Modulation of Photosynthesis and ROS Scavenging Response by Beneficial Bacteria in Olea europaea Plantlets under Salt Stress Conditions
Abstract
:1. Introduction
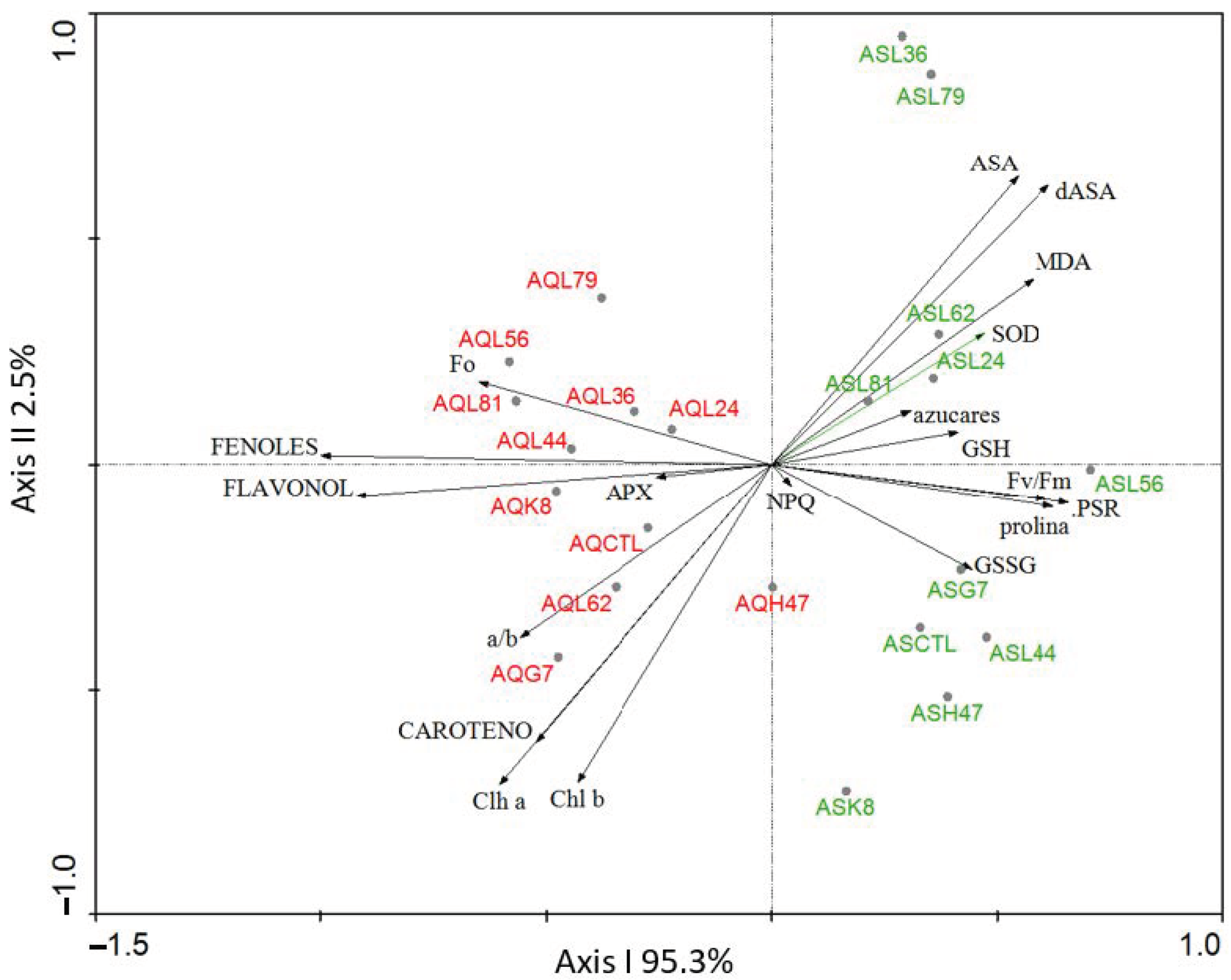
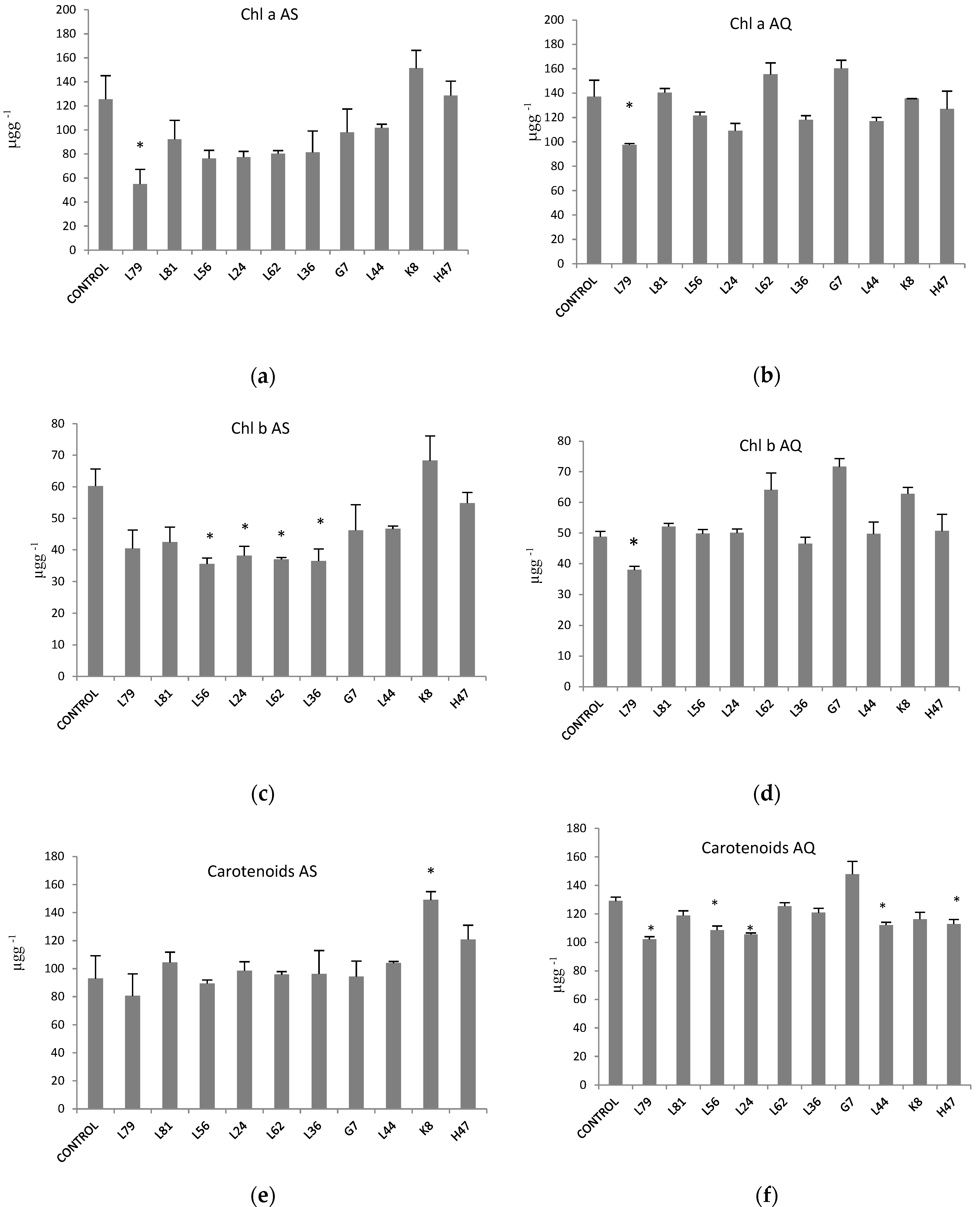
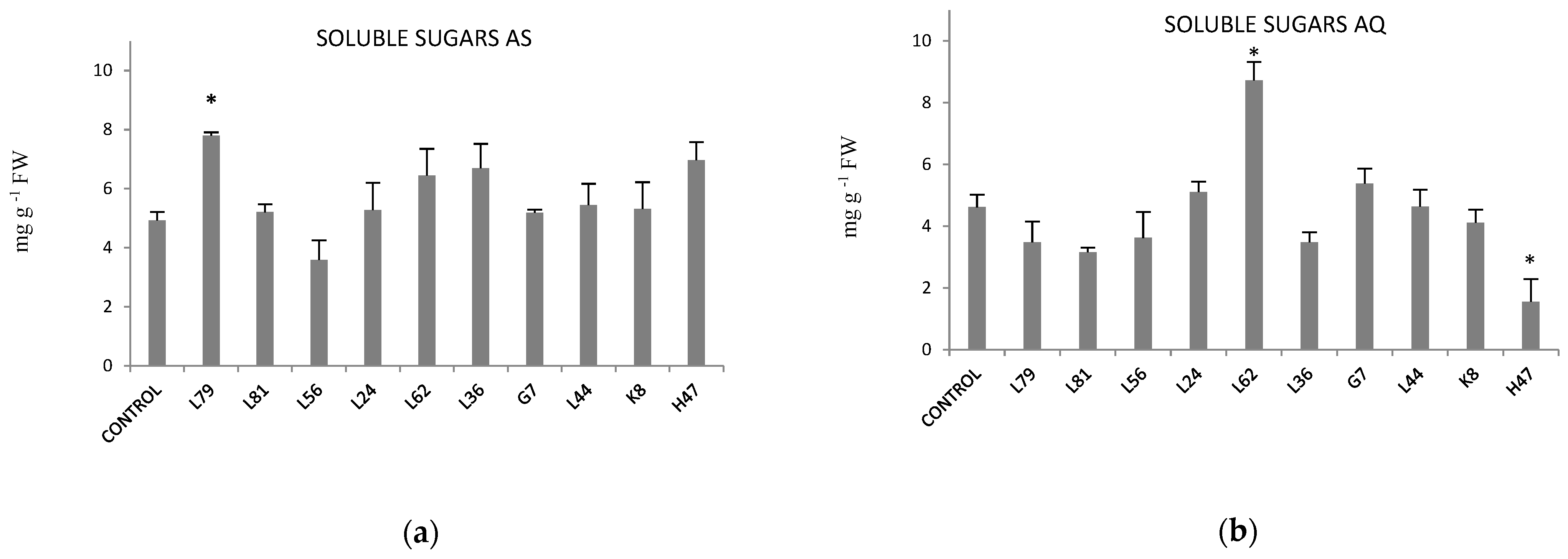
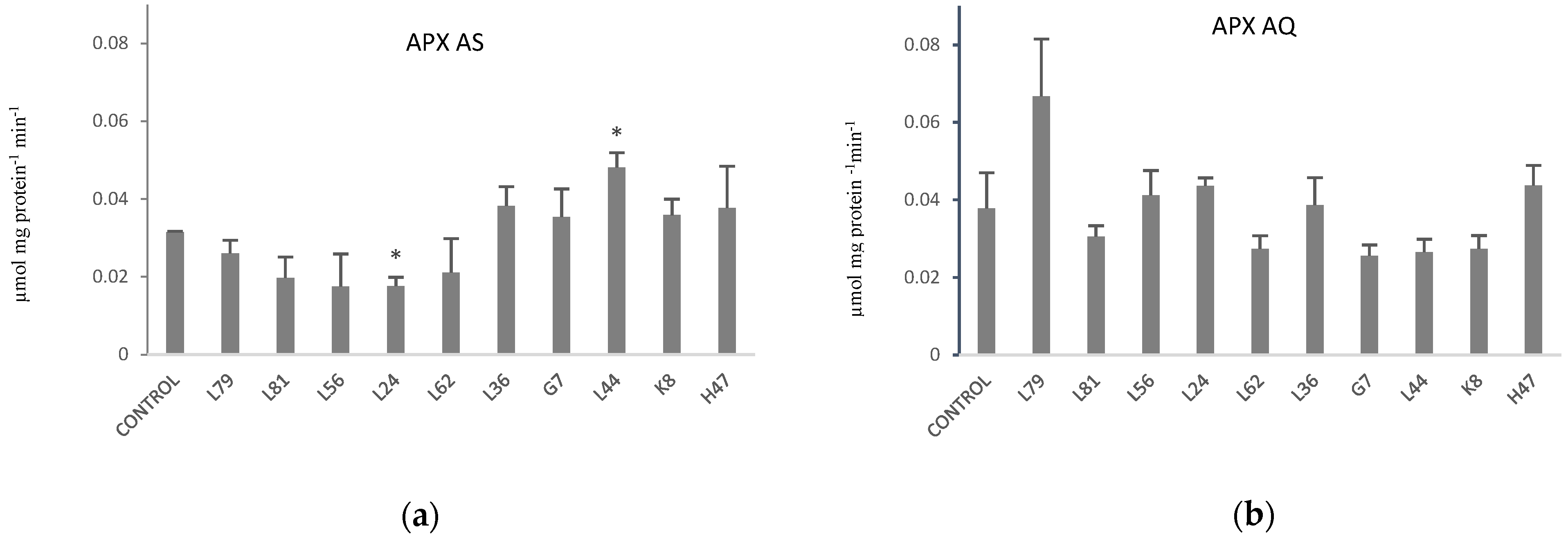
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Bacterial Strains and Inoculum Preparation
4.3. Experimental Set Up
4.4. Photosynthesis (Chlorophyll Fluorescence)
4.5. Photosynthetic Pigments: Chlorophyll a, Chlorophyll b and Carotenoids
4.6. Osmoprotectants: Proline and Soluble Sugars
4.7. Enzymatic Antioxidants: Superoxide Dismutase (SOD) and Ascorbate Peroxidase (APX)
4.8. Non-Enzymatic Antioxidants: Ascorbate, Glutathione, Phenols and Flavonols
4.9. Malondialdehyde
4.10. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Malhi, G.S.; Kaur, M.; Kaushik, P. Impact of Climate Change on Agriculture and Its Mitigation Strategies: A Review. Sustainability 2021, 13, 1318. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; De Paepe, R.; Foyer, C.H. Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 2007, 12, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid. Redox Signal. 2009, 11, 861–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foyer, C.H.; Bloom, A.J.; Queval, G.; Noctor, G. Photorespiratory metabolism: Genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 2009, 60, 455–484. [Google Scholar] [CrossRef]
- Pfannschmidt, T.; Bräutigam, K.; Wagner, R.; Dietzel, L.; Schröter, Y.; Steiner, S.; Nykytenko, A. Potential regulation of gene expression in photosynthetic cells by redox and energy state: Approaches towards better understanding. Ann. Bot. 2009, 103, 599–607. [Google Scholar] [CrossRef] [Green Version]
- Woodson, J.D.; Chory, J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008, 9, 383–395. [Google Scholar] [CrossRef]
- Karpinski, S.; Reynolds, H.; Karpinska, B.; Wingsle, G.; Creissen, G.; Mullineaux, P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 1999, 284, 654–657. [Google Scholar] [CrossRef]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef]
