lu, J.; Gao, M.; Shi, K.; Kong, Q.; Huang, Y.; Bie, Z. Redox signaling and cbf-responsive pathway are involved in salicylic acid-improved photosynthesis and growth under chilling stress in watermelon. Front. Plant Sci. 2016, 7, 1519. [Google Scholar] [CrossRef] [Green Version]
- Guang, Y.; Luo, S.; Ahammed, G.J.; Xiao, X.; Li, J.; Zhou, Y.; Yang, Y. The OPR gene family in watermelon: Genome-wide identification and expression profiling under hormone treatments and root not nematode infection. Plant Biol. 2020, 23, 80–88. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; An, M.; Xia, Z.; Bai, X.; Wu, Y. Transcriptome analysis of watermelon (Citrullus lanatus) fruits in response to Cucumber green mottle mosaic virus (CGMMV) infection. Sci. Rep. 2017, 7, 16747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, L.; Huo, H.; Zhang, F.; Hao, W.; Xiao, L.; Dong, C.; Xu, G. The components of rice and watermelon root exudates and their effects on pathogenic fungus and watermelon defense. Plant Signal. Behav. 2016, 11, e1187357. [Google Scholar] [CrossRef] [Green Version]
- Lv, F.; Li, S.; Feng, J.; Liu, P.; Gao, Z.; Yang, Y.; Xu, Y.; Wei, J. Hydrogen peroxide burst triggers accumulation of jasmonates and salicylic acid inducing sesquiterpene biosynthesis in wounded Aquilaria sinesis. J. Plant Physiol. 2019, 234, 167–175. [Google Scholar] [CrossRef]
- Li, D.; Pang, Y.; Li, H.; Guo, D.; Wang, R.; Ma, C.; Xu, W.; Wang, L.; Wang, S. Comparative analysis of the gene expression profile under two cultivation methods reveals the critical role of ABA in grape quality promotion. Sci. Hortic. 2021, 281, 109924. [Google Scholar] [CrossRef]
- Marnett, L.J. Lipid peroxidation—DNA damage by malondialdehyde. Mutat. Res. Mol. Mech. Mutagen. 1999, 424, 83–95. [Google Scholar] [CrossRef]
- Oliva, M.; Hatan, E.; Kumar, V.; Galsurker, O.; Nisim-Levi, A.; Ovadia, R.; Galili, G.; Lewinsohn, E.; Elad, Y.; Alkan, N.; et al. Increased phenylalanine levels in plant leaves reduces susceptibility to Botrytis cinerea. Plant Sci. 2020, 290, 110289. [Google Scholar] [CrossRef] [PubMed]
- Kostyn, K.; Czemplik, M.; Kulma, A.; Bortniczuk, M.; Skała, J.; Szopa, J. Genes of phenylpropanoid pathway are activated in early response to Fusarium attack in flax plants. Plant Sci. 2012, 190, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, B.; Zheng, X.Y.; Li, J.; Yang, M.; Dong, X.; He, G.; An, C.; Deng, X.W. Salicylic acid biosynthesis is enhanced and contributes to increased biotrophic pathogen resistance in Arabidopsis hybrids. Nat. Commun. 2015, 6, 7309. [Google Scholar] [CrossRef] [Green Version]
- Nagai, A.; Torres, P.B.; Duarte, L.M.L.; Chaves, A.L.R.; Macedo, A.F.; Floh, E.I.S.; de Oliveira, L.F.; Zuccarelli, R.; dos Santos, D.Y.A.C. Signaling pathway played by salicylic acid, gentisic acid, nitric oxide, polyamines and non-enzymatic antioxidants in compatible and incompatible Solanum-tomato mottle mosaic virus interactions. Plant Sci. 2020, 290, 110274. [Google Scholar] [CrossRef] [PubMed]
- Sözen, C.; Schenk, S.T.; Boudsocq, M.; Chardin, C.; Almeida-Trapp, M.; Krapp, A.; Hirt, H.; Mithöfer, A.; Colcombet, J. Wounding and Insect Feeding Trigger Two Independent MAPK Pathways with Distinct Regulation and Kinetics. Plant Cell 2020, 32, 1988–2003. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Liu, G.; Meng, X.; Liu, Y.; Ji, X.; Li, Y.; Nie, X.; Wang, Y. A WRKY gene from Tamarix hispida, ThWRKY4, mediates abiotic stress responses by modulating reactive oxygen species and expression of stress-responsive genes. Plant Mol. Biol. 2013, 82, 303–320. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 Transcription Factor: A Node of Convergence for Jasmonate-Mediated and Salicylate-Mediated Signals in Plant Defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 2018, 173, 1454–1467.e15. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.F.; Zhou, X.E.; Chen, J.; Brunzelle, J.; Griffin, P.R.; et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 2015, 525, 269–273. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Fan, M.; Yang, M.; Zhao, J.; Zhang, W.; Su, Y.; Xiao, L.; Deng, H.; Xie, D. Injury activates Ca2+/calmodulin-dependent phosphorylation of jav1-jaz8-wrky51 complex for jasmonate biosynthesis. Mol. Cell 2018, 70, 136–149.e7. [Google Scholar] [CrossRef] [Green Version]
- Santiago, J.; Dupeux, F.; Betz, K.; Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Márquez, J.A.; Rodriguez, P.L. Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs. Plant Sci. 2012, 182, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018, 23, 3340–3351.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.J.; Xiao, L.T.; Xue, H.W. Dynamic Cytology and Transcriptional Regulation of Rice Lamina Joint Development. Plant Physiol. 2017, 174, 1728–1746. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, F.; Wang, Z.; Fang, Y.; Tong, J.; Xiang, J.; Yang, K.; Wang, R. Study on the Role of Phytohormones in Resistance to Watermelon Fusarium Wilt. Plants 2022, 11, 156. https://doi.org/10.3390/plants11020156
Zhu F, Wang Z, Fang Y, Tong J, Xiang J, Yang K, Wang R. Study on the Role of Phytohormones in Resistance to Watermelon Fusarium Wilt. Plants. 2022; 11(2):156. https://doi.org/10.3390/plants11020156
Chicago/Turabian StyleZhu, Feiying, Zhiwei Wang, Yong Fang, Jianhua Tong, Jing Xiang, Kankan Yang, and Ruozhong Wang. 2022. "Study on the Role of Phytohormones in Resistance to Watermelon Fusarium Wilt" Plants 11, no. 2: 156. https://doi.org/10.3390/plants11020156





