Role of Sulfate Transporters in Chromium Tolerance in Scenedesmus acutus M. (Sphaeropleales)
Abstract
:1. Introduction
2. Results
2.1. Analysis of Plasma-Membrane Sulfate Transporter Genes
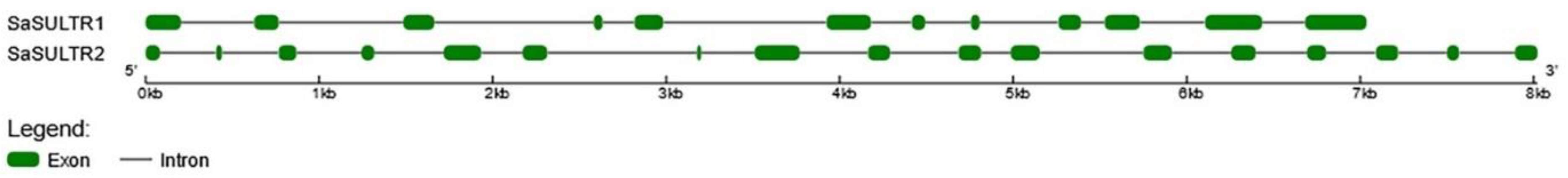
2.1.1. SaSULTRs-Type Transporters
2.1.2. SLTs-Type Transporters
2.2. Analysis of Chloroplast Sulfate Transporter Genes
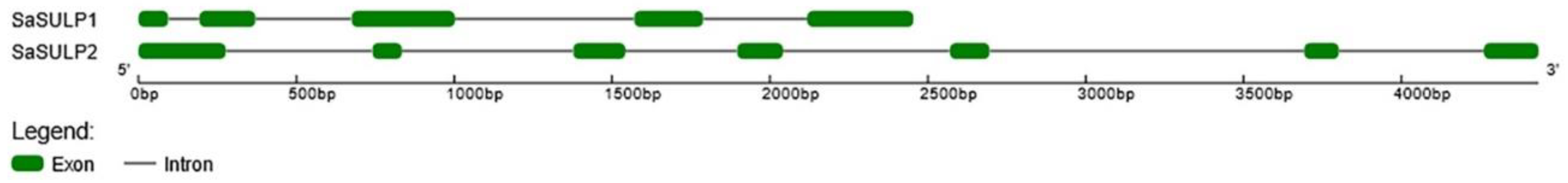
2.2.1. SaSULPs
2.2.2. SaSabc
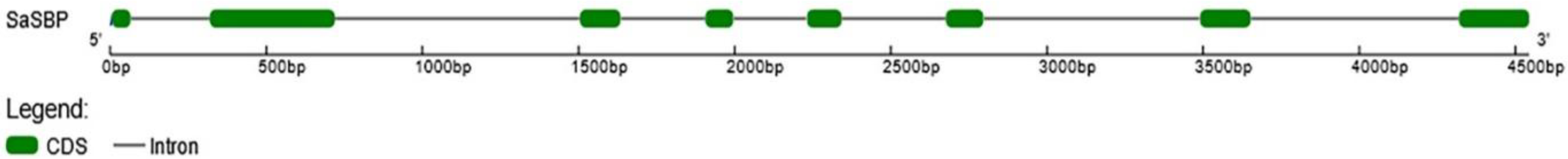
2.2.3. SaSBP
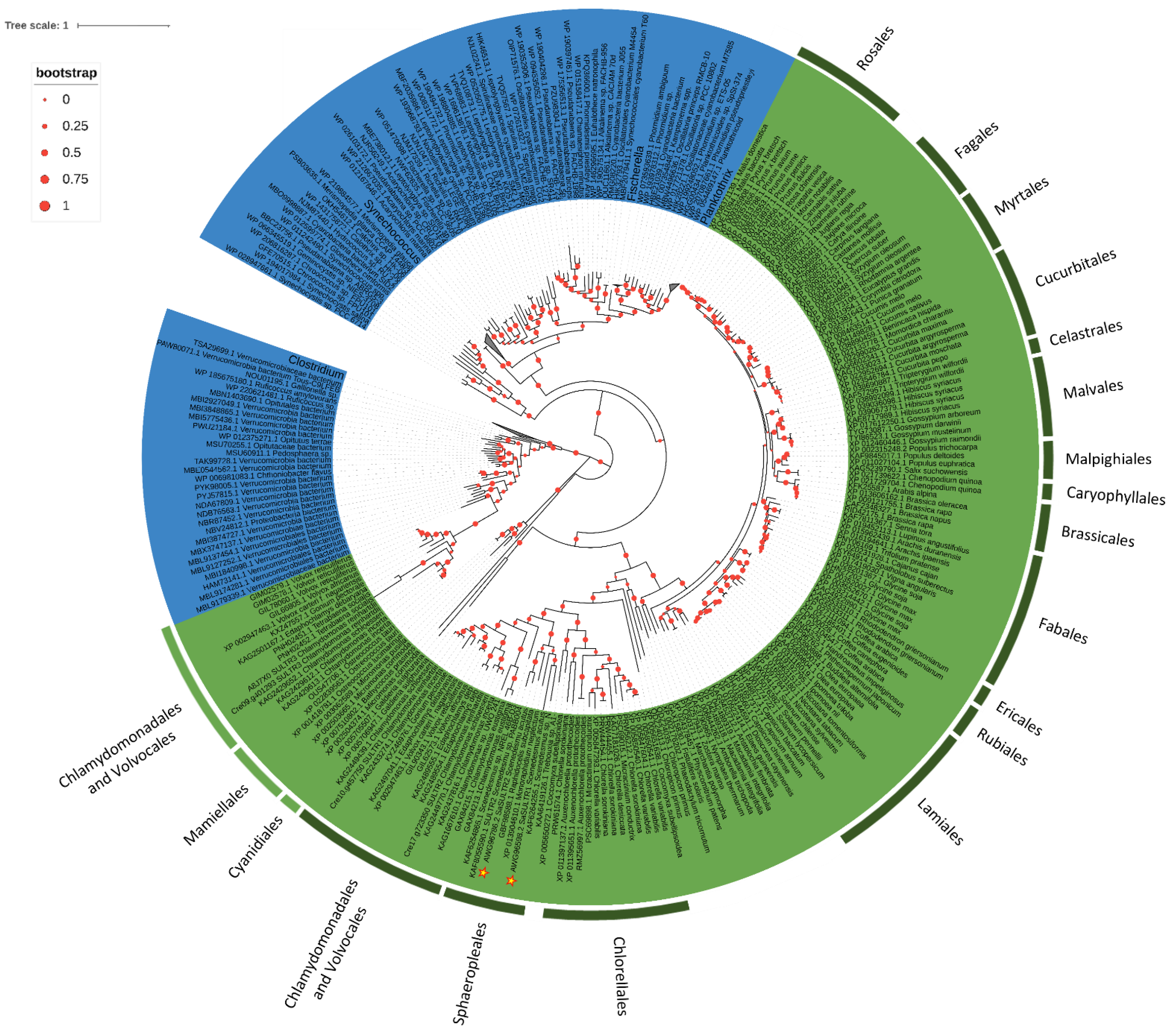
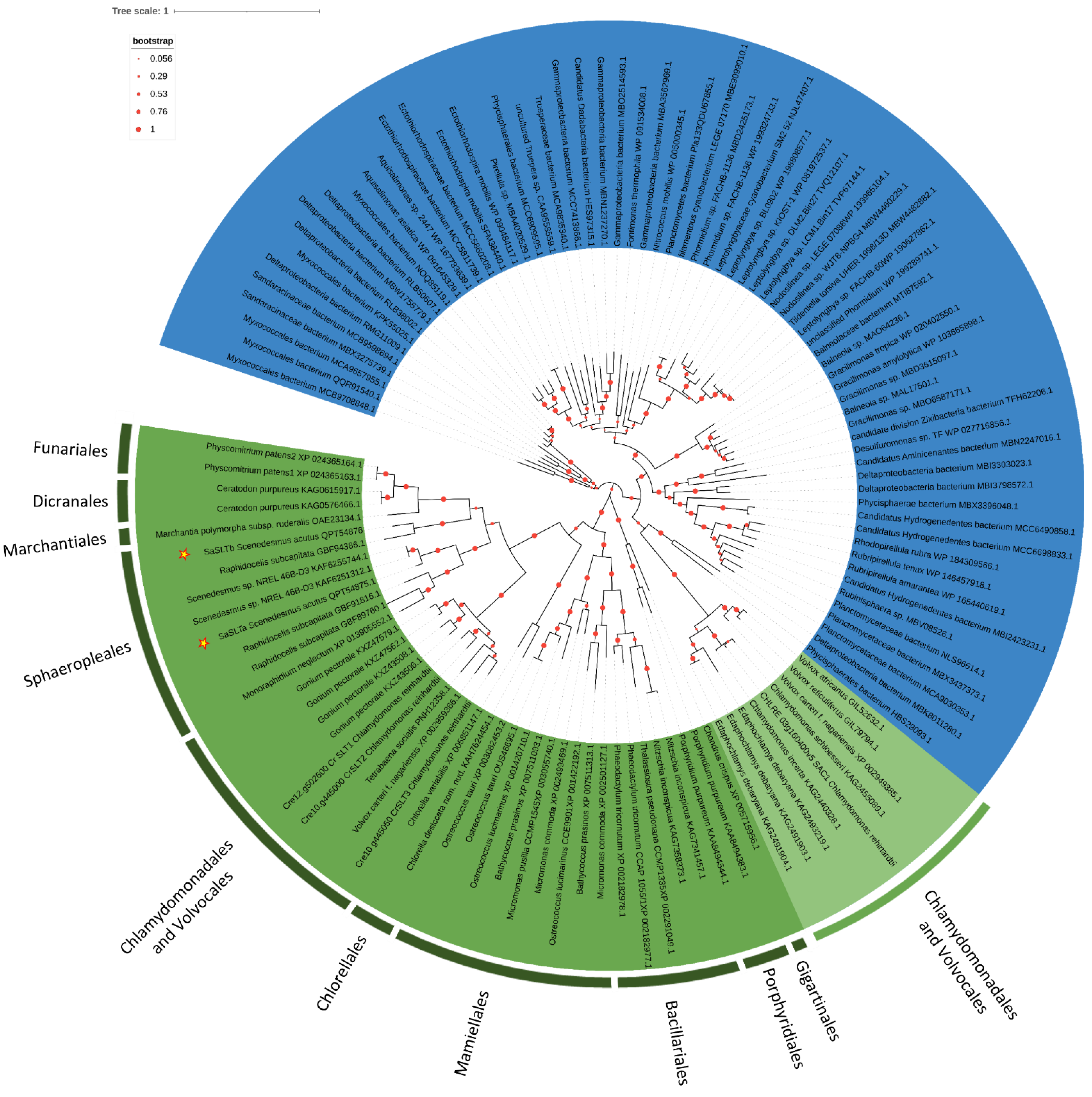
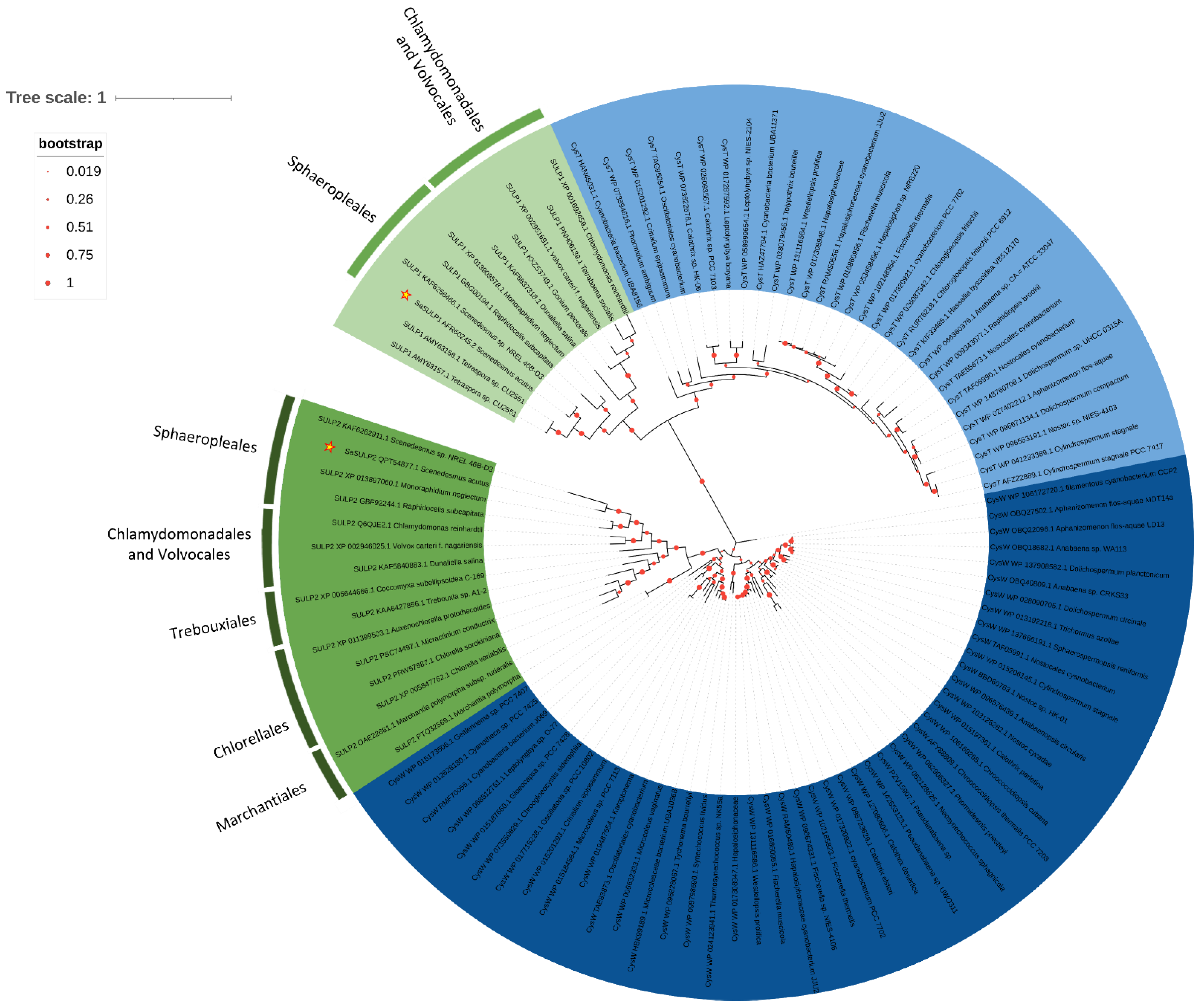
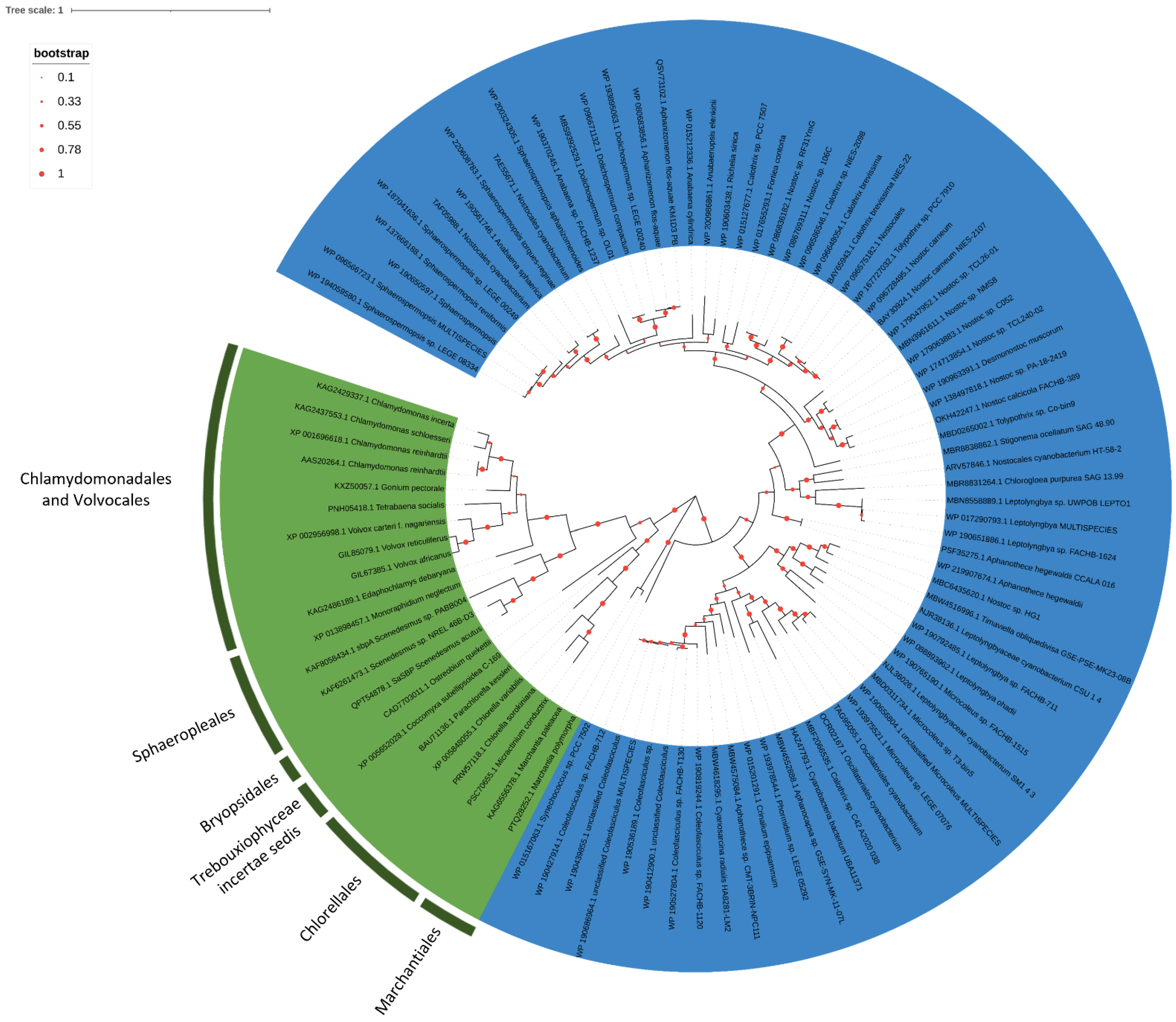
2.3. Phylogenesis Analysis of Sulfate Transporter
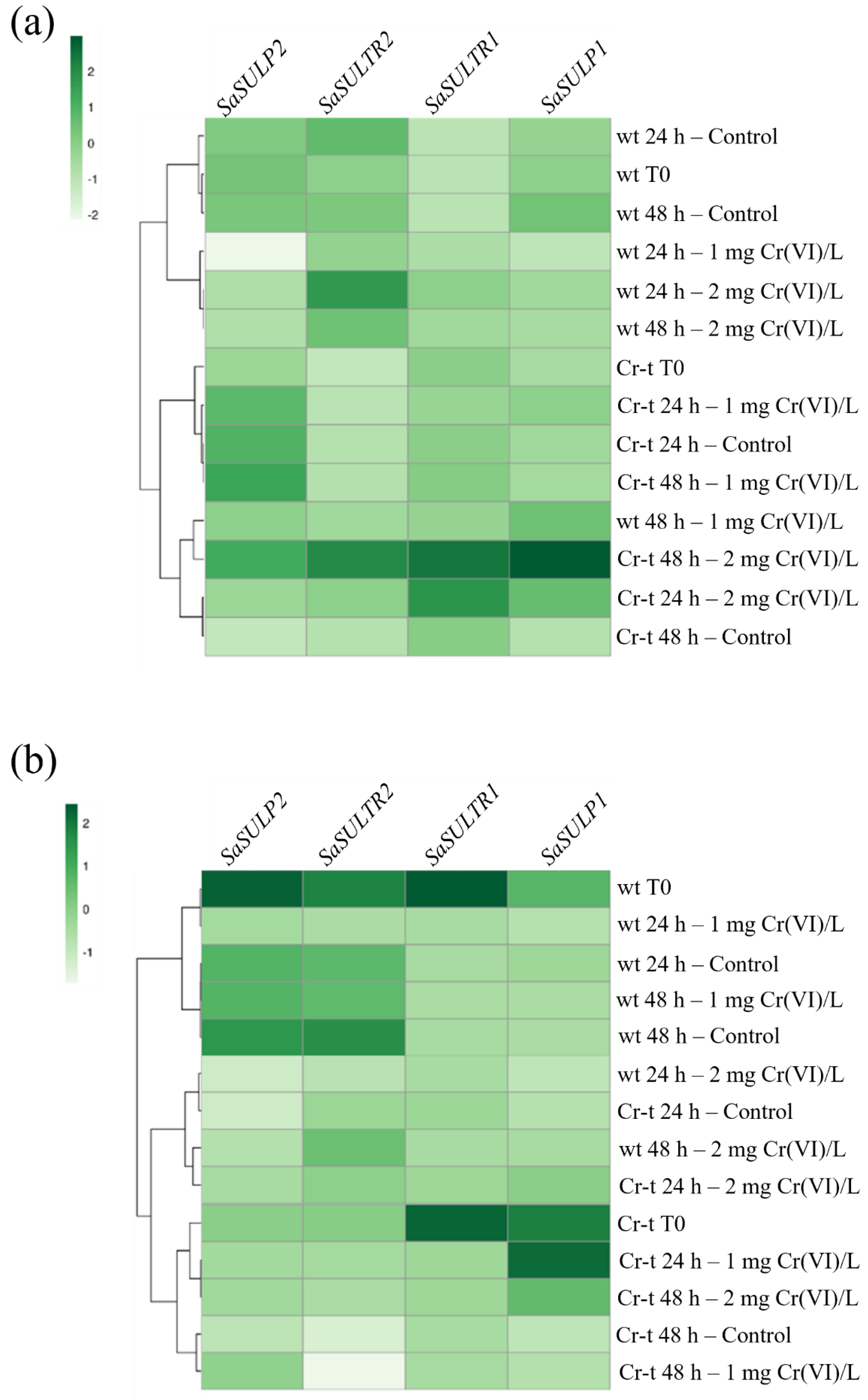
2.4. aqPCR of SULTRs and SULPs Genes
2.4.1. SULTRs Gene Expression
2.4.2. SULPs Gene Expression
3. Discussion
3.1. Analysis of Sulfate Transporter Genes
3.2. Phylogenetic Analysis of Sulfate Transporter Sequences
3.3. Analysis of SaSULTRs and SaSULPs Transcription
4. Materials and Methods
4.1. Analysis of Gene Sequences Related to Sulfate Uptake and Chloroplast Intake
Phylogenetic Analysis
4.2. In Vitro Culture of Scenedesmus Acutus
4.3. Sulfur Starvation and Chromium Treatments
4.4. Total RNA Extraction and cDNA Synthesis
4.5. Absolute Quantification Real-Time PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rausch, T.; Wachter, A. Sulfur metabolism: A versatile platform for launching defence operations. Trends Plant Sci. 2005, 10, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, A.; Bibi, I.; Niazi, N.K.; Ok, Y.S.; Murtaza, G.; Shahid, M.; Kunhikrishnan, A.; Li, D.; Mahmood, T. Chromium(vi) sorption efficiency of acid-activated banana peel over organo-montmorillonite in aqueous solutions. Int. J. Phytoremediat. 2017, 19, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Gorbi, G.; Corradi, M.G.; Invidia, M.; Bassi, M. Light intensity influences chromium bioaccumulation and toxicity in scenedesmus acutus (chlorophyceae). Ecotoxicol. Environ. Saf. 2001, 48, 36–42. [Google Scholar] [CrossRef]
- Hörcsik, Z.T.; Kovács, L.; Láposi, R.; Mészáros, I.; Lakatos, G.; Garab, G.J.P. Effect of chromium on photosystem 2 in the unicellular green alga, chlorella pyrenoidosa. Photosynthetica 2007, 45, 65–69. [Google Scholar] [CrossRef]
- Guo, X.; Feng, L.; Lemos, B.; Lou, J. DNA methylation modifications induced by hexavalent chromium. J. Environ. Sci. Health Part C Environ. Carcinog. Ecotoxicol. Rev. 2019, 37, 133–145. [Google Scholar] [CrossRef]