- Garg, N.; Manchanda, G. ROS generation in plants, boon or bane? Plant Biosyst. 2009, 143, 81–96. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Shigeoka, S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011, 155, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Noctor, G.; Foyer, C.H. Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 2016, 171, 1581–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullineaux, P.M.; Baker, N.R. Oxidative stress, antagonistic signaling for acclimation or cell death? Plant Physiol. 2010, 154, 521–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griebel, T.; Zeier, J. Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: Phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 2008, 147, 790–801. [Google Scholar] [CrossRef] [Green Version]
- Muühlenbock, P.; Szechynska-Hebda, M.; Płaszczyca, M.; Baudo, M.; Mateo, A.; Mullineaux, P.M.; Parker, J.E.; Karpińska, B.; Karpiński, S. Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 2008, 20, 2339–2356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilangumaran, G.; Smith, D.L. Plant growth promoting rhizobacteria in amelioration of salinity stress: A systems biology perspective. Front. Plant Sci. 2017, 8, 1768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Singh, S.; Gaurav, A.K.; Srivastava, S.; Verma, J.P. Plant growth-promoting bacteria, biological tools for the mitigation of salinity stress in plants. Front. Microbiol. 2020, 11, 1216. [Google Scholar] [CrossRef]
- Rojas-Tapias, D.; Moreno-Galván, A.; Pardo-Díaz, S.; Obando, M.; Rivera, D.; Bonilla, R. Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Appl. Soil Ecol. 2012, 61, 264–272. [Google Scholar] [CrossRef]
- Gutierrez Albanchez, E.; García-Villaraco, A.; Lucas, J.A.; Gutierrez, F.J.; Ramos-Solano, B. Priming fingerprint induced by Bacillus amyloliquefaciens QV15, a common pattern in Arabidopsis thaliana and in field-grown blackberry. J. Plant Interact. 2018, 13, 398–408. [Google Scholar] [CrossRef]
- Galicia-Campos, E.; Ramos-Solano, B.; Montero-Palmero, M.; Gutierrez-Mañero, F.J.; García-Villaraco, A. Management of Plant Physiology with Beneficial Bacteria to Improve Leaf Bioactive Profiles and Plant Adaptation under Saline Stress in Olea europea L. Foods 2020, 9, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamane, K.; KawAsAki, M.; Taniguchi, M.; Miyake, H. Correlation between chloroplast ultrastructure and chlorophyll fluorescence characteristics in the leaves of rice (Oryza sativa L.) grown under salinity. Plant Prod. Sci. 2008, 11, 139–145. [Google Scholar] [CrossRef]
- Lutts, S.; Kinet, J.; Bouharmont, J. NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann. Bot. 1996, 78, 389–398. [Google Scholar] [CrossRef]
- Tsai, Y.; Chen, K.; Cheng, T.; Lee, C.; Lin, S.; Tung, C. Chlorophyll fluorescence analysis in diverse rice varieties reveals the positive correlation between the seedlings salt tolerance and photosynthetic efficiency. BMC Plant Biol. 2019, 19, 403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonilla, A.; Sarria, A.; Algar, E.; Ledesma, F.M.; Solano, B.R.; Fernandes, J.; Mañero, F.G. Microbe associated molecular patterns from rhizosphere bacteria trigger germination and Papaver somniferum metabolism under greenhouse conditions. Plant Physiol. Biochem. 2014, 74, 133–140. [Google Scholar] [CrossRef]
- Guerfel, M.; Baccouri, O.; Boujnah, D.; Chaïbi, W.; Zarrouk, M. Impacts of water stress on gas exchange, water relations, chlorophyll content and leaf structure in the two main Tunisian olive (Olea europaea L.) cultivars. Sci. Hortic. 2009, 119, 257–263. [Google Scholar] [CrossRef]
- Takahashi, S.; Badger, M.R. Photoprotection in plants, a new light on photosystem II damage. Trends Plant Sci 2011, 16, 53–60. [Google Scholar] [CrossRef]