- Pereira, Y.; Lagniel, G.; Godat, E.; Baudouin-Cornu, P.; Junot, C.; Labarre, J. Chromate causes sulfur starvation in yeast. Toxicol. Sci. 2008, 106, 400–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiavon, M.; Pilon-Smits, E.A.H.; Wirtz, M.; Hell, R.; Malagoli, M. Interactions between chromium and sulfur metabolism in brassica juncea. J. Environ. Qual. 2008, 37, 1536–1545. [Google Scholar] [CrossRef] [Green Version]
- Holland, S.L.; Avery, S.V. Chromate toxicity and the role of sulfur. Metallomics 2011, 3, 1119–1123. [Google Scholar] [CrossRef]
- Ding, G.; Jin, Z.; Han, Y.; Sun, P.; Li, G.; Li, W. Mitigation of chromium toxicity in arabidopsis thaliana by sulfur supplementation. Ecotoxicol. Environ. Saf. 2019, 182, 109379. [Google Scholar] [CrossRef]
- Ackerley, D.F.; Barak, Y.; Lynch, S.V.; Curtin, J.; Matin, A. Effect of Chromate Stress on Escherichia coli K-12. J. Bacteriol. 2006, 188, 3371–3381. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.D.; Thompson, M.R.; VerBerkmoes, N.C.; Chourey, K.; Shah, M.; Zhou, J.; Hettich, R.L.; Thompson, D.K. Molecular Dynamics of the Shewanella Oneidensis Response to Chromate Stress* S. Mol. Cell. Proteom. 2006, 5, 1054–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chourey, K.; Thompson, M.R.; Morrell-Falvey, J.; Verberkmoes, N.C.; Brown, S.D.; Shah, M.; Zhou, J.; Doktycz, M.; Hettich, R.L.; Thompson, D.K. Global molecular and morphological effects of 24-h chromium(vi) exposure on shewanella oneidensis mr-1. Appl. Environ. Microbiol. 2006, 72, 6331–6344. [Google Scholar] [CrossRef] [Green Version]
- Henne, K.L.; Turse, J.E.; Nicora, C.D.; Lipton, M.S.; Tollaksen, S.L.; Lindberg, C.; Babnigg, G.; Giometti, C.S.; Nakatsu, C.H.; Thompson, D.K.; et al. Global proteomic analysis of the chromate response in Arthrobacter sp. Strain fb24. J. Proteome Res. 2009, 8, 1704–1716. [Google Scholar] [CrossRef]
- Thompson, D.K.; Chourey, K.; Wickham, G.S.; Thieman, S.B.; VerBerkmoes, N.C.; Zhang, B.; McCarthy, A.T.; Rudisill, M.A.; Shah, M.; Hettich, R.L. Proteomics reveals a core molecular response of Pseudomonas putida F1 to acute chromate challenge. BMC Genom. 2010, 11, 311. [Google Scholar] [CrossRef] [Green Version]
- Monsieurs, P.; Moors, H.; Van Houdt, R.; Janssen, P.J.; Janssen, A.; Coninx, I.; Mergeay, M.; Leys, N. Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network. BioMetals 2011, 24, 1133–1151. [Google Scholar] [CrossRef]