- Abdallah, M.B.; Trupiano, D.; Polzella, A.; De Zio, E.; Sassi, M.; Scaloni, A.; Zarrouk, M.; Youssef, N.B.; Scippa, G.S. Unraveling physiological, biochemical and molecular mechanisms involved in olive (Olea europaea L. cv. Chétoui) tolerance to drought and salt stresses. J. Plant Physiol. 2018, 220, 83–95. [Google Scholar] [CrossRef]
- Gutiérrez-Albanchez, E.; Gradillas, A.; García, A.; García-Villaraco, A.; Gutierrez-Mañero, F.J.; Ramos-Solano, B. Elicitation with Bacillus QV15 reveals a pivotal role of F3H on flavonoid metabolism improving adaptation to biotic stress in blackberry. PLoS ONE 2020, 15, e0232626. [Google Scholar]
- Trabelsi, L.; Gargouri, K.; Hassena, A.B.; Mbadra, C.; Ghrab, M.; Ncube, B.; Van Staden, J.; Gargouri, R. Impact of drought and salinity on olive water status and physiological performance in an arid climate. Agric. Water Manag. 2019, 213, 749–759. [Google Scholar] [CrossRef]
- Sgherri, C.L.; Pinzino, C.; Navari-Izzo, F. Sunflower seedlings subjected to increasing stress by water deficit: Changes in O2—Production related to the composition of thylakoid membranes. Physiol. Plant. 1996, 96, 446–452. [Google Scholar] [CrossRef]
- Boo, Y.C.; Jung, J. Water deficit—Induced oxidative stress and antioxidative defenses in rice plants. J. Plant Physiol. 1999, 155, 255–261. [Google Scholar]
- Srivalli, B.; Sharma, G.; Khanna-Chopra, R. Antioxidative defense system in an upland rice cultivar subjected to increasing intensity of water stress followed by recovery. Physiol. Plant. 2003, 119, 503–512. [Google Scholar] [CrossRef]
- Burton, G.W.; Ingold, K. β-Carotene, an unusual type of lipid antioxidant. Science 1984, 224, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Kchaou, H.; Larbi, A.; Chaieb, M.; Sagardoy, R.; Msallem, M.; Morales, F. Genotypic differentiation in the stomatal response to salinity and contrasting photosynthetic and photoprotection responses in five olive (Olea europaea L.) cultivars. Sci. Hortic. 2013, 160, 129–138. [Google Scholar] [CrossRef]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [Green Version]
- Doke, N.; Miura, Y.; Sanchez, L.M.; Kawakita, K. Involvement of superoxide in signal transduction: Responses to attack by pathogens, physical and chemical shocks, and UV irradiation. In Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants; CRC Press: Boca Raton, FL, USA, 2019; pp. 177–198. [Google Scholar]
- Ramos-Solano, B.; Algar, E.; Garcia-Villaraco, A.; Garcia-Cristobal, J.; Lucas Garcia, J.A.; Gutierrez-Mañero, F.J. Biotic elicitation of isoflavone metabolism with plant growth promoting rhizobacteria in early stages of development in Glycine max var. Osumi. J. Agric. Food Chem. 2010, 58, 1484–1492. [Google Scholar] [CrossRef]
- Dietz, K.J. Thiol-Based Peroxidases and Ascorbate Peroxidases, Why Plants Rely on Multiple Peroxidase Systems in the Photosynthesizing Chloroplast? Mol. Cells 2016, 39, 20–25. [Google Scholar]
- Arora, A.; Sairam, R.; Srivastava, G. Oxidative stress and antioxidative system in plants. Curr. Sci. 2002, 82, 1227–1238. [Google Scholar]
- Asghari, B.; Khademian, R.; Sedaghati, B. Plant growth promoting rhizobacteria (PGPR) confer drought resistance and stimulate biosynthesis of secondary metabolites in pennyroyal (Mentha pulegium L.) under water shortage condition. Sci. Hortic. 2020, 263, 109132. [Google Scholar] [CrossRef]
- Khademian, R.; Asghari, B.; Sedaghati, B.; Yaghoubian, Y. Plant beneficial rhizospheric microorganisms (PBRMs) mitigate deleterious effects of salinity in sesame (Sesamum indicum L.), physio-biochemical properties, fatty acids composition and secondary metabolites content. Ind. Crops Prod. 2019, 136, 129–139. [Google Scholar] [CrossRef]
- Chin, D.; Kumar, R.S.; Suen, C.; Chien, C.; Hwang, M.; Hsu, C.; Xuhan, X.; Lai, Z.X.; Yeh, K.-W. Plant cytosolic ascorbate peroxidase with dual catalytic activity modulates abiotic stress tolerances. iScience 2019, 16, 31–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barriuso, J.; Ramos-Solano, B.; Santamaria, C.; Daza, A.; Gutierrez-Mañero, F. Effect of inoculation with putative plant growth-promoting rhizobacteria isolated from Pinus spp. on Pinus pinea growth, mycorrhization and rhizosphere microbial communities. J. Appl. Microbiol. 2008, 105, 1298–1309. [Google Scholar] [CrossRef]