- Takahashi, H.; Buchner, P.; Yoshimoto, N.; Hawkesford, M.J.; Shiu, S.H. Evolutionary relationships and functional diversity of plant sulfate transporters. Front. Plant Sci. 2011, 2, 119. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Ballester, D.; Grossman, A.R. Sulfur: From acquisition to assimilation. In The Chlamydomonas Sourcebook; Elsevier: New York, NY, USA, 2009; pp. 159–187. [Google Scholar]
- Bromke, M.A.; Hoefgen, R.; Hesse, H. Phylogenetic aspects of the sulfate assimilation genes from Thalassiosira pseudonana. Amino Acids 2013, 44, 1253–1265. [Google Scholar] [CrossRef] [Green Version]
- Alper, S.L.; Sharma, A.K. The slc26 gene family of anion transporters and channels. Mol. Asp. Med. 2013, 34, 494–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibagaki, N.; Grossman, A.R. The role of the stas domain in the function and biogenesis of a sulfate transporter as probed by random mutagenesis. J. Biol. Chem. 2006, 281, 22964–22973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giordano, M.; Prioretti, L. Sulphur and algae: Metabolism, ecology and evolution. In The Physiology of Microalgae; Borowitzka, M.A., Beardall, J., Raven, J.A., Eds.; Springer International Publishing: Cham, Germany, 2016; pp. 185–209. [Google Scholar]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pootakham, W.; Gonzalez-Ballester, D.; Grossman, A.R. Identification and regulation of plasma membrane sulfate transporters in chlamydomonas. Plant Physiol. 2010, 153, 1653–1668. [Google Scholar] [CrossRef] [Green Version]
- Melis, A.; Chen, H.-C. Chloroplast sulfate transport in green algae—Genes, proteins and effects. Photosynth. Res. 2005, 86, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, P.; Melis, A. The chloroplast sulfate transport system in the green alga Chlamydomonas reinhardtii. Planta 2008, 228, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Giordano, M.; Norici, A.; Hell, R. Sulfur and phytoplankton: Acquisition, metabolism and impact on the environment. New Phytol. 2005, 166, 371–382. [Google Scholar] [CrossRef]
- Gorbi, G.; Torricelli, E.; Pawlik-Skowrońska, B.; di Toppi, L.S.; Zanni, C.; Corradi, M.G. Differential responses to cr(vi)-induced oxidative stress between cr-tolerant and wild-type strains of Scenedesmus acutus (chlorophyceae). Aquat. Toxicol. 2006, 79, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Gorbi, G.; Zanni, C.; Corradi, M.G. Sulfur starvation and chromium tolerance in Scenedesmus acutus: A possible link between metal tolerance and the regulation of sulfur uptake/assimilation processes. Aquat. Toxicol. 2007, 84, 457–464. [Google Scholar] [CrossRef]