- Genty, B.; Briantais, J.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta Gen. Subj. 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids, pigments of photosynthetic biomembranes. Meth. Enzymol. 1987, 148, 350–382. [Google Scholar]
- Porra, R.; Thompson, W.; Kriedemann, P. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta Bioenerg. 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Carillo, P.; Gibon, Y. Protocol: Extraction and determination of proline. Prometheuswiki 2011, 1–5. [Google Scholar]
- Yemm, E.; Willis, A. The estimation of carbohydrates in plant extracts by anthrone. Biochem. J. 1954, 57, 508–514. [Google Scholar] [CrossRef] [Green Version]
- García-Limones, C.; Hervás, A.; Navas-Cortés, J.A.; Jiménez-Díaz, R.M.; Tena, M. Induction of an antioxidant enzyme system and other oxidative stress markers associated with compatible and incompatible interactions between chickpea (Cicer arietinum L.) and Fusarium oxysporum f. sp. ciceris. Physiol. Mol. Plant Pathol. 2002, 61, 325–337. [Google Scholar] [CrossRef]
- Zhang, J.; Kirkham, M. Antioxidant responses to drought in sunflower and sorghum seedlings. New Phytol. 1996, 132, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.J.; Chang, S. A comparative study on phenolic profiles and antioxidant activities of legumes as affected by extraction solvents. J. Food Sci. 2007, 72, S159–S166. [Google Scholar] [CrossRef] [PubMed]
- Zhishen, J.; Mengcheng, T.; Jianming, W. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar] [CrossRef]
- Lucas, J.A.; Gutierrez-Albanchez, E.; Alfaya, T.; Feo-Brito, F.; Gutiérrez-Mañero, F.J. Oxidative stress in ryegrass growing under different air pollution levels and its likely effects on pollen allergenicity. Plant Physiol. Biochem. 2019, 135, 331–340. [Google Scholar] [CrossRef] [PubMed]

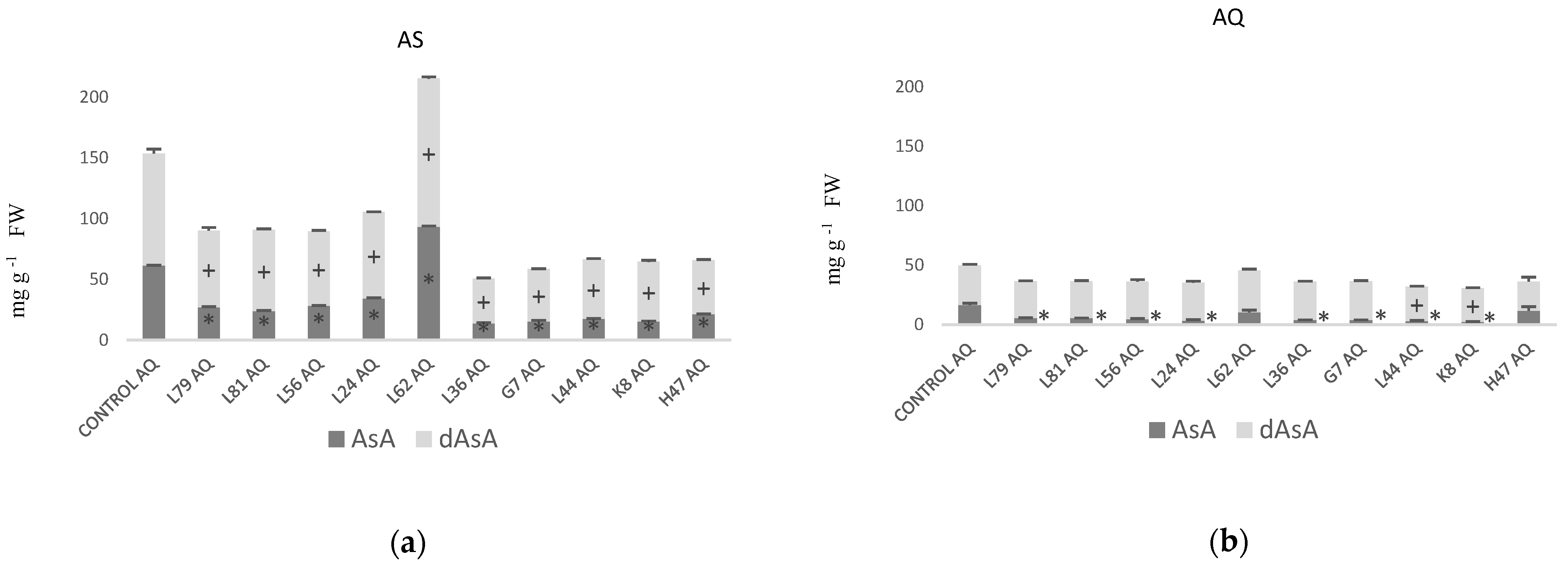

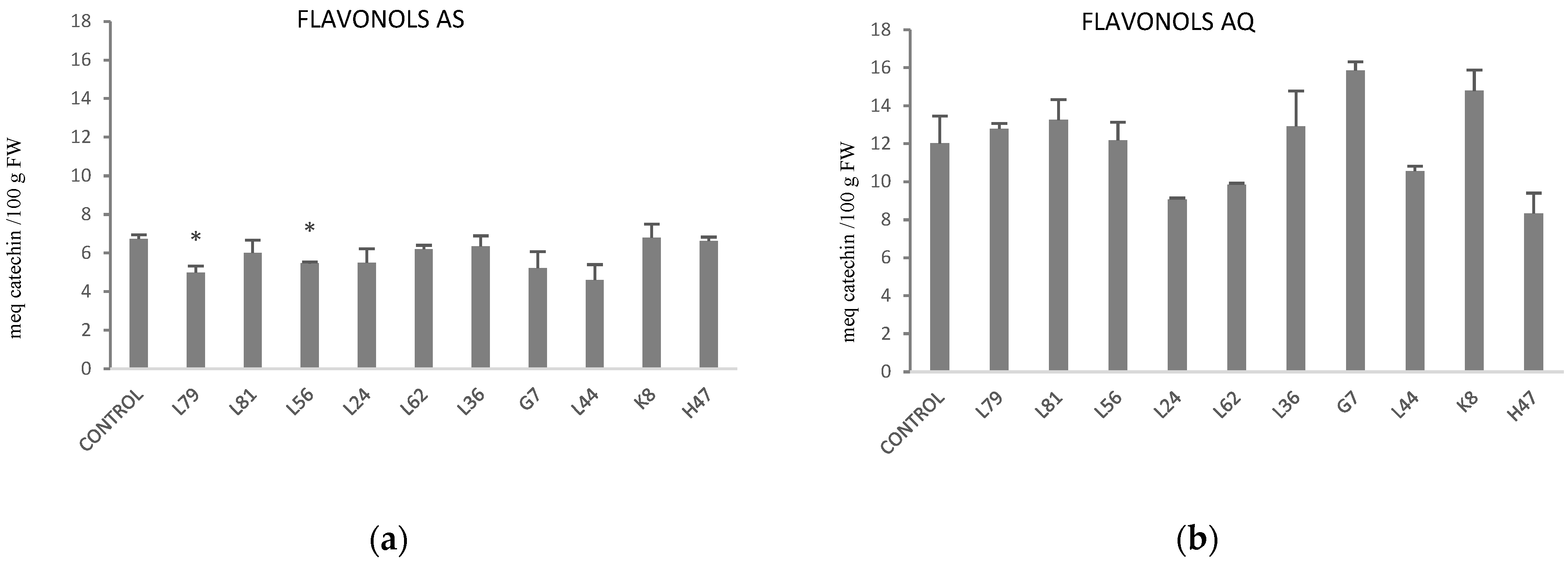