- Marieschi, M.; Gorbi, G.; Zanni, C.; Sardella, A.; Torelli, A. Increase of chromium tolerance in Scenedesmus acutus after sulfur starvation: Chromium uptake and compartmentalization in two strains with different sensitivities to cr(vi). Aquat. Toxicol. 2015, 167, 124–133. [Google Scholar] [CrossRef]
- Sardella, A.; Marieschi, M.; Mercatali, I.; Zanni, C.; Gorbi, G.; Torelli, A. The relationship between sulfur metabolism and tolerance of hexavalent chromium in Scenedesmus acutus (spheropleales): Role of atp sulfurylase. Aquat. Toxicol. 2019, 216, 105320. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, M.; Torelli, A.; Marieschi, M.; Cozza, R. Role of DNA methylation in the chromium tolerance of Scenedesmus acutus (chlorophyceae) and its impact on the sulfate pathway regulation. Plant Sci. Int. J. Exp. Plant Biol. 2020, 301, 110680. [Google Scholar] [CrossRef]
- Chen, B.-L.; Mhuantong, W.; Ho, S.-H.; Chang, J.-S.; Zhao, X.-Q.; Bai, F.-W. Genome sequencing, assembly, and annotation of the self-flocculating microalga Scenedesmus obliquus as-6-11. BMC Genom. 2020, 21, 743. [Google Scholar] [CrossRef]
- Astafyeva, Y.; Alawi, M.; Indenbirken, D.; Danso, D.; Grundhoff, A.; Hanelt, D.; Streit, W.R.; Krohn, I. Draft genome sequence of the green alga Scenedesmus acuminatus SAG 38.81. Microbiol. Resour. Announc. 2020, 9, e01278-19. [Google Scholar] [CrossRef]
- Carreres, B.M.; de Jaeger, L.; Springer, J.; Barbosa, M.J.; Breuer, G.; van den End, E.J.; Kleinegris, D.M.M.; Schäffers, I.; Wolbert, E.J.H.; Zhang, H.; et al. Draft genome sequence of the oleaginous green alga Tetradesmus obliquus UTEX 393. Genome Announc. 2017, 5, e01449-16. [Google Scholar] [CrossRef] [Green Version]
- Most, P.; Papenbrock, J. Possible roles of plant sulfurtransferases in detoxification of cyanide, reactive oxygen species, selected heavy metals and arsenate. Molecules 2015, 20, 1410–1423. [Google Scholar] [CrossRef]
- Giordano, M.; Raven, J.A. Nitrogen and sulfur assimilation in plants and algae. Aquat. Bot. 2014, 118, 45–61. [Google Scholar] [CrossRef]
- Waite, D.W.; Chuvochina, M.; Pelikan, C.; Parks, D.H.; Yilmaz, P.; Wagner, M.; Loy, A.; Naganuma, T.; Nakai, R.; Whitman, W.B.; et al. Proposal to reclassify the proteobacterial classes deltaproteobacteria and oligoflexia, and the phylum thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 2020, 70, 5972–6016. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, T.; Nagai, T.; Kapoor, M.; Sugita, M.; Ito, M.; Ito, S.; Tsudzuki, J.; Nakashima, K.; Tsudzuki, T.; Suzuki, Y.; et al. Complete nucleotide sequence of the chloroplast genome from the green alga chlorella vulgaris: The existence of genes possibly involved in chloroplast division. Proc. Natl. Acad. Sci. USA 1997, 94, 5967–5972. [Google Scholar] [CrossRef] [Green Version]