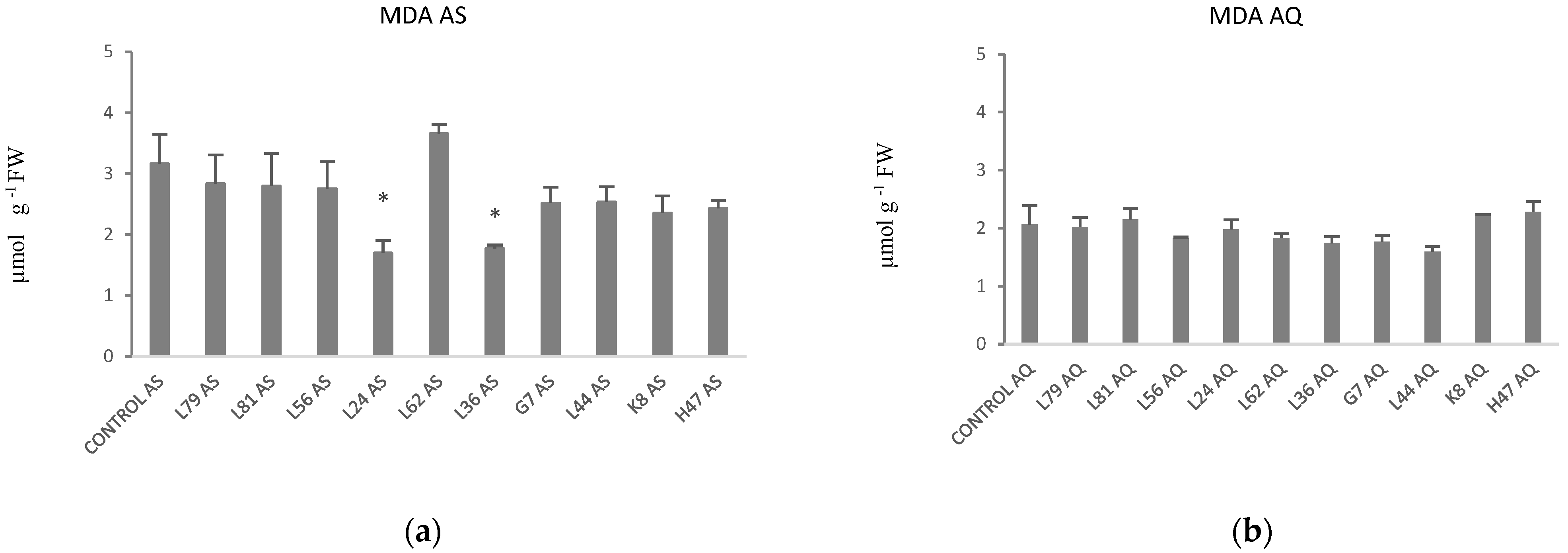
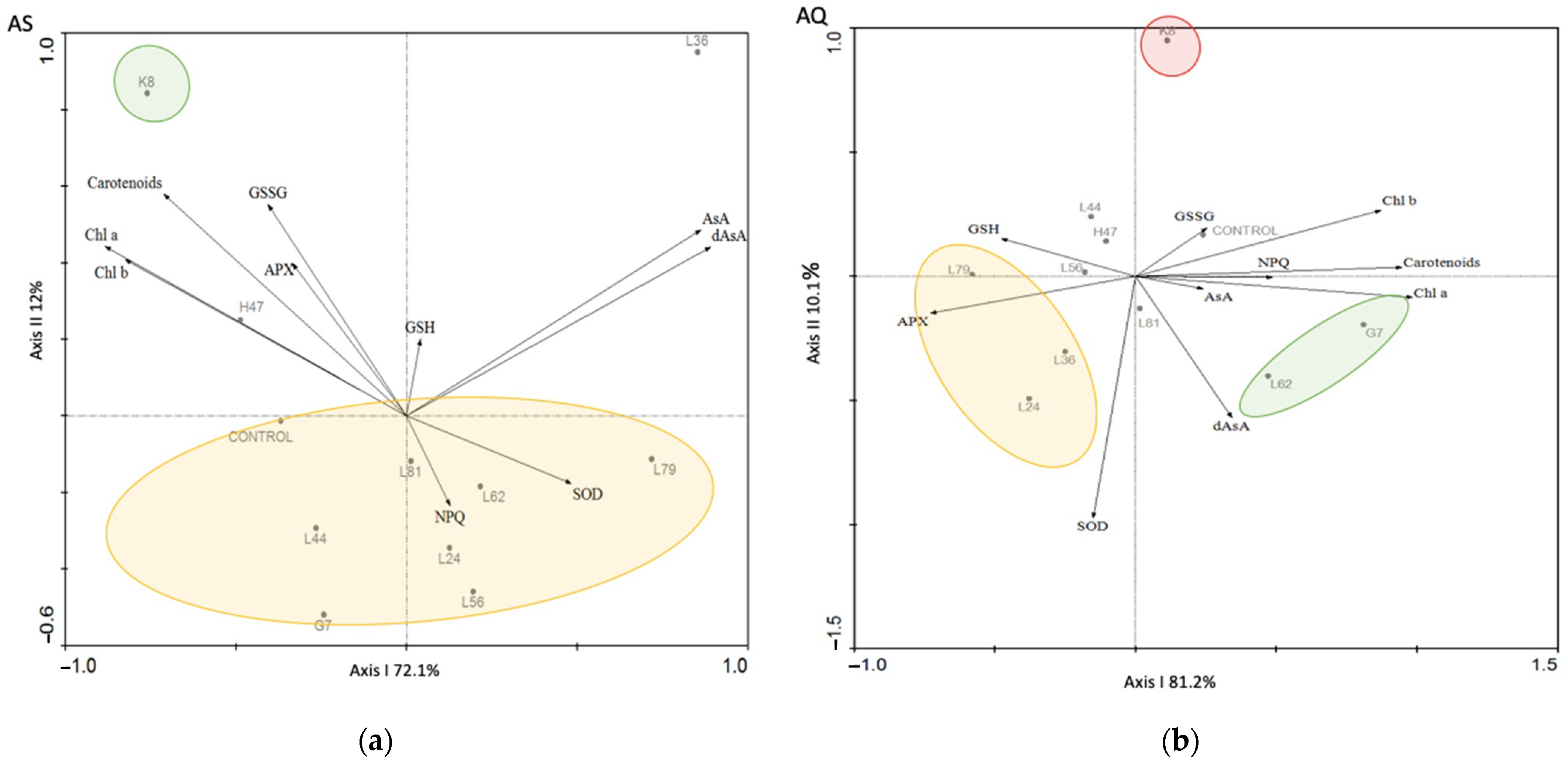
| Parameters | Control AS | Control AQ | % AS vs. AQ |
|---|---|---|---|
| F0 | 157.67 ± 8.76 | 216.78 ± 16.84 | −27% |
| Fv/Fm | 0.85 ± 0.003 | 0.77 ± 0.05 | 10% |
| PSR | 0.77 ± 0.02 | 0.74 ± 0.02 | 4% |
| NPQ | 0.10 ± 0.03 | 0.24 ± 0.02 | −59% |
| Chl a (µg g−1) | 125.54 ± 19.67 | 137.02 ± 13.65 | −8% |
| Chl b (µg g−1) | 60.25 ± 5.39 | 52.29 ± 5.15 | 15% |
| Carotenes (µg g−1) | 92.93 ± 16.35 | 134.3 ± 7.91 | −19% |
| SOD (% inhibition g−1 protein) | 102.2 ± 3.86 | 90.65 ± 0.9 | 13% |
| APX (µmol g−1 protein) | 0.03 ± 0.00 | 0.04 ± 0.01 | −17% |
| GSSG (mg g−1 FW) | 2.24 ± 0.087 | 2.55 ± 0.087 | −12% |
| GSH (mg g−1 FW) | 6.82 ± 1.05 | 1.82 ± 0.26 | 275% |
| AsA (mg g−1 FW) | 61.56 ± 0.15 | 16.64 ± 1.39 | 270% |
| dAsA (mg g−1 FW) | 92.08 ± 3.82 | 33.38 ± 0.82 | 176% |
| MDA | 3.16 ± 0.48 | 2.06 ± 0.32 | 53% |
| Phenols (meq gallic acid per 100 g FW) | 739.94 ± 70.42 | 1031.81 ± 89.63 | −28% |
| Flavonols (meq catechin per 100 g FW) | 6.73 ± 0.21 | 12.02 ± 1.45 | −44% |
| Proline (nmol g−1 FW) | 0.45 ± 0.02 | 0.39 ± 0.02 | 15% |
| Soluble sugars (mg g−1 FW) | 4.93 ± 0.60 | 4.62 ± 0.74 | 7% |
| ARBOSANA | ARBEQUINA | |||||||
|---|---|---|---|---|---|---|---|---|
| F0 | Fv/Fm | PSR | NPQ | F0 | Fv/Fm | PSR | NPQ | |
| Control | 157.67 ± 8.76 | 0.85 ± 0.003 | 0.77 ± 0.02 | 0.10 ± 0.03 | 216.78 ± 16.84 | 0.77 ± 0.05 | 0.74 ± 0.02 | 0.24 ± 0.02 |
| L79 | 195.22 ± 17.96 | 0.83 ± 0.01 | 0.77 ± 0.03 | 0.15 ± 0.02 | 211.33 ± 8.97 | 0.84 ± 0.01 | 0.7 ± 0.02 | 0.23 ± 0.02 |
| L81 | 150.67 ± 14.71 | 0.85 ± 0.01 | 0.8 ± 0.02 | 0.18 ± 0.03 | 213.67 ± 8.24 | 0.82 ± 0.01 | 0.7 ± 0.02 | 0.18 ± 0.01 |
| L56 | 163 ± 0.01 | 0.85 ± 0.01 | 0.82 ± 0 | 0.61 ± 0.01 | 185 ± 7.62 | 0.84 ± 0.01 | 0.75 ± 0.02 | 0.15 ± 0.01 * |
| L24 | 166.67 ± 0.27 | 0.87 ± 0.01 | 0.78 ± 0.02 | 0.12 ± 0.02 | 190.33 ± 4.48 | 0.85 ± 0.01 | 0.72 ± 0.02 | 0.17 ± 0.01 |
| L62 | 179.33 ± 5.46 | 0.85 ± 0.01 | 0.8 ± 0.01 | 0.1 ± 0.01 | 206.67 ± 8.45 | 0.83 ± 0.01 | 0.72 ± 0.01 | 0.23 ± 0.03 |