- Yildiz, F.H.; Davies, J.P.; Grossman, A. Sulfur availability and the SAC1 gene control adenosine triphosphate sulfurylase gene expression in chlamydomonas reinhardtii. Plant Physiol. 1996, 112, 669–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkesford, M.J. Transporter gene families in plants: The sulphate transporter gene family—Redundancy or specialization? Physiol. Plant. 2003, 117, 155–163. [Google Scholar] [CrossRef]
- Zhang, Z.; Shrager, J.; Jain, M.; Chang, C.W.; Vallon, O.; Grossman, A.R. Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot. Cell 2004, 3, 1331–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollock, S.V.; Pootakham, W.; Shibagaki, N.; Moseley, J.L.; Grossman, A.R. Insights into the acclimation of Chlamydomonas reinhardtii to sulfur deprivation. Photosynth. Res. 2005, 86, 475–489. [Google Scholar] [CrossRef]
- Appenroth, K.-J.; Luther, A.; Jetschke, G.; Gabrys, H. Modification of chromate toxicity by sulphate in duckweeds (lemnaceae). Aquat. Toxicol. 2008, 89, 167–171. [Google Scholar] [CrossRef]
- González-Ballester, D.; Casero, D.; Cokus, S.; Pellegrini, M.; Merchant, S.S.; Grossman, A.R. Rna-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 2010, 22, 2058–2084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gigolashvili, T.; Kopriva, S. Transporters in plant sulfur metabolism. Front. Plant Sci. 2014, 5, 442. [Google Scholar] [CrossRef] [Green Version]
- Schiavon, M.; Wirtz, M.; Borsa, P.; Quaggiotti, S.; Hell, R.; Malagoli, M. Chromate differentially affects the expression of a high-affinity sulfate transporter and isoforms of components of the sulfate assimilatory pathway in Zea mays (L.). Plant Biol. 2007, 9, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Shibagaki, N.; Grossman, A.R. Binding of cysteine synthase to the stas domain of sulfate transporter and its regulatory consequences. J. Biol. Chem. 2010, 285, 25094–25102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, F.W.; Hawkesford, M.J.; Ealing, P.M.; Clarkson, D.T.; Vanden Berg, P.J.; Belcher, A.R.; Warrilow, A.G. Regulation of expression of a cdna from barley roots encoding a high affinity sulphate transporter. Plant J. Cell Mol. Biol. 1997, 12, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Lappartient, A.G.; Touraine, B. Demand-driven control of root atp sulfurylase activity and SO42-uptake in intact canola (the role of phloem-translocated glutathione). Plant Physiol. 1996, 111, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Bolchi, A.; Petrucco, S.; Tenca, P.L.; Foroni, C.; Ottonello, S. Coordinate modulation of maize sulfate permease and atp sulfurylase mrnas in response to variations in sulfur nutritional status: Stereospecific down-regulation by L-cysteine. Plant Mol. Biol. 1999, 39, 527–537. [Google Scholar] [CrossRef]