| L36 | 170.67 ± 8.02 | 0.84 ± 0.01 | 0.74 ± 0.02 | 0.15 ± 0.02 | 219 ± 8.26 | 0.8 ± 0.02 | 0.67 ± 0.03 | 0.22 ± 0.03 |
| G7 | 174 ± 9.81 | 0.87 ± 0.01 | 0.78 ± 0.02 | 0.13 ± 0.01 | 212.78 ± 13.39 | 0.82 ± 0.02 | 0.69 ± 0.02 | 0.25 ± 0.03 |
| L44 | 133 ± 18.12 | 0.85 ± 0.01 | 0.77 ± 0.02 | 0.13 ± 0.03 | 160.67 ± 10.39 | 0.84 ± 0.02 | 0.82 ± 0.05 | 0.2 ± 0.04 |
| K8 | 179.67 ± 11.47 | 0.85 ± 0.01 | 0.77 ± 0.01 | 0.15 ± 0.02 | 192.33 ± 9.82 | 0.82 ± 0.01 | 0.7 ± 0.04 | 0.21 ± 0.01 |
| H47 | 148 ± 21.92 | 0.86 ± 0.01 | 0.8 ± 0.01 | 0.16 ± 0.05 | 195.75 ± 7.54 | 0.83 ± 0.01 | 0.75 ± 0.02 | 0.16 ± 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galicia-Campos, E.; García-Villaraco Velasco, A.; Montero-Palmero, M.B.; Gutiérrez-Mañero, F.J.; Ramos-Solano, B. Modulation of Photosynthesis and ROS Scavenging Response by Beneficial Bacteria in Olea europaea Plantlets under Salt Stress Conditions. Plants 2022, 11, 2748. https://doi.org/10.3390/plants11202748
Galicia-Campos E, García-Villaraco Velasco A, Montero-Palmero MB, Gutiérrez-Mañero FJ, Ramos-Solano B. Modulation of Photosynthesis and ROS Scavenging Response by Beneficial Bacteria in Olea europaea Plantlets under Salt Stress Conditions. Plants. 2022; 11(20):2748. https://doi.org/10.3390/plants11202748
Chicago/Turabian StyleGalicia-Campos, Estrella, Ana García-Villaraco Velasco, Mᵃ Belén Montero-Palmero, F. Javier Gutiérrez-Mañero, and Beatriz Ramos-Solano. 2022. "Modulation of Photosynthesis and ROS Scavenging Response by Beneficial Bacteria in Olea europaea Plantlets under Salt Stress Conditions" Plants 11, no. 20: 2748. https://doi.org/10.3390/plants11202748
APA StyleGalicia-Campos, E., García-Villaraco Velasco, A., Montero-Palmero, M. B., Gutiérrez-Mañero, F. J., & Ramos-Solano, B. (2022). Modulation of Photosynthesis and ROS Scavenging Response by Beneficial Bacteria in Olea europaea Plantlets under Salt Stress Conditions. Plants, 11(20), 2748. https://doi.org/10.3390/plants11202748