- Davidian, J.-C.; Kopriva, S. Regulation of sulfate uptake and assimilation—The same or not the same? Mol. Plant 2010, 3, 314–325. [Google Scholar] [CrossRef]
- Moseley, J.L.; Gonzalez-Ballester, D.; Pootakham, W.; Bailey, S.; Grossman, A.R. Genetic interactions between regulators of chlamydomonas phosphorus and sulfur deprivation responses. Genetics 2009, 181, 889–905. [Google Scholar] [CrossRef] [Green Version]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. Snrk2 protein kinases—Key regulators of plant response to abiotic stresses. OMICS A J. Integr. Biol. 2011, 15, 859–872. [Google Scholar] [CrossRef]
- Boudsocq, M.; Barbier-Brygoo, H.; Laurière, C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 41758–41766. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.-Y.; Chao, D.-Y.; Koprivova, A.; Danku, J.; Wirtz, M.; Müller, S.; Sandoval, F.J.; Bauwe, H.; Roje, S.; Dilkes, B.; et al. Nuclear localised more sulphur accumulation1 epigenetically regulates sulphur homeostasis in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006298. [Google Scholar] [CrossRef]
- Cozza, D.; Torelli, A.; Veltri, A.; Ferrari, M.; Marieschi, M.; Cozza, R. Ultrastructural features, chromium content and in situ immunodetection of 5-methyl-cytosine following cr (vi) treatment in two strains of Scenedesmus acutus M. (chlorophyceae) with different chromium sensitivity. Eur. J. Phycol. 2016, 51, 294–306. [Google Scholar] [CrossRef] [Green Version]
- Wirtz, M.; Hell, R. Dominant-negative modification reveals the regulatory function of the multimeric cysteine synthase protein complex in transgenic tobacco. Plant Cell 2007, 19, 625–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, F.H.; Heeg, C.; Queiroz, R.; Bauer, A.; Wirtz, M.; Hell, R. Mitochondrial serine acetyltransferase functions as a pacemaker of cysteine synthesis in plant cells. Plant Physiol. 2008, 148, 1055–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heeg, C.; Kruse, C.; Jost, R.; Gutensohn, M.; Ruppert, T.; Wirtz, M.; Hell, R. Analysis of the arabidopsis o-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell 2008, 20, 168–185. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Kusano, M.; Oikawa, A.; Fukushima, A.; Noji, M.; Saito, K. Physiological roles of the β-substituted alanine synthase gene family in arabidopsis. Plant Physiol. 2007, 146, 310–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, M.; Mochida, K.; Kato, T.; Tabata, S.; Yoshimoto, N.; Noji, M.; Saito, K. Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis. Plant Cell 2008, 20, 2484–2496. [Google Scholar] [CrossRef] [Green Version]
- Krueger, S.; Niehl, A.; Martin, M.C.L.; Steinhauser, D.; Donath, A.; Hildebrandt, T.; Romero, L.C.; Hoefgen, R.; Gotor, C.; Hesse, H. Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopsis. Plant Cell Environ. 2009, 32, 349–367. [Google Scholar] [CrossRef]
- Carfagna, S.; Salbitani, G.; Vona, V.; Esposito, S. Changes in cysteine and o-acetyl-l-serine levels in the microalga chlorella sorokiniana in response to the s-nutritional status. J. Plant Physiol. 2011, 168, 2188–2195. [Google Scholar] [CrossRef] [PubMed]
- Allmer, J.; Naumann, B.; Markert, C.; Zhang, M.; Hippler, M. Mass spectrometric genomic data mining: Novel insights into bioenergetic pathways in chlamydomonas reinhardtii. Proteomics 2006, 6, 6207–6220. [Google Scholar] [CrossRef] [PubMed]
- Ravina, C.G.; Chang, C.-I.; Tsakraklides, G.P.; McDermott, J.P.; Vega, J.M.; Leustek, T.; Gotor, C.; Davies, J.P. The sac mutants of Chlamydomonas reinhardtii reveal transcriptional and posttranscriptional control of cysteine biosynthesis. Plant Physiol. 2002, 130, 2076–2084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. Weblogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; von Heijne, G. Chlorop, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. A Publ. Protein Soc. 1999, 8, 978–984. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. CABIOS Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Corradi, M.G.; Gorbi, G.; Bassi, M. Hexavalent chromium induces gametogenesis in the freshwater alga Scenedesmus acutus. Ecotoxicol. Environ. Saf. 1995, 30, 106–110. [Google Scholar] [CrossRef]
- Torricelli, E.; Gorbi, G.; Pawlik-Skowronska, B.; Di Toppi, L.S.; Corradi, M.G. Cadmium tolerance, cysteine and thiol peptide levels in wild type and chromium-tolerant strains of Scenedesmus acutus (chlorophyceae). Aquat. Toxicol. 2004, 68, 315–323. [Google Scholar] [CrossRef]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leong, D.T.; Gupta, A.; Bai, H.F.; Wan, G.; Yoong, L.F.; Too, H.-P.; Chew, F.T.; Hutmacher, D.W. Absolute quantification of gene expression in biomaterials research using real-time PCR. Biomaterials 2007, 28, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Whelan, J.A.; Russell, N.B.; Whelan, M.A. A method for the absolute quantification of cdna using real-time PCR. J. Immunol. Methods 2003, 278, 261–269. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. Clustvis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]


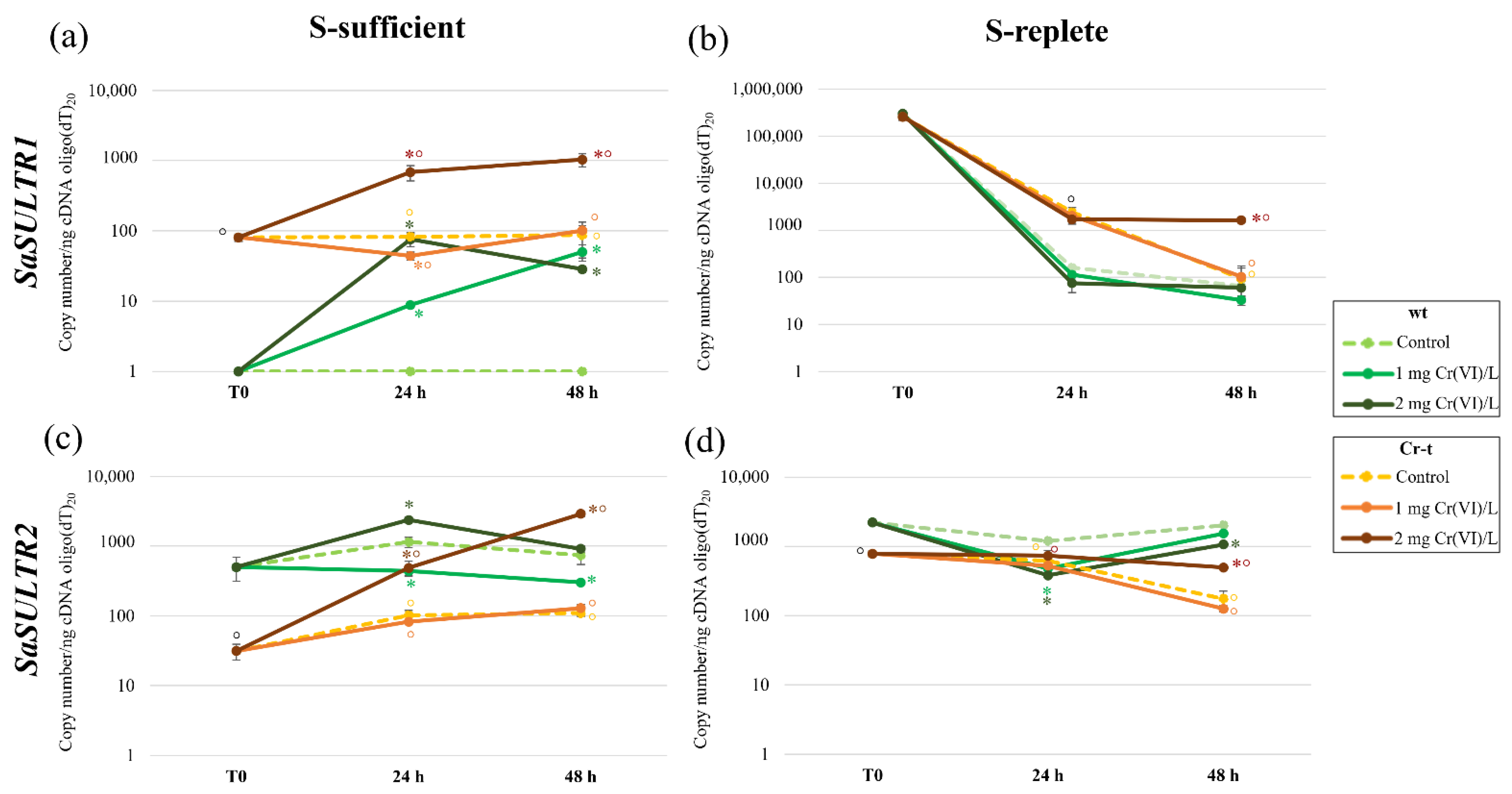

| Condition | Strain | Copy Number/ng cDNA Oligo(dT)20 | |||
|---|---|---|---|---|---|
| SaSULTR1 | SaSULTR2 | SUM of SaSULTR1 and SaSULTR2 | |||
| S-sufficient | T0 | wt | 0 ± 0 e | 500 ± 54 e | 500 ± 54 h |
| Cr-t | 81 ± 9 cd | 31 ± 8 h | 112 ± 7 g | ||
| 24 h | wt | 0 ± 0 e | 1150 ± 64 bc | 1150 ± 64 de | |
| Cr-t | 82 ±13 cd | 102 ± 18 g | 184 ± 28 g | ||
| 48 h | wt | 0 ± 0 e | 737 ± 98 de | 737 ± 98 ef | |
| Cr-t | 87 ± 32 cd | 109 ± 11 g | 196 ± 87 g | ||
| S-replete | T0 | wt | 294,000 ± 2777 a | 2210 ± 60 a | 296,200 ± 2834 a |
| Cr-t | 256,000 ± 39,939 a | 780 ± 72 cd | 256,780 ± 39,981 a | ||
| 24 h | wt | 161 ± 14 c | 1195 ± 17 b | 1356 ± 27 cd | |
| Cr-t | 2409 ± 614 b | 619 ± 71 de | 3029 ± 680 b | ||
| 48 h | wt | 65 ± 20 d | 2011 ± 36 a | 2077 ± 55 bc | |
| Cr-t | 94 ± 49 cd | 174 ± 49 f | 268 ± 87 g | ||
| Gene | Sequence | Efficiency (%) | |
|---|---|---|---|
| SaSULTR1 | FW | 5′-TGGCTACCCTTCCAGTATGTTG-3′ | 94.8 |
| BW | 5′-GGACGTGGACTCAAGCATGT-3′ | ||
| SaSULTR2 | FW | 5′-AAGGTGATCCAGGTGGCATT-3′ | 90.2 |
| BW | 5′-CGCCCCGCTGGTGAA-3′ | ||
| SaSULP1 | FW | 5′-CGAGTTTGGCAGCATTGTCA-3′ | 97.7 |
| BW | 5′-ACTGCTCAAGGCACTGGAAGA-3′ | ||
| SaSULP2 | FW | 5′-GCCAACCCTCTGCAGGTATT-3′ | 90.9 |
| BW | 5′-TTGGTCAGGATGACGCCATA-3′ | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrari, M.; Cozza, R.; Marieschi, M.; Torelli, A. Role of Sulfate Transporters in Chromium Tolerance in Scenedesmus acutus M. (Sphaeropleales). Plants 2022, 11, 223. https://doi.org/10.3390/plants11020223
Ferrari M, Cozza R, Marieschi M, Torelli A. Role of Sulfate Transporters in Chromium Tolerance in Scenedesmus acutus M. (Sphaeropleales). Plants. 2022; 11(2):223. https://doi.org/10.3390/plants11020223
Chicago/Turabian StyleFerrari, Michele, Radiana Cozza, Matteo Marieschi, and Anna Torelli. 2022. "Role of Sulfate Transporters in Chromium Tolerance in Scenedesmus acutus M. (Sphaeropleales)" Plants 11, no. 2: 223. https://doi.org/10.3390/plants11020223
APA StyleFerrari, M., Cozza, R., Marieschi, M., & Torelli, A. (2022). Role of Sulfate Transporters in Chromium Tolerance in Scenedesmus acutus M. (Sphaeropleales). Plants, 11(2), 223. https://doi.org/10.3390/plants11020223






